MaSMG7-Mediated Degradation of MaERF12 Facilitates Fusarium oxysporum f. sp. cubense Tropical Race 4 Infection in Musa acuminata
Abstract
1. Introduction
2. Results
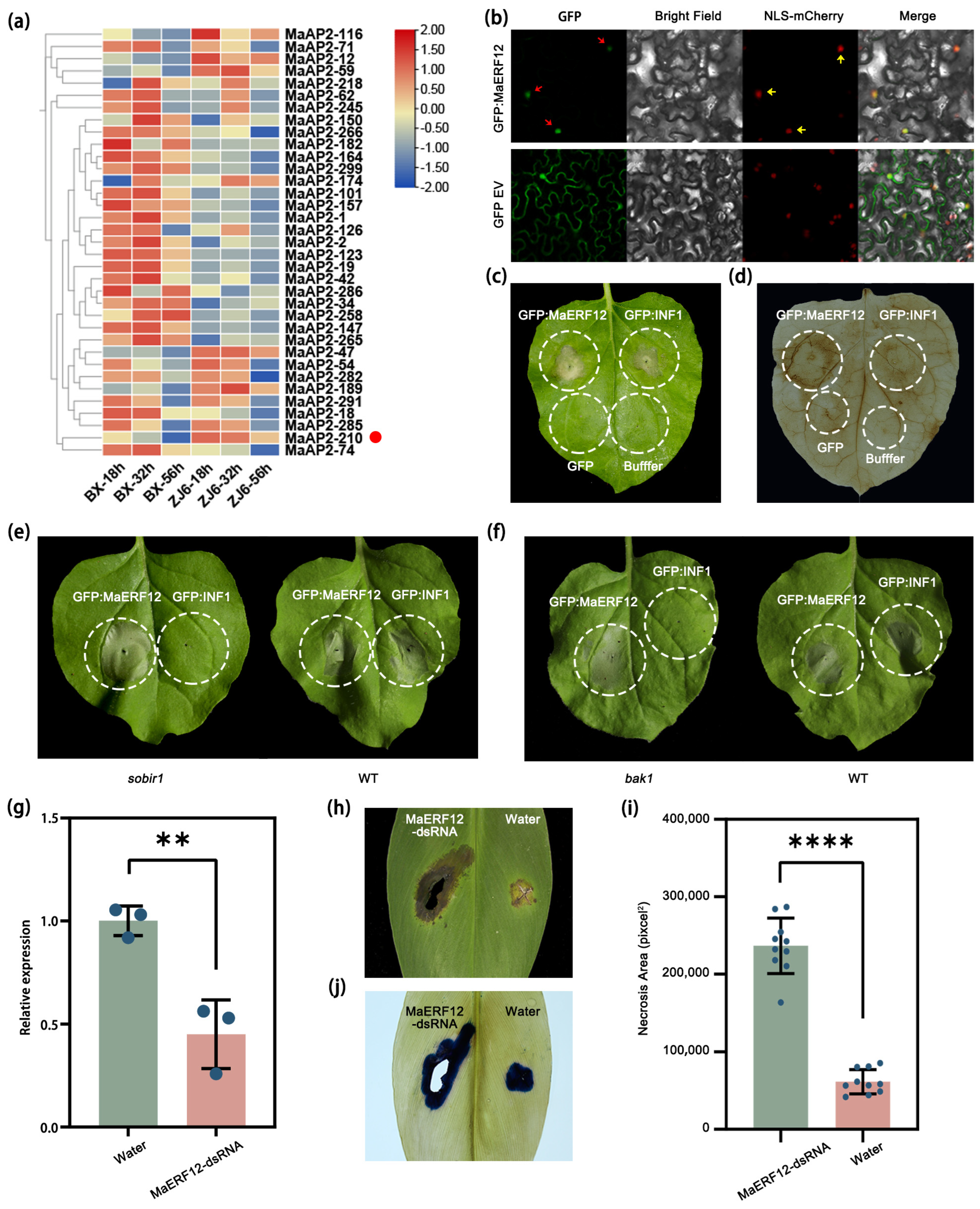
2.1. MaERF12 Positively Regulates Foc TR4 Resistance in Banana
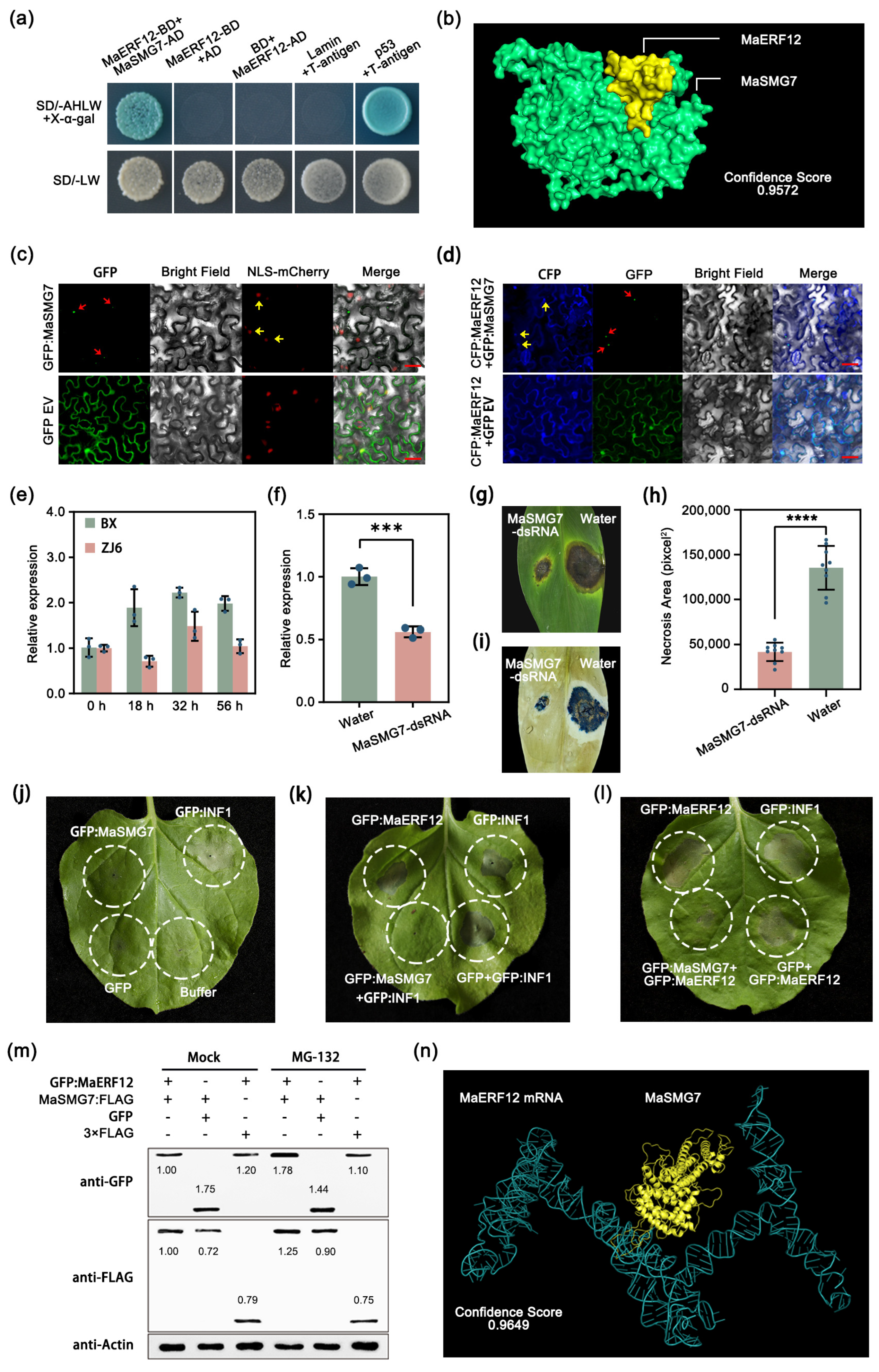
2.2. MaSMG7 Degrades MaERF12 and Negatively Regulates Banana Disease Resistance to Foc TR4
3. Discussion
4. Materials and Methods
4.1. Plant and Fungi Materials, Constructs, and Bacterial Strains
4.2. Identification of AP2/ERF Gene Family
4.3. Plasmid Construction
4.4. RNA Extraction and RT-qPCR
4.5. Transcriptomic Analysis of Banana Genes
4.6. Transient Gene Expression in N. Benthamiana
4.7. dsRNA-Mediated Gene Silencing in Banana
4.8. Trypan Blue Staining
4.9. Yeast Two-Hybrid Assays
4.10. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bubici, G.; Kaushal, M.; Prigigallo, M.I.; Gómez-Lama Cabanás, C.; Mercado-Blanco, J. Biological control agents against Fusarium wilt of banana. Front. Microbiol. 2019, 10, 616. [Google Scholar] [CrossRef] [PubMed]
- Staver, C.; Pemsl, D.E.; Scheerer, L.; Perez Vicente, L.; Dita, M. Ex ante assessment of returns on research investments to address the impact of Fusarium wilt tropical race 4 on global banana production. Front. Plant Sci. 2020, 11, 844. [Google Scholar] [CrossRef]
- Ploetz, R.C. Fusarium Wilt of Banana. Phytopathology 2015, 105, 1512–1521. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Bai, P.; Ning, Y.; Wang, J.; Shi, X.; Xiong, Y.; Zhang, K.; He, F.; Zhang, C.; Wang, R.; et al. The monocot-specific receptor-like kinase SDS2 controls cell death and immunity in rice. Cell Host Microbe 2018, 23, 498–510.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 361, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Hu, L.; Li, Y.; Chen, X.; Zhang, Z.; Liu, B.; Li, P.; Gong, X.; Ma, F. MdUGT88F1-mediated phloridzin biosynthesis regulates apple development and Valsa Canker resistance. Plant Physiol. 2019, 180, 2290–2305. [Google Scholar] [CrossRef] [PubMed]
- Koseoglou, E.; van der Wolf, J.M.; Visser, R.G.F.; Bai, Y. Susceptibility reversed: Modified plant susceptibility genes for resistance to bacteria. Trends Plant Sci. 2022, 27, 69–79. [Google Scholar] [CrossRef]
- Wang, N.; Tang, C.; Fan, X.; He, M.; Gan, P.; Zhang, S.; Hu, Z.; Wang, X.; Yan, T.; Shu, W.; et al. Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 2022, 185, 2961–2974.e19. [Google Scholar] [CrossRef]
