Gastric Cancer in the Era of Epigenetics
Abstract
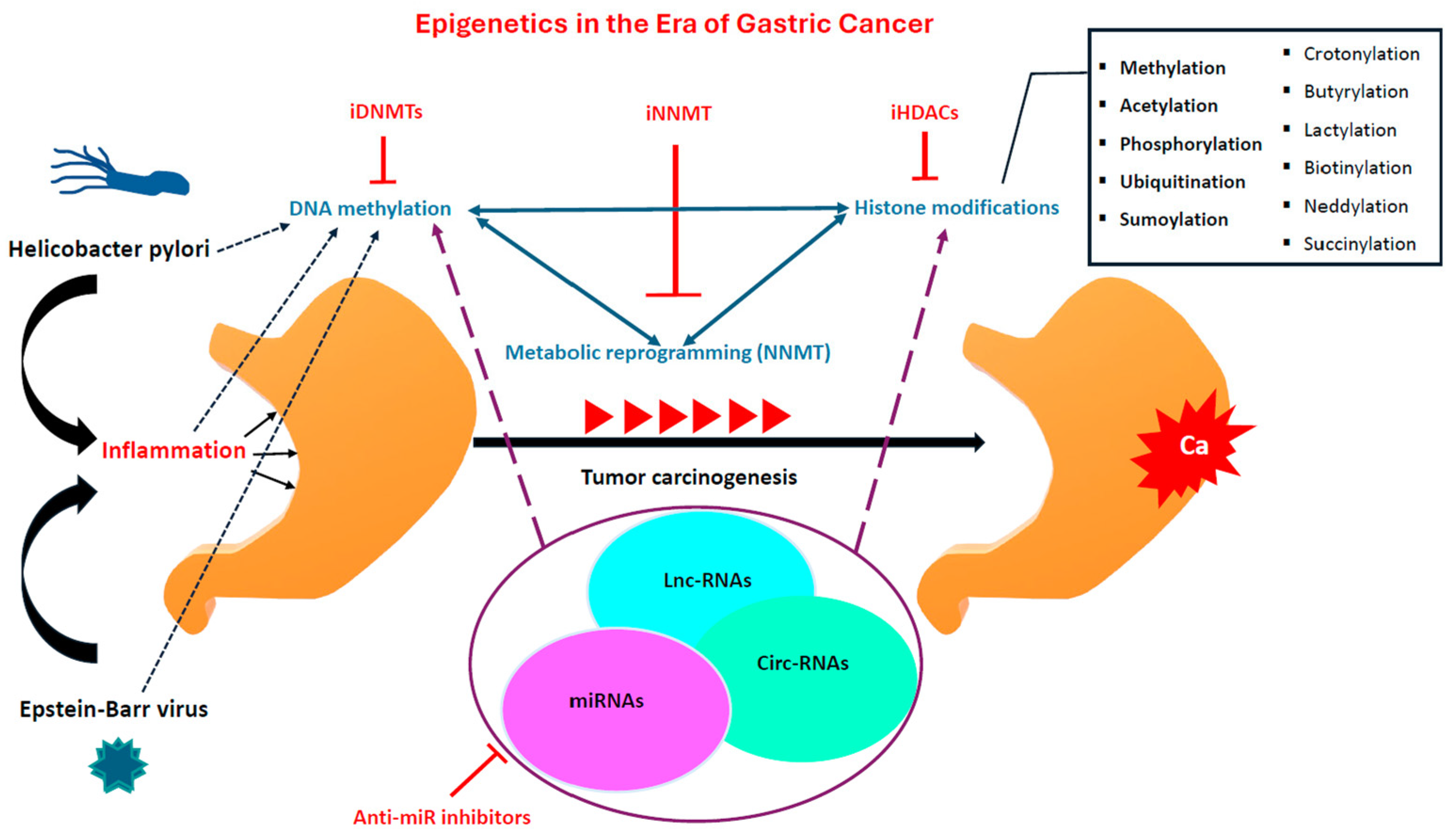
1. Introduction
2. Methods
3. Epigenetics Background (DNA Methylation, Histone Modifications, and Non-Coding RNAs)
3.1. DNA Methylation
3.2. Histone Modifications
3.3. Non-Coding RNAs
4. Gastric-Cancer Clinical Management
5. Discussion
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Arnold, M.; Park, J.Y.; Camargo, M.C.; Lunet, N.; Forman, D.; Soerjomataram, I. Is Gastric Cancer Becoming a Rare Disease? A Global Assessment of Predicted Incidence Trends to 2035. Gut 2020, 69, 823–829. [Google Scholar] [CrossRef]
- Rawla, P.; Barsouk, A. Epidemiology of Gastric Cancer: Global Trends, Risk Factors and Prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- Toh, J.W.T.; Wilson, R.B. Pathways of Gastric Carcinogenesis, Helicobacter Pylori Virulence and Interactions with Antioxidant Systems, Vitamin C and Phytochemicals. Int. J. Mol. Sci. 2020, 21, 6451. [Google Scholar] [CrossRef]
- Puneet; Kazmi, H.R.; Kumari, S.; Tiwari, S.; Khanna, A.; Narayan, G. Epigenetic Mechanisms and Events in Gastric Cancer-Emerging Novel Biomarkers. Pathol. Oncol. Res. 2018, 24, 757–770. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Cristescu, R.; Lee, J.; Nebozhyn, M.; Kim, K.-M.; Ting, J.C.; Wong, S.S.; Liu, J.; Yue, Y.G.; Wang, J.; Yu, K.; et al. Molecular Analysis of Gastric Cancer Identifies Subtypes Associated with Distinct Clinical Outcomes. Nat. Med. 2015, 21, 449–456. [Google Scholar] [CrossRef]
- Hu, X.; Wang, Z.; Wang, Q.; Chen, K.; Han, Q.; Bai, S.; Du, J.; Chen, W. Molecular Classification Reveals the Diverse Genetic and Prognostic Features of Gastric Cancer: A Multi-Omics Consensus Ensemble Clustering. Biomed. Pharmacother. 2021, 144, 112222. [Google Scholar] [CrossRef]
- Wang, H.; Ding, Y.; Chen, Y.; Jiang, J.; Chen, Y.; Lu, J.; Kong, M.; Mo, F.; Huang, Y.; Zhao, W.; et al. A Novel Genomic Classification System of Gastric Cancer via Integrating Multidimensional Genomic Characteristics. Gastric. Cancer 2021, 24, 1227–1241. [Google Scholar] [CrossRef]
- Li, B.; Zhang, F.; Niu, Q.; Liu, J.; Yu, Y.; Wang, P.; Zhang, S.; Zhang, H.; Wang, Z. A Molecular Classification of Gastric Cancer Associated with Distinct Clinical Outcomes and Validated by an XGBoost-Based Prediction Model. Mol. Ther. Nucleic Acids 2023, 31, 224–240. [Google Scholar] [CrossRef]
- Weng, S.; Li, M.; Deng, J.; Xu, H.; Ren, Y.; Zhou, Z.; Wang, L.; Zhang, Y.; Xing, Z.; Li, L.; et al. Epigenetically regulated gene expression profiles decipher four molecular subtypes with prognostic and therapeutic implications in gastric cancer. Clin. Epigenetics 2023, 15, 64. [Google Scholar] [CrossRef]
