Abstract
Piscirickettsia salmonis is the pathogen that most affects the salmon industry in Chile. Large quantities of antibiotics have been used to control it. In search of alternatives, we have developed [Cu(NN1)2]ClO4 where NN1 = 6-((quinolin-2-ylmethylene)amino)-2H-chromen-2-one. The antibacterial capacity of [Cu(NN1)2]ClO4 was determined. Subsequently, the effect of the administration of [Cu(NN1)2]ClO4 on the growth of S. salar, modulation of the immune system and the intestinal microbiota was studied. Finally, the ability to protect against a challenge with P. salmonis was evaluated. The results obtained showed that the compound has an MIC between 15 and 33.9 μg/mL in four isolates. On the other hand, the compound did not affect the growth of the fish; however, an increase in the transcript levels of IFN-γ, IL-12, IL-1β, CD4, lysozyme and perforin was observed in fish treated with 40 μg/g of fish. Furthermore, modulation of the intestinal microbiota was observed, increasing the genera of beneficial bacteria such as Lactobacillus and Bacillus as well as potential pathogens such as Vibrio and Piscirickettsia. Finally, the treatment increased survival in fish challenged with P. salmonis by more than 60%. These results demonstrate that the compound is capable of protecting fish against P. salmonis, probably by modulating the immune system and the composition of the intestinal microbiota.
1. Introduction
The aquaculture industry occupies an important place in the economy of Chie, positioning the country as the second largest producer of salmonids worldwide after Norway [1]. Favorable climatic and environmental conditions have facilitated rapid growth in this productive sector in Chile, with Atlantic salmon, coho salmon and rainbow trout being the main species produced [2]. However, this explosive growth has generated an increase in outbreaks of infectious diseases, because fish are exposed to constant stress due to handling, transportation, higher density, changes in temperature and salinity, among others, where these conditions increase their susceptibility to different types of pathogens, which cause significant economic losses to the industry [3]. The pathogens that most affect the salmon industry are bacteria, mainly Piscirickettsia salmonis, Tenacibaculum dicentrarchi and Flavobacterium psycrhophilum [2]. In the case of P. salmonis, this bacterium has been the infectious agent that has caused the most deaths in salmonids for more than 30 years; it is estimated that the losses associated with this pathogen range between USD 700 and USD 800 million annually [4]. In 2022, P. salmonis was the cause of 50% of pathogen mortality in Atlantic salmon and rainbow trout [2]. P. salmonis is the etiological agent of salmonid rickettsial syndrome (SRS); it is a Gram-negative, pleomorphic but generally coccoid bacteria, with a variable size between 0.2 μm and 1.5 μm [5] and facultative intracellular [6]. P. salmonis is able to survive and replicate within replicative vacuoles in phagocytic cells such as macrophages and monocyte-like cells [7] by evading phagosome-lysosome fusion [8]. On the other hand, this bacterium is capable of modulating different biological processes in the host, including the cellular and humoral immune response [9,10]. Fish infected with P. salmonis exhibit lethargy, anorexia, darkening of the skin, swimming at the surface, and respiratory distress. It is possible to observe external lesions such as perianal and periocular hemorrhages, abdominal petechiae, superficial hemorrhagic ulcers of variable size ranging from 0.5 to 2 cm in diameter and white nodules of more than 1 cm in diameter [11]. Internally, the presence of yellow nodules in the liver is observed [12], as are generalized internal pallor, inflammation and multifocal pale areas in the kidney and spleen [11], affecting the brain, heart, skeletal muscle, gills and ovaries [5]. To control the disease caused by this bacterium, vaccines and antibiotics are mainly used. However, due to limited knowledge of the immune system in teleosts, these vaccines have not been effective [4,13]. Since vaccines are not sufficient to control this disease, the Chilean salmon industry uses a large amount of antibiotics. In 2022, 341.5 tons of antibiotics were used, of which 91.28% (approximately 304.6 tons) of the antibiotics were used against P. salmonis [14]. This large amount of antibiotics used has caused various problems, such as the development of P. salmonis isolates resistant to antibiotics [15,16]. On the other hand, environmental pollution is generated, because antibiotics are administered to the salmon through food, so it leaks from the cages into the water and marine sediment in which they are found. In addition, there is a flow of traces of antibiotics present in salmon urine and feces into the marine environment [17,18]. Another problem of excessive use of antibiotics, reported in recent years, is the effect on the composition of the intestinal microbiota of salmonids. Antibiotics generate dysbiosis in the intestine of fish, which causes changes in various biological processes, including the immune response [19,20,21]. One of the alternatives is the use of natural compounds, which, when administered in diets, causes positive effects in the fish organism. For example, a wide range of polyphenols have been studied as dietary supplements for fish, observing that they have antioxidant effects, modulate the immune response, generate protection from diseases, and improve reproduction and growth in a wide variety of fish. In addition, coumarin derivatives have attracted considerable attention due to their biological functions in fish, including antiparasitic, antibacterial and antiviral activities [22,23,24]. On the other hand, currently, there is great interest in copper coordination compounds based on ligands with the N- donor atom. This interest arises from its antiproliferative, antibacterial, antiviral, and cytotoxic properties [25], as well as its ability to modulate the immune response [26,27]. In the aquaculture industry, copper has historically been used as a treatment against different pathogens such as Vibrio, Aeromonas and Flavobacterium, among others, where copper (II) sulfate, CuSO4, is the most widely used form of copper supply. A study carried out with the marine pathogens Aeromonas hydrophila and Flavobacterium columnare showed that CuSO4 had an MIC of 83.2 ± 0 mg/L towards A. hydrophila, while the IC50 of CuSO4 towards F. columnare was 4.8 ± 0.3 mg/L, and the minimum bactericidal concentration of CuSO4 towards F. columnare was 25.0 ± 0 mg/L [28,29].
The adaptability of transition metal compounds, in particular, Cu(I) coordination complexes with bidentate nitrogen donor ligands, is well known. Therefore, several studies have been conducted towards the controlled obtaining of its electrochemical properties through the selection of its ligands [30]. Considering the need to use alternatives to antibiotics, in addition to the positive effects of natural compounds and their derivatives, added to the properties that transition metal complexes possess, our working group has synthesized a new [Cu(NN1)2]ClO4 complex, where NN1 is a 6-((quinolin-2-ylmethylene)amine)-2H imine ligand-chrome-2-one, a derivative of the natural compound coumarin. This complex showed a better antibacterial effect in vitro against F. psychrophilum than its precursors, coumarin and copper (I) salt [31], and when administering 60 μg/g of fish to rainbow trout, it generates protection against F. psychrophilum [32]. Considering the previous results of the effect of this compound on F. psychrophilum, in the present work, we analyze its antibacterial potential on P. salmonis and the effect of its administration in Atlantic salmon on the immune response, its modulation of the intestinal microbiota and its ability to generate protection against infection with a P. salmonis isolate such as LF-89. The results obtained in this work showed the different effects of the compound, such as the effects of antibacterial activity on P. salmonis and the ability to modulate the immune response and intestinal microbiota in Atlantic salmon. These multiple effects of the compound could be the mechanisms by which it generates protection against P. salmonis in Atlantic salmon. These are promising results for this compound as an alternative treatment for Piscirickettsiosis.
2. Results
2.1. Synthesis of Cu (I) Coordination Complex
Previously, in our laboratory, we have synthesized the coordination complex [Cu(NN1)2]ClO4, where NN1 = 6-((quinolin-2-ylmethylene)amino)-2H-chromen-2-one, a ligand derivate from natural product coumarin 1-benzopyran-2-one [31]. In this paper, we account for the changes made in the experimental procedure to improve the process. On this occasion, synthesis was easily achieved using the template condensation method from equimolar amounts of reagents, as shown in Figure 1, which allowed the reaction time to be reduced to 1 h, increasing the reaction yield to 90%, at room temperature and without requiring pressure systems, substantially improving the energy and economic cost of the process.
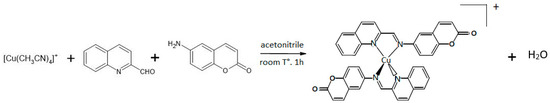
Figure 1.
Synthesis scheme of the complex [Cu(NN1)2]ClO4 using the template method.