- Sun, K.; Schipper, D.; Jacobsen, E.; Visser, R.G.F.; Govers, F.; Bouwmeester, K.; Bai, Y. Silencing susceptibility genes in potato hinders primary infection of Phytophthora infestans at different stages. Hortic. Res. 2022, 9, uhab058. [Google Scholar] [CrossRef]
- Chen, C.; Zhao, Y.; Tabor, G.; Nian, H.; Phillips, J.; Wolters, P.; Yang, Q.; Balint-Kurti, P. A leucine-rich repeat receptor kinase gene confers quantitative susceptibility to maize southern leaf blight. New Phytol. 2023, 238, 1182–1197. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Hou, X. Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 23, 573. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Hou, H.; Wu, Z.; Zhao, J.; Zhang, F.; Teng, R.; Chen, F.; Teng, N. Chrysanthemum embryo development is negatively affected by a novel ERF transcription factor, CmERF12. J. Exp. Bot. 2022, 73, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B. AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol. 2009, 150, 1916–1929. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Zhang, C.L.; Wang, G.L.; Wang, Y.X.; Qi, C.H.; You, C.X.; Li, Y.Y.; Hao, Y.J. Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis. Planta 2019, 249, 1627–1643. [Google Scholar] [CrossRef] [PubMed]
- Gutterson, N.; Reuber, T.L. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 2004, 7, 465–471. [Google Scholar] [CrossRef]
- Jia, Y.L.; Zhang, T.; Zhi, J.R.; Tuo, L.; Yue, W.B.; Li, D.Y.; Liu, L. Combined jasmonic acid and ethylene treatment induces resistance effect in faba bean plants against Frankliniella occidentalis (Pergande) (Thysanoptera: Thripidae). Insects 2022, 13, 1073. [Google Scholar] [CrossRef]
- Dong, N.; Liu, X.; Lu, Y.; Du, L.; Xu, H.; Liu, H.; Xin, Z.; Zhang, Z. Overexpression of TaPIEP1, a pathogen-induced ERF gene of wheat, confers host-enhanced resistance to fungal pathogen Bipolaris sorokiniana. Funct. Integr. Genom. 2010, 10, 215–226. [Google Scholar] [CrossRef]
- Yoshihisa, A.; Yoshimura, S.; Shimizu, M.; Sato, S.; Matsuno, S.; Mine, A.; Yamaguchi, K.; Kawasaki, T. The rice OsERF101 transcription factor regulates the NLR Xa1-mediated immunity induced by perception of TAL effectors. New Phytol. 2022, 236, 1441–1454. [Google Scholar] [CrossRef]
- Wang, J.H.; Gu, K.D.; Han, P.L.; Yu, J.Q.; Wang, C.K.; Zhang, Q.Y.; You, C.X.; Hu, D.G.; Hao, Y.J. Apple ethylene response factor MdERF11 confers resistance to fungal pathogen Botryosphaeria dothidea. Plant Sci. 2020, 291, 110351. [Google Scholar] [CrossRef]