- Yousefi, B.; Mohammadlou, M.; Abdollahi, M.; Salek Farrokhi, A.; Karbalaei, M.; Keikha, M.; Kokhaei, P.; Valizadeh, S.; Rezaiemanesh, A.; Arabkari, V.; et al. Epigenetic Changes in Gastric Cancer Induction by Helicobacter pylori. J. Cell. Physiol. 2019, 234, 21770–21784. [Google Scholar] [CrossRef]
- Ushijima, T.; Watanabe, N.; Shimizu, K.; Miyamoto, K.; Sugimura, T.; Kaneda, A. Decreased fidelity in replicating CpG methylation patterns in cancer cells. Cancer Res. 2005, 65, 11–17. [Google Scholar] [CrossRef]
- Ushijima, T.; Okochi-Takada, E. Aberrant methylations in cancer cells: Where do they come from? Cancer Sci. 2005, 96, 206–211. [Google Scholar] [CrossRef]
- Lee, T.L.; Leung, W.K.; Chan, M.W.; Ng, E.K.; Tong, J.H.; Lo, K.W.; Chung, S.C.; Sung, J.J.; To, K.F. Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin. Cancer Res. 2002, 8, 1761–1766. [Google Scholar]
- Miyamoto, K.; Ushijima, T. Diagnostic and therapeutic applications of epigenetics. Jpn. J. Clin. Oncol. 2005, 35, 293–301. [Google Scholar] [CrossRef]
- Yamada, H.; Shinmura, K.; Goto, M.; Iwaizumi, M.; Konno, H.; Kataoka, H.; Yamada, M.; Ozawa, T.; Tsuneyoshi, T.; Tanioka, F.; et al. Absence of germline mono-allelic promoter hypermethylation of the CDH1 gene in gastric cancer patients. Mol. Cancer 2009, 8, 63. [Google Scholar] [CrossRef]
- Zeng, W.; Zhu, J.; Shan, L.; Han, Z.; Aerxiding, P.; Quhai, A.; Zeng, F.; Wang, Z.; Li, H. The Clinicopathological Significance of CDH1 in Gastric Cancer: A Meta-Analysis and Systematic Review. Drug Des. Devel. Ther. 2015, 9, 2149–2157. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Lu, Y.; Herman, J.G.; Brock, M.V.; Zhao, P.; Guo, M. Predictive Value of CHFR and MLH1 Methylation in Human Gastric Cancer. Gastric. Cancer 2015, 18, 280–287. [Google Scholar] [CrossRef]
- Dong, C.-X.; Deng, D.-J.; Pan, K.-F.; Zhang, L.; Zhang, Y.; Zhou, J.; You, W.-C. Promoter Methylation of P16 Associated with Helicobacter pylori Infection in Precancerous Gastric Lesions: A Population-Based Study. Int. J. Cancer 2009, 124, 434–439. [Google Scholar] [CrossRef]
- He, D.; Zhang, Y.; Zhang, N.; Zhou, L.; Chen, J.; Jiang, Y.; Shao, C. Aberrant Gene Promoter Methylation of P16, FHIT, CRBP1, WWOX, and DLC-1 in Epstein-Barr Virus-Associated Gastric Carcinomas. Med. Oncol. 2015, 32, 92. [Google Scholar] [CrossRef]
- Wang, X.; Hou, Q.; Zhou, X. LAMA4 Expression Is Activated by Zinc Finger E-box-binding Homeobox 1 and Independently Predicts Poor Overall Survival in Gastric Cancer. Oncol. Rep. 2018, 40, 1725–1733. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, M.; Guo, Y.; Shen, S.; Guo, X.; Dong, Z. FBXO32, a New TGF-β/Smad Signaling Pathway Target Gene, Is Epigenetically Inactivated in Gastric Cardia Adenocarcinoma. Neoplasma 2015, 62, 646–657. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yuan, S.; Li, L.; Xiang, S.; Jia, H.; Luo, T. Cadherin-11 Is Inactivated Due to Promoter Methylation and Functions in Colorectal Cancer as a Tumour Suppressor. Cancer Manag. Res. 2019, 11, 2517–2529. [Google Scholar] [CrossRef]