2.2. Antibacterial Activity of [Cu(NN1)2]ClO4
The antibacterial activity of [Cu(NN1)2]ClO4 was evaluated in four isolates of P. salmonis. Isolate 8149 was the most sensitive with an MIC value of 15.0 + 7.1 and an IC50 value of 5.3 + 0.8 μg/mL. On the other hand, isolates CGRO2, 12,201 and 727 were more resistant to the compound with MIC values between 29 and 34 μg/mL and IC50 values between 11 and 15 μg/mL, approximately (Table 1).

Table 1.
MIC and IC50 of [Cu(NN1)2]ClO4 on four P. salmonis isolates (n = 3).
2.3. Effect of [Cu(NN1)2]ClO4 on the Growth of Atlantic Salmon
The safety of the administration of [Cu(NN1)2]ClO4 for 60 days in Atlantic salmon was evaluated. During feeding, no negative effects on salmonid behavior were observed. Similarly, no changes were observed in the growth of the salmonids during the 60 days of experimentation (Figure 2).
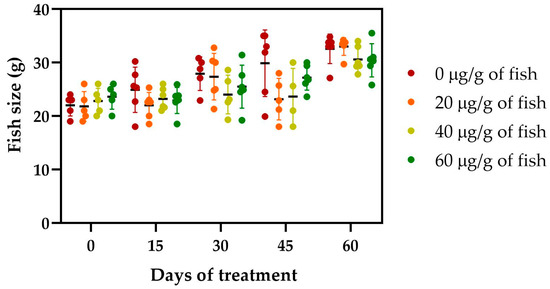
Figure 2.
Effect of the [Cu(NN1)2]ClO4 on the growth of Atlantic salmon during the 60 days of experimentation. Every 15 days, 6 fish per condition were sampled and weighted. The legend indicates the doses tested, 0 µg/g of fish (Control), 20 µg of [Cu(NN1)2]ClO4/g of fish of, 40 µg of [Cu(NN1)2]ClO4/g of fish, and 60 µg of [Cu(NN1)2]ClO4/g of fish. The significance was analyzed using the Mann–Whitney test.
2.4. Evaluation of the Administration of [Cu(NN1)2]ClO4 on Immune Status of Atlantic Salmon
The immunostimulation capacity of natural compounds has been widely studied in various organisms, including coumarins. Because our compound, [Cu(NN1)2]ClO4, which has coumarin as a natural ligand, its ability to stimulate the immune system was evaluated. Fifteen days after the administration of [Cu(NN1)2]ClO4, in fish treated with 40 μg/g of fish of the compound, an increase in INF-γ transcript levels close to 7-fold, an increase in IL-1β close to 5-fold, an increase in CD4 close to 3-fold, an increase in perforin close to 2-fold, and a 2.4-fold increase in lysozyme was observed. On the other hand, in fish treated with 60 μg/g of fish, only an increase in IL-1β transcript levels close to three-fold was observed (Figure 3a). At 30 days of [Cu(NN1)2]ClO4 administration, IFN-γ transcript levels increased five-fold, while TNF-α increased close to nine-fold in fish treated with 40 μg/g of fish (Figure 3b). After 45 days of administration of [Cu(NN1)2]ClO4, no statistically significant changes were observed in the transcript levels of the genes under study (Figure 3c). Finally, after 60 days of [Cu(NN1)2]ClO4 administration, the fish treated with 20 μg/g of fish showed an increase close to four times in the INF-γ transcript levels; on the other hand, a decrease in the CD4 and lysozyme transcript levels close to 4- and 50-fold, respectively, were observed. Fish treated with 40 μg/g fish of [Cu(NN1)2]ClO4 showed a close to six-fold increase in IL-1β transcript levels, while a close to two-fold decrease was observed in CD4. On the other hand, fish treated with 60 μg/g of fish of [Cu(NN1)2]ClO4, showed a decrease in the transcript levels of CD4, lysozyme, and perforin, between 2-fold and 3-fold (Figure 3d).
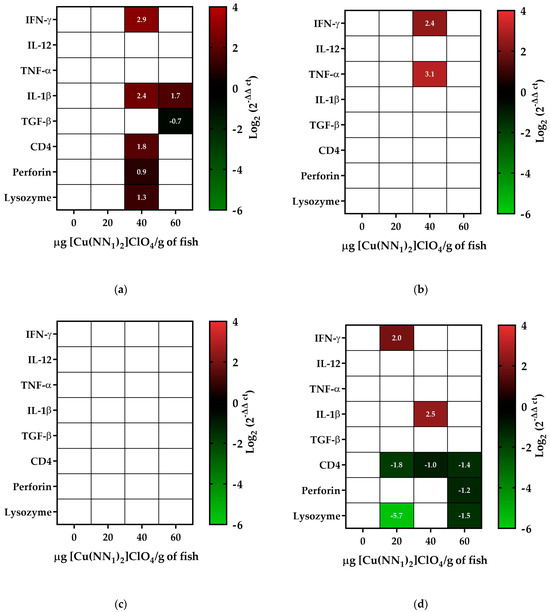
Figure 3.
Effect of oral administration of [Cu(NN1)2]ClO4 on the immune system of Atlantic salmon. The figure shows the relative transcript levels of genes encoding for Interferon-γ (IFNγ), Interleukin 12 (IL-12), Tumon Necrotic Factor α(TNF-α), Interleukin 1β (IL-1β), Tumon Growth Factor-β (TGF-β), CD4, Perforin and lysozyme. The fish were treated with 0 (Ctrl), 20 µg, 40 µg and 60 µg of [Cu(NN1)2]ClO4 per gram of fish during 15 (a), 30 (b), 45 (c), and 60 (d) days of administration. The level of expression was determined by RT-qPCR, normalized with respect to the expression housekeeping gene (EF1-α). The statistically significant difference was analyzed using the Mann–Whitney test (<0.05), comparing the control with respect to the treatments. Colors represent significant difference; white cells represent non-significant difference.
2.5. Effect of the Administration of [Cu(NN1)2]ClO4 on the Composition of the Intestinal Microbiota of Atlantic Salmon
The intestinal microbiota of the experimental fish feed with [Cu(NN1)2]ClO4 showed a different pattern to the intestinal microbiota of the control group. In control fish, the intestinal microbiota was composed of bacteria belonging to the Phylum Proteobacteria (97.8%), followed by Actinobacteria (0.65%), Firmicutes (0.38%) and Bacteroidetes (0.37%). At the phylum level, the administration of the compound [Cu(NN1)2]ClO4 increased the proportion of Proteobacteria to 98.9% and the presence of an ASV of bacterial origin that could not be classified at the Phylum level to 0.22%, while the phylum Firmicutes (0.3%) and Bacteroidetes (0.22%) showed a slight decrease in their percentage of relative abundance (Figure 4). At the genus level, 24 different genera were found to be present in both groups of fish, while 32 genera were present only in control fish, and 53 only in the treated fish. When analyzing the bacterial genera with an abundance greater than 0.05%, we found that 20 different genera were present in both groups, 24 only in the control group and 8 only in the treated group. Furthermore, administration of the compound decreased the relative abundance of 14 different bacterial genera, while it increased the relative abundance of 5 bacterial genera. The bacterial genus with the highest abundance in control and treated fish was Photobacterium with 95.86% and 97.32%, respectively. In fish treated with [Cu(NN1)2]ClO4, it was possible to find bacterial genera that were not found in the control fish, such as Lactococcus (0.09%), Lactobacillus (0.05%) and Bacillus (0.05%), that have the potential to be used as bacterial probiotics. On the other hand, it was also possible to find pathogenic bacteria of the genus Piscirickettsia (0.11%) (Figure 5).
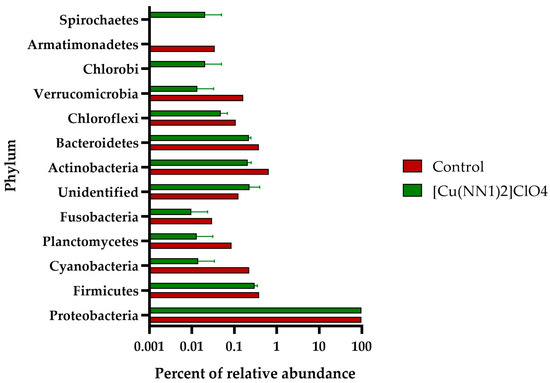
Figure 4.