- Liu, D.; Chen, X.; Liu, J.; Ye, J.; Guo, Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 2012, 63, 3899–3911. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, Y.; Yamoto, N.; Suzuki, Y.; Chiba, Y.; Yamazaki, K.; Sato, T.; Yamaguchi, J. The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Sci. 2013, 213, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.Y.; Ohta, M.; Usui, A.; Shinshi, H.; Ohme-Takagi, M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 2000, 12, 393–404. [Google Scholar] [PubMed]
- Tian, Z.; He, Q.; Wang, H.; Liu, Y.; Zhang, Y.; Shao, F.; Xie, C. The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. Plant Cell Physiol. 2015, 56, 992–1005. [Google Scholar] [CrossRef] [PubMed]
- Heese, A.; Hann, D.R.; Gimenez-Ibanez, S.; Jones, A.M.; He, K.; Li, J.; Schroeder, J.I.; Peck, S.C.; Rathjen, J.P. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl. Acad. Sci. USA 2007, 104, 12217–12222. [Google Scholar] [CrossRef] [PubMed]
- Albert, I.; Böhm, H.; Albert, M.; Feiler, C.E.; Imkampe, J.; Wallmeroth, N.; Brancato, C.; Raaymakers, T.M.; Oome, S.; Zhang, H.; et al. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants. 2015, 1, 15140. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Nomura, Y.; Egusa, M.; Hamada, T.; Hyon, G.S.; Kaminaka, H.; Watanabe, Y.; Ueda, T.; Trujillo, M.; Shirasu, K.; et al. The GYF domain protein PSIG1 dampens the induction of cell death during plant-pathogen interactions. PLoS Genet. 2017, 13, e1007037. [Google Scholar] [CrossRef] [PubMed]
- Zang, Z.; Lv, Y.; Liu, S.; Yang, W.; Ci, J.; Ren, X.; Wang, Z.; Wu, H.; Ma, W.; Jiang, L.; et al. A novel ERF transcription factor, ZmERF105, positively regulates maize resistance to Exserohilum turcicum. Front. Plant Sci. 2020, 11, 850. [Google Scholar] [CrossRef]
- Dita, M.; Barquero, M.; Heck, D.; Mizubuti, E.S.G.; Staver, C.P. Fusarium wilt of banana: Current knowledge on epidemiology and research needs toward sustainable disease management. Front. Plant Sci. 2018, 9, 1468. [Google Scholar] [CrossRef]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef]
- Huysmans, M.; Lema, A.S.; Coll, N.S.; Nowack, M.K. Dying two deaths—Programmed cell death regulation in development and disease. Curr. Opin. Plant Biol. 2017, 35, 37–44. [Google Scholar] [CrossRef]