- Lin, Y.-L.; Gui, S.-L.; Ma, J.-G. Aberrant Methylation of CDH11 Predicts a Poor Outcome for Patients with Bladder Cancer. Oncol. Lett. 2015, 10, 647–652. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, J.; Kim, W.H.; Byeon, S.-J.; Lee, B.L.; Kim, M.A. Epigenetic Downregulation and Growth Inhibition of IGFBP7 in Gastric Cancer. Asian Pac. J. Cancer Prev. 2018, 19, 667–675. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, W.; Chen, S.; Chen, Y.; Chen, L.; Zhang, S. Methylation of the Claudin-3 Promoter Predicts the Prognosis of Advanced Gastric Adenocarcinoma. Oncol. Rep. 2018, 40, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, G.; He, D.; Yang, F.; He, G.; He, L.; Zhang, H.; Deng, Y.; Fan, M.; Shen, L.; et al. Telomerase Reverse Transcriptase Methylation Predicts Lymph Node Metastasis and Prognosis in Patients with Gastric Cancer. Onco Targets Ther. 2016, 9, 279–286. [Google Scholar] [CrossRef][Green Version]
- Vahidi, S.; Norollahi, S.E.; Agah, S.; Samadani, A.A. DNA Methylation Profiling of hTERT Gene Alongside with the Telomere Performance in Gastric Adenocarcinoma. J. Gastrointest. Cancer 2020, 51, 788–799. [Google Scholar] [CrossRef]
- Zhu, T.; Hu, Z.; Wang, Z.; Ding, H.; Li, R.; Sun, J.; Wang, G. Epigenetically Silenced PD-L1 Confers Drug Resistance to Anti-PD1 Therapy in Gastric Cardia Adenocarcinoma. Int. Immunopharmacol. 2020, 82, 106245. [Google Scholar] [CrossRef]
- Lingohr, P.; Dohmen, J.; Semaan, A.; Branchi, V.; Dietrich, J.; Bootz, F.; Kalff, J.C.; Matthaei, H.; Dietrich, D. Clinicopathological, Immune and Molecular Correlates of PD-L2 Methylation in Gastric Adenocarcinomas. Epigenomics 2019, 11, 639–653. [Google Scholar] [CrossRef] [PubMed]
- Brasacchio, D.; Busuttil, R.A.; Noori, T.; Johnstone, R.W.; Boussioutas, A.; Trapani, J.A. Down-Regulation of a pro-Apoptotic Pathway Regulated by PCAF/ADA3 in Early Stage Gastric Cancer. Cell Death Dis. 2018, 9, 442. [Google Scholar] [CrossRef] [PubMed]
- Ramezankhani, R.; Solhi, R.; Es, H.A.; Vosough, M.; Hassan, M. Novel Molecular Targets in Gastric Adenocarcinoma. Pharmacol. Ther. 2021, 220, 107714. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.J.; Arai, H.; Kitayama, Y.; Igarashi, H.; Hemmi, H.; Arai, T.; Hanai, H.; Sugimura, H. Microsatellite instability of papillary subtype of human gastric adenocarcinoma and hMLH1 promoter hypermethylation in the surrounding mucosa. Pathol. Int. 2001, 51, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Li, H.; Sun, M.; Yuan, Y.; Sun, L.-P. DNMT1 Overexpression Predicting Gastric Carcinogenesis, Subsequent Progression and Prognosis: A Meta and Bioinformatic Analysis. Oncotarget 2017, 8, 96396–96408. [Google Scholar] [CrossRef] [PubMed]