Relative abundance of Phyla identified in the gut of Atlantic salmon. The figure shows the relative abundance of the Phylum identified in the intestine of Atlantic salmon specimens from fish fed with a control diet and treated with [Cu(NN1)2]ClO4.
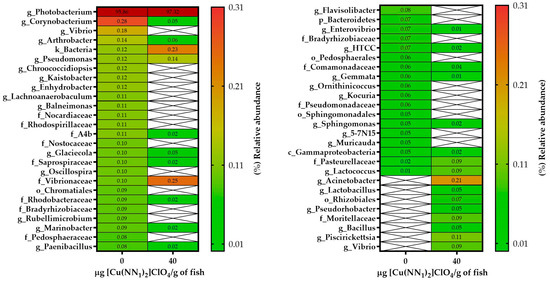
Figure 5.
Relative abundance of genera identified in the gut of Atlantic salmon. The figure shows the relative abundance greater than 0.05% of the genera identified in the intestine of Atlantic salmon specimens from fish fed with a control diet and treated with [Cu(NN1)2]ClO4 (40 μg/g of fish) during 15 days. White cells with crosses mean a relative abundance equal to 0.
2.6. Evaluation of the Protective Effect of [Cu(NN1)2]ClO4 against a Challenge with P. salmonis
The effect of administering 40 μg/g of [Cu(NN1)2]ClO4 to fish during a challenge with P. salmonis was evaluated. The results showed that the compound administered in the feed generated protection in Atlantic salmon when challenged with P. salmonis. A decrease in mortality could be observed in the fish treated with the compound, with 90% of the treated fish surviving, while only 30% of the untreated fish survived during 30 days of experimentation. The survival analysis was carried out using the Kaplan–Meier estimate, performing a log-rank test (Mantel–Cox), obtaining a statistically significant difference with a value of p < 0.0001 (Figure 6a). The analysis of bacterial loads in dead fish from both treated and untreated fish showed similar values, between 1 × 102 and 1 × 104 the number of copies/50 ng of sample. On the other hand, in the surviving fish, those treated with [Cu(NN1)2]ClO4, it was not possible to detect the gene of P. salmonis in any of the fish, while in those without treatment, two of the fish had a bacterial load close to 1 × 102 copy number/50 ng of sample (Figure 6b). These results show that the administration of [Cu(NN1)2]ClO4 in Atlantic salmon reduces the mortality generated by P. salmonis and decreases the amount of pathogen in the fish organism.
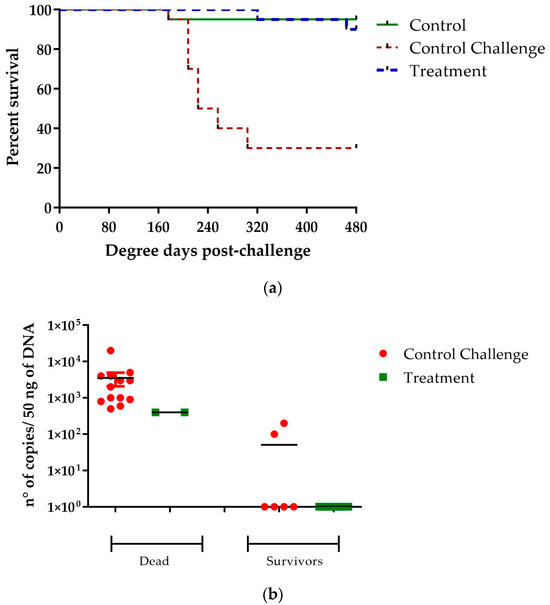
Figure 6.
Challenge assay with P. salmonis. The figure shows the effect of [Cu(NN1)2]ClO4 on the survival (a) and bacterial load (b) of fish challenge with P. salmonis. The panel (a) shows the percentage of survival after an intraperitoneal injection with P. salmonis of fish feed with normal food (control) and fish feed with food supplemented with [Cu(NN1)2]ClO4 at 40 μg/g of fish. Panel (b) shows the bacterial load present in the survival and dead fish of the group control and treated with [Cu(NN1)2]ClO4. Statistically significant differences were determined by comparing the curves of treatment versus challenge control, using a long-rank test (Mantel–Cox) and Gehan–Breslow–Wilcoxon test (p < 0.0001).
3. Discussion
The aquaculture industry in Chile is one of the most important economic sectors. However, the rapid growth of this productive sector has generated the continuous appearance of pathogens that affect the different production species. To face this problem, the Chilean aquaculture industry has used a large amount of antibiotics, generating various environmental problems and devaluing the Chilean product. In search of new alternatives, our working group synthesized the Cu (I) complex [Cu(NN1)2]ClO4 using coumarin as a ligand, achieving improvements in the antibacterial capacity against F. psycrhophilum compared to its precursor molecules (coumarin and [Cu(CH3CN)4]ClO4) [31] and generating protection for rainbow trout against a challenge with F. psycrhophilum [32]. Considering these previous works, in this article, we evaluate the antibacterial capacity of this compound against P. salmonis. As well as its ability to stimulate the immune response in Atlantic salmon and generate protection against a challenge with P. salmonis. The importance of working with this bacterium is because it is the most relevant in the Chilean salmon industry, being responsible for the highest mortalities in the industry for more than 30 years.
The results obtained showed that the compound is capable of reducing the growth of different isolates of P. salmonis, in concentrations similar to what was previously reported against F. psychrophilum [31]. On the other hand, no considerable differences were observed in the antibacterial activity of the compound between the P. salmonis-like LF-89 and P. salmonis-like EM-90 isolates. Although only four isolates were used in this work, it is relevant to identify compounds that have a similar effect between different isolates and genogroups of P. salmonis. Genotypical and phenotypical differences at the level of isolates and genogroups have been widely studied in P. salmonis [33]. These differences affect the sensitivity of the isolates to different antibiotics, making it even more difficult to find a treatment effective against several P. salmonis outbreaks [16,34]. Considering these results, the [Cu(NN1)2]ClO4 complex is a promising compound for a common treatment effective against a wide spectrum of isolates from different genogroups; however, it is necessary to perform additional experiments using a wider range of P. salmonis isolates to confirm this property.
The administration of the compound through feeding for 60 days did not appear to affect the growth of Atlantic salmon. These results are similar to those previously reported for this compound in rainbow trout [32]. On the other hand, the same effects have been observed in other nutritional supplements for salmonids based on natural products [35,36].