- Govrin, E.M.; Levine, A. The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 2000, 10, 751–757. [Google Scholar] [CrossRef]
- Faris, J.D.; Friesen, T.L. Plant genes hijacked by necrotrophic fungal pathogens. Curr. Opin. Plant Biol. 2020, 56, 74–80. [Google Scholar] [CrossRef]
- Sha, G.; Sun, P.; Kong, X.; Han, X.; Sun, Q.; Fouillen, L.; Zhao, J.; Li, Y.; Yang, L.; Wang, Y.; et al. Genome editing of a rice CDP-DAG synthase confers multipathogen resistance. Nature 2023, 618, 1017–1023. [Google Scholar] [CrossRef]
- Riehs, N.; Akimcheva, S.; Puizina, J.; Bulankova, P.; Idol, R.A.; Siroky, J.; Schleiffer, A.; Schweizer, D.; Shippen, D.E.; Riha, K. Arabidopsis SMG7 protein is required for exit from meiosis. J. Cell Sci. 2008, 121 Pt 13, 2208–2216. [Google Scholar] [CrossRef]
- Riehs-Kearnan, N.; Gloggnitzer, J.; Dekrout, B.; Jonak, C.; Riha, K. Aberrant growth and lethality of Arabidopsis deficient in nonsense-mediated RNA decay factors is caused by autoimmune-like response. Nucleic Acids Res. 2012, 40, 5615–5624. [Google Scholar] [CrossRef] [PubMed]
- Mérai, Z.; Benkovics, A.H.; Nyikó, T.; Debreczeny, M.; Hiripi, L.; Kerényi, Z.; Kondorosi, É.; Silhavy, D. The late steps of plant nonsense-mediated mRNA decay. Plant J. 2013, 73, 50–62. [Google Scholar] [CrossRef]
- Xing, A.; Gao, Y.; Ye, L.; Zhang, W.; Cai, L.; Ching, A.; Llaca, V.; Johnson, B.; Liu, L.; Yang, X.; et al. A rare SNP mutation in Brachytic2 moderately reduces plant height and increases yield potential in maize. J. Exp. Bot. 2015, 66, 3791–3802. [Google Scholar] [CrossRef]
- Li, N.; Lin, B.; Wang, H.; Li, X.; Yang, F.; Ding, X.; Yan, J.; Chu, Z. Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat. Genet. 2019, 51, 1540–1548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Yan, X.; Gebrewahid, T.W.; Zhou, Y.; Yang, E.; Xia, X.; He, Z.; Li, Z.; Liu, D. Genome-wide association mapping of leaf rust and stripe rust resistance in wheat accessions using the 90K SNP array. Theor. Appl. Genet. 2021, 134, 1233–1251. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Hwang, I.S.; Choi, D.S.; Kim, N.H.; Kim, D.S.; Hwang, B.K. Pathogenesis-related protein 4b interacts with leucine-rich repeat protein 1 to suppress PR4b-triggered cell death and defense response in pepper. Plant J. 2014, 77, 521–533. [Google Scholar] [CrossRef]