- Fattahi, S.; Kosari-Monfared, M.; Ghadami, E.; Golpour, M.; Khodadadi, P.; Ghasemiyan, M.; Akhavan-Niaki, H. Infection-Associated Epigenetic Alterations in Gastric Cancer: New Insight in Cancer Therapy. J. Cell. Physiol. 2018, 233, 9261–9270. [Google Scholar] [CrossRef]
- Zhou, X.; Jiao, D.; Dou, M.; Zhang, W.; Hua, H.; Chen, J.; Li, Z.; Li, L.; Han, X. Association of APC Gene Promoter Methylation and the Risk of Gastric Cancer: A Meta-Analysis and Bioinformatics Study. Medicine 2020, 99, e19828. [Google Scholar] [CrossRef]
- Du, L.; Fakih, M.G.; Rosen, S.T.; Chen, Y. SUMOylation of E2F1 Regulates Expression of EZH2. Cancer Res. 2020, 80, 4212–4223. [Google Scholar] [CrossRef]
- Kishore, C. Epigenetic Regulation and Promising Therapies in Colorectal Cancer. Curr. Mol. Pharmacol. 2021, 14, 838–852. [Google Scholar] [CrossRef]
- Wan, J.; Liu, H.; Ming, L. Lysine Crotonylation Is Involved in Hepatocellular Carcinoma Progression. Biomed. Pharmacother. 2019, 111, 976–982. [Google Scholar] [CrossRef]
- Yang, G.; Yuan, Y.; Yuan, H.; Wang, J.; Yun, H.; Geng, Y.; Zhao, M.; Li, L.; Weng, Y.; Liu, Z.; et al. Histone Acetyltransferase 1 Is a Succinyltransferase for Histones and Non-Histones and Promotes Tumorigenesis. EMBO Rep. 2021, 22, e50967. [Google Scholar] [CrossRef]
- Jarrold, J.; Davies, C.C. PRMTs and Arginine Methylation: Cancer’s Best-Kept Secret? Trends Mol. Med. 2019, 25, 993–1009. [Google Scholar] [CrossRef]
- He, L.-J.; Cai, M.-Y.; Xu, G.-L.; Li, J.-J.; Weng, Z.-J.; Xu, D.-Z.; Luo, G.-Y.; Zhu, S.-L.; Xie, D. Prognostic Significance of Overexpression of EZH2 and H3k27me3 Proteins in Gastric Cancer. Asian Pac. J. Cancer Prev. 2012, 13, 3173–3178. [Google Scholar] [CrossRef]
- Hatakeyama, M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer 2004, 4, 688–694. [Google Scholar] [CrossRef]
- Hayashi, Y.; Tsujii, M.; Wang, J.; Kondo, J.; Akasaka, T.; Jin, Y.; Li, W.; Nakamura, T.; Nishida, T.; Lijima, H.; et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut 2013, 62, 1536–1546. [Google Scholar] [CrossRef]
- Herz, H.M.; Garruss, A.; Shilatifard, A. SET for life: Biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem. Sci. 2013, 38, 621–639. [Google Scholar] [CrossRef]
- Reyes, D.A.; Sarría, V.M.S.; Salazar-Viedma, M.; D’Afonseca, V. Histone Methyltransferases Useful in Gastric Cancer Research. Cancer Inform. 2021, 20, 11769351211039862. [Google Scholar] [CrossRef]