Although the compound was initially designed as an antibacterial, in this work, we also studied the capacity of [Cu(NN1)2]ClO4 to stimulate the cellular immune response in Atlantic salmon and its effect on the composition of the intestine microbiota. The administration of [Cu(NN1)2]ClO4 modulated the transcription levels of all the immune system markers analyzed in their different concentrations and times tested. [Cu(NN1)2]ClO4 at a concentration of 40 μg/g of fish generated the most important immunostimulating effect in this work, increasing the levels of lysozyme, perforin, IFN-γ, TNF-α, IL-1β and CD4. In the case of perforin, a glycoprotein that forms pores in the cell membrane of target cells [37], and which, in mammals, is expressed in NK cells, CD8+ T lymphocytes and CD4+ T lymphocytes, its antibacterial and antiviral in fish widely studied. For example, in Atlantic salmon, an increase in perforin is observed in the inhibition of Rock bream iridovirus (RBIV) replication [38]. Similar results were observed in Atlantic salmon against Pilchard orthomyxovirus (POMV) [39] and rohu and common carp against Aeromonas hydrophila [40,41]. On the other hand, the increase in lysozyme expression levels observed by treatment with [Cu(NN1)2]ClO4 is also related to an antibacterial effect. This enzyme hydrolyzes the peptidoglycan layer of the bacterial cell wall by cleaving the beta-1,4 glycosidic bonds between N-acetylmuramic acid and N-acetylglucosamine [42,43]. Lysozyme is the first line of defense against different types of pathogens such as fungi, parasites, bacteria and viruses [43,44,45]. Several studies show that feed supplementation with different secondary metabolites such as tea polyphenols increases lysozyme activity in common carp [46] and Asian sea bass [47], as well as flavonoids in northern snakehead fish [48]. In the case of cytokine markers, administration of [Cu(NN1)2]ClO4 markedly increased INF-γ transcript levels. The functions of IFN-γ are several, among which the activation of macrophages and CD4+ T helper lymphocytes has been studied, a fundamental process for eliminating intracellular pathogens, in addition to promoting inflammation and the presentation of antigens, a process for the inhibition of viral replication [49,50]. On the other hand, IFN-γ activates macrophages towards the “M1” phenotype, which increases the expression of pro-inflammatory cytokines such as IL-12, IL-1β and TNF-α, in addition to reactive oxygen species [51]. Moreover, IL-12, IL-1β and IL-8 promote the synthesis of IFN-γ, while TGF-β represses the expression of IFN-γ [52]. Various cell types such as Natural Killer (NK), CD4+ T helper (Th) cells, and cytotoxic CD8+ cells, among others, produce IFN-γ [52,53]. The transcript levels of IL-1β, TNF-α and CD4 were also increased in fish treated with [Cu(NN1)2]ClO4, demonstrating a relationship in the ability to stimulate the cellular immune response of this compound. Various studies, in which different molecules are administered in fish, have demonstrated a relationship between the increase in the transcription levels of pro-inflammatory and anti-inflammatory cytokines and protection against different pathogens. For example, caffeic acid and ferulic acid increase the transcript levels of IFN-γ, IL-1β and TNF-α in Nile Tilapia, generating protection against A. veronii [54,55]. Trans-cinnamic acid increases IFN-γ, IL-1β, IL-8, TGF-β and TNF-α, increasing the survival of rainbow trout against Y. ruckeri [56]. Coumarin increases IFN-γ levels, generating protection in zebrafish infected with spring viraemia of carp virus (SVCV) [23,57]. The immunomodulatory effect of the compound could be the mechanism of action through which the compound is capable of generating protection against P. salmonis. Similar mechanisms of action were reported for treatments based on natural compounds against P. salmonis [35,36].
[Cu(NN1)2]ClO4 also showed the ability to modify the intestinal microbiota composition of Atlantic salmon. The fish used in this study had a microbiota composed mainly of Proteobacteria, particularly dominated by Photobacterium. This composition deviates somewhat from the commonly observed fish microbiota, which is typically dominated by Firmicutes and Proteobacteria [58]. This discrepancy could be attributed to the cultivation conditions favoring the presence of Photobacterium in the Atlantic salmon specimens used. High relative abundance values have also been reported in freshwater Atlantic salmon specimens, with values of 40 ± 25% [59]. On the other hand, the administration of [Cu(NN1)2]ClO4 generated a decrease in the abundance of bacteria of the Corynebacterium genus, associated with opportunistic pathogens that strongly affect the salmon industry. In addition, it was observed that it modulates the abundance of bacteria of the genus Vibrio, in which both pathogenic bacteria and bacteria with probiotic potential can be found [60]. Moreover, various bacterial genera widely studied for their probiotic potential, such as Pseudomonas, Lactobacillus, Lactoccocus and Bacillus, increased their abundance [61,62,63]. This effect of modulating the composition of the intestinal microbiota, through the incorporation of different nutrients in the fish diet, has been widely studied. For example, administration of the prebiotic Selectovit in Atlantic salmon increases the abundance of Bacillus and Mycoplasma [64]. In turbot, the administration of daidzein has a dual effect, increasing the abundance of lactic acid bacteria but also potential pathogens such as Prevotella copri [65]. Similar results were reported in tilapia fed with resveratrol, modulating the taxa of potential pathogenic and commensal bacteria, depending on the concentration [66]. Finally, it was observed that the administration of the compound surprisingly increased the abundance of bacteria of the genus Piscirickettsia, to which P. salmonis belongs. However, it has been reported that this bacterium is part of the intestinal microbiota of healthy fish, and that an imbalance in the composition of the microbiota and the interaction between different metabolites of various bacteria generates the appearance of SRS [67]. These data are relevant when interpreting our results, because although the compound generated an increase in this bacterial genus, the administration of [Cu(NN1)2]ClO4 during the challenge with P. salmonis generated protection, considerably increasing the survival of Atlantic salmon. This suggests that the compound generates a balance in the intestinal microbiota at the time of challenge. Finally, considering that in vitro the compound has an antibacterial effect, and that in vivo it can modulate the immune response and the composition of the intestinal microbiota. The protective effect observed may be multifactorial, attacking different fronts to increase the survival of Atlantic salmon against P. salmonis.
4. Materials and Methods
4.1. Synthesis of Cu (I) Coordination Complex
The synthesis of Cu(I) coordination compound [Cu(NN1)2]ClO4, where NN1 is the ligand 6-((quinolin-2-ylmethylene)amino)-2H-chromen-2-one was carried out using the template condensation method [30]. To 3 mmol of the precursor reagent [Cu(CH3CN)4]ClO4 dissolved in 50 mL of acetonitrile, double the amounts of the reactants 2-quinoline-carboxaldehyde (6 mmol) and 6-amino-chromen-2-one (6 mmol), dissolved in acetonitrile, were added and maintained at room temperature and constant stirring for 1 h, forming a colored solution. The volume of solution was reduced in a rotary evaporator and the concentrate was precipitated with cold ethyl ether. The microcrystalline precipitate was recrystallized from an ethyl ether/acetonitrile (9:1) mixture and finally washed with ethyl ether. The chemical structure was confirmed by comparing the signals obtained from the NMR1H spectrum of the complex obtained, with what was previously reported [31].
4.2. Fish and Maintenance
Pre-smolt Atlantic salmon (S. salar) weighing between 25 and 35 g (Blumar, Los Angeles, Chile) was used. One week before the start of the experiments, the fish were acclimatized in ponds with a biomass of 14 g/L and a temperature of 12 °C and fed daily at 1% of body weight (Golden Optima, Biomar, Chiloé, Chile). The water parameters were monitored daily, maintaining the pH at between 6.6 and 7, while the salinity was adjusted to 6 PSU. On the other hand, total ammonia was monitored to maintain values below 0.02 mg/L. Each day, changing the water in the ponds, measuring pH, salinity, and ammonia, and feeding the fish were performed manually. The experiments were carried out in accordance with the ethical standards of the Institutional Ethics Committee of the University of Santiago de Chile and the relevant current legislation. The authorization from the Ethics Committee of the University of Santiago de Chile to carry out the experiments with fish in the FONDEF VIU project was approved with number 354.
4.3. Antibacterial Activity
To determine the MIC and IC50 of [Cu(NN1)2]ClO4 on P. salmonis, the microdilution method was used [68] with some modifications. For this experiment, four isolates of P. salmonis were used (CGRO2, 12,201 and 8149 provided by Dr. Veronica Cambiazo, bioinformatics and gene expression laboratory, INTA, University of Chile and 727 provided by Dr. Aldo Gaggero, virology laboratory, ICBM, Universidad de Chile), which were grown in cell-free medium (Austral-SRS medium) [6]. The bacteria were incubated on plates with Austral-SRS agar medium for 5 days at 18 °C. Subsequently, the bacteria were collected and inoculated in 5 mL of Austral-SRS medium and incubated for 3 days at 18 °C with shaking at 180 rpm. Bacteria were adjusted to OD600 = 0.1 and inoculated again into 10% Austral-SRS medium in a final volume of 5 mL and incubated for 3 days at 18 °C with shaking at 180 rpm. Subsequently, P. salmonis isolates were inoculated at an OD600 = 0.1 in a 10% cell suspension in AUSTRAL-SRS medium in 96-well plates (SPL), and treated with [Cu(NN1)2]ClO4 in serial concentrations between 128 µg/mL and 2 µg/mL. The plates were incubated for 96 h at 16 °C with shaking at 180 rpm. To calculate the MIC and IC50 of [Cu(NN1)2]ClO4 in the four P. salmonis isolates, the OD600 nm of each well was measured using Nanoquant Infinite M200 Pro (TECAN, Grödig, Austria). Data were analyzed using GraphPad Prism 8.0 software. Concentrations were log (10) transformed, OD600 nm was normalized as a percentage, and nonlinear regression was performed to calculate MIC and IC50.