- Wang, N.; Yin, Z.; Zhao, Y.; Wang, J.; Pei, Y.; Ji, P.; Daly, P.; Li, Z.; Dou, D.; Wei, L. An F-box protein attenuates fungal xylanase-triggered immunity by destabilizing LRR-RLP NbEIX2 in a SOBIR1-dependent manner. New Phytol. 2022, 236, 2202–2215. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Y.; Zhang, X.; Chen, Z.; Xia, Y.; Wang, L.; Sun, Y.; Zhang, M.; Xiao, Y.; Han, Z.; et al. Plant receptor-like protein activation by a microbial glycoside hydrolase. Nature 2022, 610, 335–342. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.; Sun, Y.; Wang, H.; Qi, J.; Wan, B.; Ye, W.; Lin, Y.; Shao, Y.; Dong, S.; et al. Leucine-rich repeat receptor-like gene screen reveals that Nicotiana RXEG1 regulates glycoside hydrolase 12 MAMP detection. Nat. Commun. 2018, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Tian, Y.; Huo, Y.; Liu, Y.; Yang, W.; Li, Y.; Zhuo, M.; Xiang, D.; Li, C.; Yi, G.; et al. The autophagy-related Musa acuminata protein MaATG8F interacts with MaATG4B, regulating banana disease resistance to Fusarium oxysporum f. sp. cubense tropical race 4. J. Fungi 2024, 10, 91. [Google Scholar] [CrossRef]
- Fernández-Bautista, N.; Domínguez-Núñez, J.A.; Moreno, M.M.C.; Berrocal-Lobo, M. Plant tissue trypan blue staining during phytopathogen infection. Bio-protocol 2016, 6, e2078. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, H.; Liu, S.; Huo, Y.; Tian, Y.; Liu, Y.; Yi, G.; Li, C. MaSMG7-Mediated Degradation of MaERF12 Facilitates Fusarium oxysporum f. sp. cubense Tropical Race 4 Infection in Musa acuminata. Int. J. Mol. Sci. 2024, 25, 3420. https://doi.org/10.3390/ijms25063420
Huang H, Liu S, Huo Y, Tian Y, Liu Y, Yi G, Li C. MaSMG7-Mediated Degradation of MaERF12 Facilitates Fusarium oxysporum f. sp. cubense Tropical Race 4 Infection in Musa acuminata. International Journal of Molecular Sciences. 2024; 25(6):3420. https://doi.org/10.3390/ijms25063420
Chicago/Turabian StyleHuang, Huoqing, Siwen Liu, Yile Huo, Yuzhen Tian, Yushan Liu, Ganjun Yi, and Chunyu Li. 2024. "MaSMG7-Mediated Degradation of MaERF12 Facilitates Fusarium oxysporum f. sp. cubense Tropical Race 4 Infection in Musa acuminata" International Journal of Molecular Sciences 25, no. 6: 3420. https://doi.org/10.3390/ijms25063420
APA StyleHuang, H., Liu, S., Huo, Y., Tian, Y., Liu, Y., Yi, G., & Li, C. (2024). MaSMG7-Mediated Degradation of MaERF12 Facilitates Fusarium oxysporum f. sp. cubense Tropical Race 4 Infection in Musa acuminata. International Journal of Molecular Sciences, 25(6), 3420. https://doi.org/10.3390/ijms25063420