- Donner, I.; Kiviluoto, T.; Ristimäki, A.; Aaltonen, L.A.; Vahteristo, P. Exome sequencing reveals three novel candidate predisposition genes for diffuse gastric cancer. Fam. Cancer 2015, 14, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Torres, I.O.; Kuchenbecker, K.M.; Nnadi, C.I.; Fletterick, R.J.; Kelly, M.J.; Fujimori, D.G. Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat. Commun. 2015, 6, 6204. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.W.; Xia, R.; Lu, K.; Xie, M.; Yang, F.; Sun, M.; De, W.; Wang, C.; Ji, G. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol. Cancer 2017, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Nishikawaji, T.; Akiyama, Y.; Shimada, S.; Kojima, K.; Kawano, T.; Eishi, Y.; Yuasa, Y.; Tanaka, S. Oncogenic roles of the SETDB2 histone methyltransferase in gastric cancer. Oncotarget 2016, 7, 67251–67265. [Google Scholar] [CrossRef]
- Wisnieski, F.; Calcagno, D.Q.; Leal, M.F.; Santos, L.C.; Gigek, C.O.; Chen, E.S.; Demachki, S.; Artigiani, R.; Assumpção, P.P.; Lourenço, L.G.; et al. CDKN1A Histone Acetylation and Gene Expression Relationship in Gastric Adenocarcinomas. Clin. Exp. Med. 2017, 17, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Wisnieski, F.; Leal, M.F.; Calcagno, D.Q.; Santos, L.C.; Gigek, C.O.; Chen, E.S.; Artigiani, R.; Demachki, S.; Assumpção, P.P.; Lourenço, L.G.; et al. BMP8B Is a Tumor Suppressor Gene Regulated by Histone Acetylation in Gastric Cancer. J. Cell. Biochem. 2017, 118, 869–877. [Google Scholar] [CrossRef]
- Song, J.; Noh, J.H.; Lee, J.H.; Eun, J.W.; Ahn, Y.M.; Kim, S.Y.; Lee, S.H.; Park, W.S.; Yoo, N.J.; Lee, J.Y.; et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS 2005, 113, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Noh, J.H.; Lee, J.H.; Eun, J.W.; Ahn, Y.M.; Kim, S.Y.; Lee, S.H.; Park, W.S.; Yoo, N.J.; Lee, J.Y.; et al. Association of patterns of class I histone deacetylase expression with patient prognosis in gastric cancer: A retrospective analysis. Lancet Oncol. 2008, 9, 139–148. [Google Scholar]
- Xia, G.; Schneider-Stock, R.; Diestel, A.; Habold, C.; Krueger, S.; Roessner, A.; Naumann, M.; Lendeckel, U. Helicobacter pylori regulates p21(WAF1) by histone H4 acetylation. Biochem. Biophys. Res. Commun. 2008, 369, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Ren, J.; Zhang, J.; Sun, Y.; Ma, F.; Liu, Z.; Yu, H.; Jia, J.; Li, W. JMJD2B is required for Helicobacter pylori-induced gas-tric carcinogenesis via regulating COX-2 expression. Oncotarget 2016, 7, 38626–38637. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Jin, M.Y.; Kim, Y.J.; Yook, J.H.; Kim, B.S.; Jang, S.J. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann. Surg. Oncol. 2008, 15, 1968–1976. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.Y.; Zhou, P.J.; Meng, Y.; Zeng, F.R.; Deng, G.T. Gastric cancer: An epigenetic view. World J. Gastrointest. Oncol. 2022, 14, 90–109. [Google Scholar] [CrossRef]