4.4. Evaluation of the Immune Response and Modulation of the Intestinal Microbiota
The evaluation of the immune system and the composition of the intestinal microbiota was carried out using 120 Atlantic salmon of 25 ± 2 g, which were divided into 5 groups, each group with 24 fish divided into 2 aquariums. Group A contained untreated fish (Ctrl), group B fish were treated with 20 µg/g of [Cu(NN1)2]ClO4 (20), group C fish were treated with 40 µg/g of [Cu(NN1)2]ClO4 (40), and group D fish were treated with 60 µg/g of [Cu(NN1)2]ClO4 (60). All the fish were fed for 60 days with commercial pellets plus the respective treatments, mixed with commercial oil, while the control fish were only fed with the commercial pellet plus oil. Three fish were sacrificed per aquarium at 15, 30, 45, and 60 days of experimentation (n = 6 per treatment); then, the fish were weighed and the head kidney, and intestine were removed. The samples were stored at −80 °C.
4.5. Growth of P. salmonis in Cell Culture
The challenge experiment was performed with P. salmonis isolate 727 (like LF-89). For the growth of this bacteria, the SHK-1 cell line (ECACC) was used, which was grown in L-15 medium (Cytiva, Hyclone, South Logan, UT, USA) supplemented with 10% fetal bovine serum (Cytiva, Hyclone, South Logan, UT, USA), 4 mM L-glutamine (Mediatech, Corning, Manassas, VA, USA) and 40 μM β-mercaptoethanol (Life technologies, Gibco, New York, NY, USA), in T175 cell culture bottles (SPL). The cells at 80% confluence were incubated with P. salmonis for 24 h; later, the cells were washed twice with PBS 1X (Cytiva, Hyclone, South Logan, UT, USA) and incubated for 2 h with 50 μg/mL gentamicin. Subsequently, the cells were washed twice with PBS 1X and incubated with fresh L-15 medium, described previously, for 12 days at 16 °C. The cells were collected using a cell scraper and centrifuged at 1000× g to precipitate the cells, the supernatant was collected and centrifuged at 5000× g to collect the bacteria, the supernatant was discarded, and the pellet containing the bacteria was resuspended in 1 mL of physiological serum.
The quantification of P. salmonis was performed using the kit LIVE/DEAD BacLight bacterial viability and counting kit (Life technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions.
4.6. Evaluation of the Protective Effect of [Cu(NN1)2]ClO4
The ability of [Cu(NN1)2]ClO4 to protect Atlantic salmon against P. salmonis was evaluated. For this experiment, 60 Atlantic salmon 35 ± 2 g were used, divided into 3 different groups with 10 fish per pond in duplicate (total n = 20). Group A: Fish fed with pellets mixed with commercial oil (untreated). Group B: Fish fed with pellet mixed with commercial oil (challenge control). Group C: Fish fed with [Cu(NN1)2]ClO4 at 40 µg/g of fish, mixed in the pellet with the commercial oil. All fish were fed with their respective treatments for 15 days before being challenged with 1 × 107 P. salmonis bacteria in 100 µL, via intraperitoneal injection (previously anesthetized with 30 mg/L benzocaine) in the case of group B and C, while group A was injected with 100 µL of physiological serum. Subsequently, all fish were fed throughout the experiment with their respective treatments. Daily mortality was recorded for 30 days after challenge.
4.7. RNA Extraction
To extract the RNA from the kidneys of the fish, the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used, following the manufacturer’s instructions, with some modifications (Ref parra 2020). The RT reaction was performed with the All-In-One 5X RT MasterMix kit (ABM, Richmond, BC, Canada), using 2 µg of RNA, 4 µL of the master mix and nuclease-free H2O to a final volume of 20 µL. The thermal profile used was 30 min at 37 °C, 10 min at 60 °C and 3 min at 95 °C.
4.8. DNA Extraction
DNA extraction from the intestine and anterior kidney was performed using Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
4.9. Real-Time PCR
The quantification of the transcript levels of the immunological genes was carried out using real-time PCR. Reactions were performed in 96-well plates (Thermoscientific, Waltham, MA, USA) using the PikoReal 96 Real-Time PCR System (Thermoscientific). The reaction mixture consisted of 5 μL of SsoAdvanced Universal™ SYBR® Green Supermix (Biorad, South Granville, Australia), 0.5 μL of each primer (10 uM), 1 μL of cDNA (80 ng) and 3 μL of ultrapure water (Invitrogen) to complete 10 μL. Subsequently, the transcript levels of the genes (IL-12, IFN-γ, IL-1β, TNF-α, TGF-β, CD4, Perforin and Lysozyme) were quantified. The thermal profile used was 1 cycle at 95 °C for 2 min and 40 cycles at 95 °C for 5 s, 60 °C for 15 s, and 72 °C for 15 s. To analyze the change in the level of transcripts of each gene under study, elongation factor 1α (ef1a) was used to normalize the expression of target genes using the ΔΔCT method. [69]. Statistically significant differences were determined by comparing the control with each treatment, using a one-way nonparametric t-test (Mann–Whitney) (* p < 0.05, ** p < 0.01, *** p < 0.001).
The quantification of the bacterial load was carried out by detecting the 16S rRNA gene of P. salmonis. The reaction was carried out according to the aforementioned protocol, adding 50 ng of DNA. The thermal profile used was 1 cycle at 95 °C for 2 min, 35 cycles at 95 °C for 5 s, 60 °C for 15 s, and 72 °C for 15 s. To calculate the number of copies of the gene, a previously made calibration curve was used. The primers used in these experiments are listed in Supplementary Table S1.
4.10. 16 S Ribosomal Sequencing
The 16 S rRNA amplification and high-throughput sequencing was performed by Molecular Research LP (MR DNA; Shallowater, TX, USA) following the protocol described by Vargas et al. 2023 [70]. Briefly, each sample of DNA was diluted to 25 ng/μL. The DNA of three fish per pond were mixed in equal volumes to form a pool. For each condition, two pools were generated. Each DNA pool was used as a template to amplify the V4 variable region using primers 515-F and 806-R [71] with a HotStarTaq Plus Master Mix Kit (Qiagen, Hilden, Germany). The PCR conditions were set up to 94 °C (3 min), 30 cycles of 94 °C (30 s), 53 °C (40 s), and 72 °C (60 s), with a final elongation to 72 °C (5 min). PCR products were analyzed in a 2% agarose gel, purified using Ampure XP beads. Illumina DNA library using purified PCR products was prepared with a TruSeq Nano kit. MiSeq reagent kit v3 on the Illumina MiSeq platform (2 × 300-bp paired ends [PE]) was used for high-throughput sequencing of amplicons following the manufacturer’s guidelines.
4.11. Bioinformatic Analysis
The output data from sequencing were processed following the pipeline described by Vargas et at 2023 [70]. Briefly, data were processed using QIIME2 version 2020.2 [72]. DADA2 [73] was used for quality filtering and prediction of Amplicon Sequence Variants (ASV). The reads (forward and reverse) were truncated to 250 nts. ASVs/features were taxonomically classified using a pre-trained Naive Bayes taxonomy classifier, Greengenes 13,899% ASVs [74]. We obtained a mean of 108,898 individual sequencing reads per sample (min = 5166; max = 55,565). After data processing, the average number of sequences for each sample passing through ASV classification was 185276 (SD: 105,416). The average number of ASVs per sample was 30,381 (SD: 19,314). Diversity analyses were performed using the same number of random reads obtained by rarefaction with a sampling depth of 26.200 per sample. The very low-abundance taxa (<0.05%) and taxa not represented in at least half the samples were removed for further analysis.
5. Conclusions
[Cu(NN1)2]ClO4, a copper (I) complex developed in our laboratory, is capable of inhibiting the growth of P. salmonis isolates like LF-89 and EM-90, demonstrating its broad effect on this bacterium. On the other hand, the administration of the compound through diet is capable of modulating both the intestinal microbiota and the cellular immune response in Atlantic salmon; however, we cannot determine whether the effect of the compound modulates the microbiota, which in turn modulates the immune response, or whether they are independent effects. Future work is needed to determine this. On the other hand, these could be the mechanisms of action by which the compound is capable of generating protection against a challenge against P. salmonis. Finally, the results obtained in this work demonstrated the potential of using [Cu(NN1)2]ClO4 as a treatment against P. salmonis, an intracellular pathogen of great relevance in the Chilean salmon industry, representing a promising alternative to the use of antibiotics. However, it is necessary to continue carrying out tests and studies to increase the characterization of the effect of the compound, such as the evaluation of toxicity on other aquatic organisms, its accumulation on the seabed, and synergism effect with antibiotics, among other experiments.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25073700/s1.
Author Contributions
Conceptualization, M.P., B.M. and M.T. Methodology, M.P. Investigation, M.P., S.S.-A., M.A. and P.A. Resources, M.A., J.G. and B.M. Writing—original draft preparation, M.P. Writing, review and editing, M.P., M.T., B.M. and R.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by VIU 21P0017 to M.A., DICYT022341MC to B.M., DICYT022343TR to M.T.
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board of Universidad de Santiago de Chile (protocol code No. 354/2021, 4 October 2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Materials.
Acknowledgments
The authors thank Aldo Gaggero and Verónica Cambiazo for providing the P. salmonis isolates used in this work. The authors also acknowledge Carolina Zavala for the technical support in handling the fish in this work. M.P. gives thanks to National Doctorate Scholarship No.: 21212170 (ANID, Government of Chile). This research was supported by the high-performance computing system PIDi-UTEM (SCC-PIDi-UTEM—CONICYT—FONDEQUIP—EQM180180). The researchers thank Daniel Conejeras for technical support.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- FAO. The State of World Fisheries and Aquaculture 2022; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- SERNAPESCA. Informe Sanitario Con Información Sanitaria de Agua Dulce y Mar Año 2022; Informe Sanitario; Ministerio de Economía, Fomento y Turismo: Valparaíso, Chile, 2023. [Google Scholar]
- Mohapatra, S.; Chakraborty, T.; Kumar, V.; Deboeck, G.; Mohanta, K.N. Aquaculture and Stress Management: A Review of Probiotic Intervention. J. Anim. Physiol. Anim. Nutr. 2013, 97, 405–430. [Google Scholar] [CrossRef] [PubMed]
- Maisey, K.; Montero, R.; Christodoulides, M. Vaccines for Piscirickettsiosis (Salmonid Rickettsial Septicaemia, SRS): The Chile Perspective. Expert. Rev. Vaccines 2017, 16, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Rozas, M.; Enríquez, R. Piscirickettsiosis and Piscirickettsia salmonis in Fish: A Review. J. Fish. Dis. 2014, 37, 163–188. [Google Scholar] [CrossRef]
- Yañez, A.J.; Valenzuela, K.; Silva, H.; Retamales, J.; Romero, A.; Enriquez, R.; Figueroa, J.; Claude, A.; Gonzalez, J.; Avendaño-Herrera, R.; et al. Broth Medium for the Successful Culture of the Fish Pathogen Piscirickettsia salmonis. Dis. Aquat. Organ. 2012, 97, 197–205. [Google Scholar] [CrossRef]
- Rojas, V.; Galanti, N.; Bols, N.C.; Jiménez, V.; Paredes, R.; Marshall, S.H. Piscirickettsia salmonis Induces Apoptosis in Macrophages and Monocyte-like Cells from Rainbow Trout. J. Cell Biochem. 2010, 110, 468–476. [Google Scholar] [CrossRef]
- Gómez, F.A.; Tobar, J.A.; Henríquez, V.; Sola, M.; Altamirano, C.; Marshall, S.H. Evidence of the Presence of a Functional Dot/Icm Type IV-B Secretion System in the Fish Bacterial Pathogen Piscirickettsia salmonis. PLoS ONE 2013, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rozas-Serri, M.; Peña, A.; Arriagada, G.; Enríquez, R.; Maldonado, L. Comparison of Gene Expression in Post-Smolt Atlantic Salmon Challenged by LF-89-like and EM-90-like Piscirickettsia salmonis Isolates Reveals Differences in the Immune Response Associated with Pathogenicity. J. Fish. Dis. 2018, 41, 539–552. [Google Scholar] [CrossRef]
- Rozas-Serri, M.; Peña, A.; Maldonado, L. Transcriptomic Profiles of Post-Smolt Atlantic Salmon Challenged with Piscirickettsia salmonis Reveal a Strategy to Evade the Adaptive Immune Response and Modify Cell-Autonomous Immunity. Dev. Comp. Immunol. 2018, 81, 348–362. [Google Scholar] [CrossRef]
- Almendras, F.E.; Fuentealba, I.C. Salmonid Rickettsial Septicemia Caused by Piscirickettsia salmonis: A Review. Dis. Aquat. Organ. 1997, 29, 137–144. [Google Scholar] [CrossRef]
- Cvitanich, J.D. The Isolation of a Rickettsia-like Organism Causing Disease and Mortality in Chilean Salmonids and Its Confirmation by Koch’s Postulate. J. Fish. Dis. 1991, 14, 2–45. [Google Scholar] [CrossRef]
- Figueroa, C.; Torrealba, D.; Morales-Lange, B.; Mercado, L.; Dixon, B.; Conejeros, P.; Silva, G.; Soto, C.; Gallardo, J.A. Commercial Vaccines Do Not Confer Protection against Two Genogroups of Piscirickettsia salmonis, LF-89 and EM-90, in Atlantic Salmon. Biology 2022, 11, 993. [Google Scholar] [CrossRef]
- SERNAPESCA. Informe Sobre Uso de Antimicrobianos En La Salmonicultura Nacional; Informe Sanitario; Ministerio de Economía, Fomento y Turismo: Valparaíso, Chile, 2023. [Google Scholar]
- Henríquez, P.; Kaiser, M.; Bohle, H.; Bustos, P.; Mancilla, M. Comprehensive Antibiotic Susceptibility Profiling of Chilean Piscirickettsia salmonis Field Isolates. J. Fish. Dis. 2016, 39, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, J.; Hernandez, N.; Osses, A.; Castillo, A.; Cancino, A.; Grothusen, H.; Navas, E.; Henriquez, P.; Bohle, H.; Bustamante, F.; et al. Prevalence, Geographic Distribution and Phenotypic Differences of Piscirickettsia salmonis EM-90-like Isolates. J. Fish. Dis. 2017, 40, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Habibullah-Al-Mamun, M.; Nagano, I.; Masunaga, S.; Kitazawa, D.; Matsuda, H. Antibiotics, Antibiotic-Resistant Bacteria, and Resistance Genes in Aquaculture: Risks, Current Concern, and Future Thinking. Environ. Sci. Pollut. Res. 2022, 29, 11054–11075. [Google Scholar] [CrossRef] [PubMed]
- Quiñones, R.A.; Fuentes, M.; Montes, R.M.; Soto, D.; León-Muñoz, J. Environmental Issues in Chilean Salmon Farming: A Review. Rev. Aquac. 2019, 11, 375–402. [Google Scholar] [CrossRef]
- Donati, V.L.; Madsen, L.; Middelboe, M.; Strube, M.L.; Dalsgaard, I. The Gut Microbiota of Healthy and Flavobacterium psychrophilum-Infected Rainbow Trout Fry Is Shaped by Antibiotics and Phage Therapies. Front. Microbiol. 2022, 13, 771296. [Google Scholar] [CrossRef] [PubMed]
- Huyben, D.; Jarau, M.; MacInnes, J.; Stevenson, R.; Lumsden, J. Impact of Infection with Flavobacterium psychrophilum and Antimicrobial Treatment on the Intestinal Microbiota of Rainbow Trout. Pathogens 2023, 12, 454. [Google Scholar] [CrossRef]
- Xavier, R.; Severino, R.; Silva, S.M. Signatures of Dysbiosis in Fish Microbiomes in the Context of Aquaculture. Rev. Aquac. 2023, 16, 706–731. [Google Scholar] [CrossRef]
- Hu, Y.; Shen, Y.; Wu, X.; Tu, X.; Wang, G.X. Synthesis and Biological Evaluation of Coumarin Derivatives Containing Imidazole Skeleton as Potential Antibacterial Agents. Eur. J. Med. Chem. 2018, 143, 958–969. [Google Scholar] [CrossRef]
- Liu, G.; Wang, C.; Wang, H.; Zhu, L.; Zhang, H.; Wang, Y.; Pei, C.; Liu, L. Antiviral Efficiency of a Coumarin Derivative on Spring Viremia of Carp Virus In Vivo. Virus Res. 2019, 268, 11–17. [Google Scholar] [CrossRef]
- Liu, G.L.; Hu, Y.; Chen, X.H.; Wang, G.X.; Ling, F. Synthesis and Anthelmintic Activity of Coumarin–Imidazole Hybrid Derivatives against Dactylogyrus intermedius in Goldfish. Bioorg Med. Chem. Lett. 2016, 26, 5039–5043. [Google Scholar] [CrossRef]
- Krasnovskaya, O.; Naumov, A.; Guk, D.; Gorelkin, P.; Erofeev, A.; Beloglazkina, E.; Majouga, A. Copper Coordination Compounds as Biologically Active Agents. Int. J. Mol. Sci. 2020, 21, 3965. [Google Scholar] [CrossRef] [PubMed]
- Djoko, K.Y.; S Achard, M.E.; Phan, M.-D.; Lo, A.W.; Miraula, M.; Prombhul, S.; Hancock, S.J.; Peters, K.M.; Sidjabat, H.E.; Harris, P.N.; et al. Copper Ions and Coordination Complexes as Novel Carbapenem Adjuvants. Antimicrob. Agents Chemother. 2018, 62, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, J.; Xu, L.; Wang, B.; Lin, W.; Luo, Y. Copper Regulation of Immune Response and Potential Implications for Treating Orthopedic Disorders. Front. Mol. Biosci. 2022, 9, 1065265. [Google Scholar] [CrossRef] [PubMed]
- Dawood, M.A.O. Dietary Copper Requirements for Aquatic Animals: A Review. Biol. Trace Elem. Res. 2022, 200, 5273–5282. [Google Scholar] [CrossRef] [PubMed]
- Schrader, K.; Harries, M.D.; Darwish, A.M. In vitro comparisons of the inhibitory activity of florfenicol copper sulfate and potassium permanganate towards Aeromonas hydrophila and Flavobacterium columnare. Aquac. Res. 2013, 44, 212–219. [Google Scholar] [CrossRef]
- Cáceres-Vásquez, J.; Jara, D.H.; Costamagna, J.; Martínez-Gómez, F.; Silva, C.P.; Lemus, L.; Freire, E.; Baggio, R.; Vera, C.; Guerrero, J. Effect of Non-Covalent Self-Dimerization on the Spectroscopic and Electrochemical Properties of Mixed Cu(i) Complexes. RSC Adv. 2023, 13, 825–838. [Google Scholar] [CrossRef]
- Aldabaldetrecu, M.; Parra, M.; Soto, S.; Arce, P.; Tello, M.; Guerrero, J.; Modak, B. New Copper(I) Complex with a Coumarin as Ligand with Antibacterial Activity against Flavobacterium psychrophilum. Molecules 2020, 25, 3183. [Google Scholar] [CrossRef]
- Aldabaldetrecu, M.; Parra, M.; Soto-Aguilera, S.; Arce, P.; de la Vega Quiroz, A.P.; Segura, R.; Tello, M.; Guerrero, J.; Modak, B. Protective Effect of [Cu(NN1)2](ClO4) Complex in Rainbow Trout Challenged against Flavobacterium psychrophilum. Microorganisms 2022, 10, 2296. [Google Scholar] [CrossRef]
- Figueroa, J.; Castro, D.; Lagos, F.; Cartes, C.; Isla, A.; Yáñez, A.J.; Avendaño-Herrera, R.; Haussmann, D. Analysis of Single Nucleotide Polymorphisms (SNPs) Associated with Antibiotic Resistance Genes in Chilean Piscirickettsia salmonis Strains. J. Fish. Dis. 2019, 42, 1645–1655. [Google Scholar] [CrossRef]
- Tandberg, J.I.; Lagos, L.X.; Langlete, P.; Berger, E.; Rishovd, A.L.; Roos, N.; Varkey, D.; Paulsen, I.T.; Winther-Larsen, H.C. Comparative Analysis of Membrane Vesicles from Three Piscirickettsia salmonis Isolates Reveals Differences in Vesicle Characteristics. PLoS ONE 2016, 11, e0165099. [Google Scholar] [CrossRef] [PubMed]
- Hernández, A.J.; Romero, A.; Gonzalez-Stegmaier, R.; Dantagnan, P. The Effects of Supplemented Diets with a Phytopharmaceutical Preparation from Herbal and Macroalgal Origin on Disease Resistance in Rainbow Trout against Piscirickettsia salmonis. Aquaculture 2016, 454, 109–117. [Google Scholar] [CrossRef]
- Romero, A.; Pérez, T.; Santibáñez, N.; Vega, M.; Miranda, P. Phytogenic Feed Additive (PFA) Standardized in Labdane Diterpens Have a Protective Effect in Salmo salar against Piscirickettsia salmonis. Aquaculture 2021, 533, 736170. [Google Scholar] [CrossRef]
- Lichtenheld, M.G.; Olsen, K.J.; Lu, P.; Lowrey, D.M.; Hameed, A.; Hengartner, H.; Podack, E.R. Structure and Function of Human Perforin. Nature 1988, 335, 448–451. [Google Scholar] [CrossRef] [PubMed]
- Jung, M.H.; Jung, S.J. CpG ODN 1668 Induce Innate and Adaptive Immune Responses in Rock Bream (Oplegnathus fasciatus) against Rock Bream Iridovirus (RBIV) Infection. Fish Shellfish Immunol. 2017, 69, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Samsing, F.; Alexandre, P.; Rigby, M.; Taylor, R.S.; Chong, R.; Wynne, J.W. Transcriptome Response of Atlantic Salmon (Salmo salar) to a New Piscine Orthomyxovirus. Pathogens 2020, 9, 807. [Google Scholar] [CrossRef] [PubMed]
- Robinson, N.; Baranski, M.; Mahapatra, K.D.; Saha, J.N.; Das, S.; Mishra, J.; Das, P.; Kent, M.; Arnyasi, M.; Sahoo, P.K. A Linkage Map of Transcribed Single Nucleotide Polymorphisms in Rohu (Labeo rohita) and QTL Associated with Resistance to Aeromonas hydrophila. BMC Genom. 2014, 15, 541. [Google Scholar] [CrossRef][Green Version]
- Li, T.; Wang, L.; Zhang, Y.; Guo, X.; Chen, X.; Zhang, F.; Yang, G.; Wen, W.; Li, H. Molecular Characterization of Three Novel Perforins in Common Carp (Cyprinus carpio L.) and Their Expression Patterns during Larvae Ontogeny and in Response to Immune Challenges. BMC Vet. Res. 2018, 14, 299. [Google Scholar] [CrossRef]
- Saurabh, S.; Sahoo, P.K. Lysozyme: An Important Defence Molecule of Fish Innate Immune System. Aquac. Res. 2008, 39, 223–239. [Google Scholar] [CrossRef]
- Song, Q.; Xiao, Y.; Xiao, Z.; Liu, T.; Li, J.; Li, P.; Han, F. Lysozymes in Fish. J. Agric. Food Chem. 2021, 69, 15039–15051. [Google Scholar] [CrossRef]
- Buonocore, F.; Randelli, E.; Trisolino, P.; Facchiano, A.; de Pascale, D.; Scapigliati, G. Molecular Characterization, Gene Structure and Antibacterial Activity of a g-Type Lysozyme from the European Sea Bass (Dicentrarchus labrax L.). Mol. Immunol. 2014, 62, 10–18. [Google Scholar] [CrossRef]
- Wei, S.; Huang, Y.; Huang, X.; Cai, J.; Wei, J.; Li, P.; Ouyang, Z.; Qin, Q. Molecular Cloning and Characterization of a New G-Type Lysozyme Gene (Ec-LysG) in Orange-Spotted Grouper, Epinephelus coioides. Dev. Comp. Immunol. 2014, 46, 401–412. [Google Scholar] [CrossRef]
- Jahazi, M.A.; Hoseinifar, S.H.; Jafari, V.; Hajimoradloo, A.; Van Doan, H.; Paolucci, M. Dietary Supplementation of Polyphenols Positively Affects the Innate Immune Response, Oxidative Status, and Growth Performance of Common Carp, Cyprinus carpio L. Aquaculture 2020, 517, 734709. [Google Scholar] [CrossRef]
- Ahmadi, A.; Bagheri, D.; Hoseinifar, S.H.; Morshedi, V.; Paolucci, M. Beneficial Role of Polyphenols as Feed Additives on Growth Performances, Immune Response and Antioxidant Status of Lates calcarifer (Bloch, 1790) Juveniles. Aquaculture 2022, 552, 737955. [Google Scholar] [CrossRef]
- Li, M.; Zhu, X.; Tian, J.; Liu, M.; Wang, G. Dietary Flavonoids from Allium mongolicum Regel Promotes Growth, Improves Immune, Antioxidant Status, Immune-Related Signaling Molecules and Disease Resistance in Juvenile Northern Snakehead Fish (Channa argus). Aquaculture 2019, 501, 473–481. [Google Scholar] [CrossRef]
- Boehm, U.; Klamp, T.; Groot, M.; Howard, J.C. 40 Annual Reviews AR026-28 AR26-28. Annu. Rev. Immunol. 1997, 8, 749–795. [Google Scholar] [CrossRef] [PubMed]
- Green, D.S.; Young, H.A.; Valencia, J.C. Current Prospects of Type II Interferon γ Signaling & Autoimmunity. J. Biol. Chem. 2017, 292, 13925–13933. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage Plasticity and Polarization: In Vivo Veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Pereiro, P.; Figueras, A.; Novoa, B. Insights into Teleost Interferon-Gamma Biology: An Update. Fish. Shellfish. Immunol. 2019, 90, 150–164. [Google Scholar] [CrossRef]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-γ: An Overview of Signals, Mechanisms and Functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef]
- Yilmaz, S. Effects of Dietary Caffeic Acid Supplement on Antioxidant, Immunological and Liver Gene Expression Responses, and Resistance of Nile Tilapia, Oreochromis niloticus to Aeromonas veronii. Fish. Shellfish. Immunol. 2019, 86, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Dawood, M.A.O.; Metwally, A.E.S.; El-Sharawy, M.E.; Ghozlan, A.M.; Abdel-Latif, H.M.R.; Van Doan, H.; Ali, M.A.M. The Influences of Ferulic Acid on the Growth Performance, Haemato-Immunological Responses, and Immune-Related Genes of Nile Tilapia (Oreochromis niloticus) Exposed to Heat Stress. Aquaculture 2020, 525, 735320. [Google Scholar] [CrossRef]
- Yılmaz, S.; Ergün, S. Trans-Cinnamic Acid Application for Rainbow Trout (Oncorhynchus mykiss): I. Effects on Haematological, Serum Biochemical, Non-Specific Immune and Head Kidney Gene Expression Responses. Fish. Shellfish. Immunol. 2018, 78, 140–157. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.F.; Liu, L.; Feng, C.Z.; Hu, Y.; Chen, C.; Wang, G.X.; Zhu, B. Synthesis and Antiviral Activity of a New Coumarin Derivative against Spring Viraemia of Carp Virus. Fish. Shellfish. Immunol. 2018, 81, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.S.; Shin, N.R.; Lee, J.B.; Kim, M.S.; Whon, T.W.; Hyun, D.W.; Yun, J.H.; Jung, M.J.; Kim, J.Y.; Bae, J.W. Host Habitat Is the Major Determinant of the Gut Microbiome of Fish. Microbiome 2021, 9, 166. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jaramillo-Torres, A.; Li, Y.; Kortner, T.M.; Gajardo, K.; Brevik, Ø.J.; Jakobsen, J.V.; Krogdahl, Å. Microbiota in Intestinal Digesta of Atlantic Salmon (Salmo salar), Observed from Late Freshwater Stage until One Year in Seawater, and Effects of Functional Ingredients: A Case Study from a Commercial Sized Research Site in the Arctic Region. Anim. Microbiome 2021, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Kuebutornye, F.K.A.; Abarike, E.D.; Lu, Y.; Hlordzi, V.; Sakyi, M.E.; Afriyie, G.; Wang, Z.; Li, Y.; Xie, C.X. Mechanisms and the Role of Probiotic Bacillus in Mitigating Fish Pathogens in Aquaculture. Fish. Physiol. Biochem. 2020, 46, 819–841. [Google Scholar] [CrossRef]
- Soltani, M.; Pakzad, K.; Taheri-Mirghaed, A.; Mirzargar, S.; Shekarabi, S.P.H.; Yosefi, P.; Soleymani, N. Dietary Application of the Probiotic Lactobacillus plantarum 426951 Enhances Immune Status and Growth of Rainbow Trout (Oncorhynchus mykiss) Vaccinated Against Yersinia ruckeri. Probiotics Antimicrob. Proteins 2019, 11, 207–219. [Google Scholar] [CrossRef]
- Yi, Y.; Zhang, Z.; Zhao, F.; Liu, H.; Yu, L.; Zha, J.; Wang, G. Probiotic Potential of Bacillus velezensis JW: Antimicrobial Activity against Fish Pathogenic Bacteria and Immune Enhancement Effects on Carassius auratus. Fish. Shellfish. Immunol. 2018, 78, 322–330. [Google Scholar] [CrossRef]
- Xia, Y.; Lu, M.; Chen, G.; Cao, J.; Gao, F.; Wang, M.; Liu, Z.; Zhang, D.; Zhu, H.; Yi, M. Effects of Dietary Lactobacillus rhamnosus JCM1136 and Lactococcus lactis Subsp. Lactis JCM5805 on the Growth, Intestinal Microbiota, Morphology, Immune Response and Disease Resistance of Juvenile Nile Tilapia, Oreochromis niloticus. Fish. Shellfish. Immunol. 2018, 76, 368–379. [Google Scholar] [CrossRef]
- Baumgärtner, S.; James, J.; Ellison, A. The Supplementation of a Prebiotic Improves the Microbial Community in the Gut and the Skin of Atlantic Salmon (Salmo salar). Aquac. Rep. 2022, 25, 101204. [Google Scholar] [CrossRef]
- Ou, W.; Hu, H.; Yang, P.; Dai, J.; Ai, Q.; Zhang, W.; Zhang, Y.; Mai, K. Dietary Daidzein Improved Intestinal Health of Juvenile Turbot in Terms of Intestinal Mucosal Barrier Function and Intestinal Microbiota. Fish. Shellfish. Immunol. 2019, 94, 132–141. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, W.; Hu, G.; Qiu, L.; Meng, S.; Song, C.; Fan, L.; Zhao, Z.; Bing, X.; Chen, J. Gut Microbiota Analysis of Juvenile Genetically Improved Farmed Tilapia (Oreochromis niloticus) by Dietary Supplementation of Different Resveratrol Concentrations. Fish. Shellfish. Immunol. 2018, 77, 200–207. [Google Scholar] [CrossRef]
- Coca, Y.; Godoy, M.; Pontigo, J.P.; Caro, D.; Maracaja-Coutinho, V.; Arias-Carrasco, R.; Rodríguez-Córdova, L.; de Oca, M.M.; Sáez-Navarrete, C.; Burbulis, I. Bacterial Networks in Atlantic Salmon with Piscirickettsiosis. Sci. Rep. 2023, 13, 17321. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, M.P. Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Clinical and Laboratory Standars Institute: Wyne, NJ, USA, 2020; pp. 1–332. ISBN 9781684400324. [Google Scholar]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, 2001–2007. [Google Scholar] [CrossRef] [PubMed]
- Vargas, R.A.; Soto-Aguilera, S.; Parra, M.; Herrera, S.; Santibañez, A.; Kossack, C.; Saavedra, C.P.; Mora, O.; Pineda, M.; Gonzalez, O.; et al. Analysis of Microbiota-Host Communication Mediated by Butyrate in Atlantic Salmon. Comput. Struct. Biotechnol. J. 2023, 21, 2558–2578. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S RRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a Chimera-Checked 16S RRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).