- Murakami, Y. Phosphorylation of Repressive Histone Code Readers by Casein Kinase 2 Plays Diverse Roles in Heterochromatin Regulation. J. Biochem. 2019, 166, 3–6. [Google Scholar] [CrossRef]
- Khan, S.A.; Amnekar, R.; Khade, B.; Barreto, S.G.; Ramadwar, M.; Shrikhande, S.V.; Gupta, S. P38-MAPK/MSK1-Mediated Overexpression of Histone H3 Serine 10 Phosphorylation Defines Distance-Dependent Prognostic Value of Negative Resection Margin in Gastric Cancer. Clin. Epigenetics 2016, 8, 88. [Google Scholar] [CrossRef]
- Takahashi, H.; Murai, Y.; Tsuneyama, K.; Nomoto, K.; Okada, E.; Fujita, H.; Takano, Y. Overexpression of Phosphorylated Histone H3 Is an Indicator of Poor Prognosis in Gastric Adenocarcinoma Patients. Appl. Immunohistochem. Mol. Morphol. 2006, 14, 296–302. [Google Scholar] [CrossRef]
- Kim, S.R.; Kim, K.B.; Chae, Y.C.; Park, J.W.; Seo, S.B. H3S10 phosphorylation-mediated transcriptional regulation by Aurora kinase, A. Biochem. Biophys. Res. Commun. 2016, 469, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Dar, A.A.; Zaika, A.; Piazuelo, M.B.; Correa, P.; Koyama, T.; Belkhiri, A.; Washington, K.; Castells, A.; Pera, M.; El-Rifai, W. Frequent overexpression of Aurora Kinase A in upper gastrointestinal adenocarcinomas correlates with potent antiapoptotic functions. Cancer 2008, 112, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Kamran, M.; Long, Z.J.; Xu, D.; Lv, S.S.; Liu, B.; Wang, C.L.; Xu, J.; Lam, E.W.-F.; Liu, Q. Aurora kinase A regulates survivin stability through targeting FBXL7 in gastric cancer drug resistance and prognosis. Oncogenesis 2017, 6, e298. [Google Scholar] [CrossRef]
- Xu, X.; Xiao, J.; Wei, S.; Xie, X.; Huang, Y.; Tian, X.; Wang, C. Relationship between expression of Aurka and clinicopathological characteristics in gastric cancer patients. Sci. Res. 2014, 6, 243–249. [Google Scholar] [CrossRef]
- Yang, T.; Cao, N.; Zhang, H.; Wei, J.; Song, X.; Yi, D.; Chao, S.; Zhang, L.; Kong, L.; Han, S.; et al. Helicobacter pylori infection-induced H3Ser10 phosphorylation in stepwise gastric carcinogenesis and its clinical implications. Helicobacter 2018, 23, e12486. [Google Scholar] [CrossRef]
- Fehri, L.F.; Rechner, C.; Janßen, S.; Mak, T.N.; Holland, C.; Bartfeld, S.; Brüggemann, H.; Meyer, T.F. Helicobacter pylori-induced modification of the histone H3 phosphorylation status in gastric epithelial cells reflects its impact on cell cycle regulation. Epigenetics 2009, 4, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qiu, Z.; Wu, Y. Ubiquitin Regulation: The Histone Modifying Enzyme’s Story. Cells 2018, 7, 118. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin Modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Wang, Z.-J.; Yang, J.-L.; Wang, Y.-P.; Lou, J.-Y.; Chen, J.; Liu, C.; Guo, L.-D. Decreased Histone H2B Monoubiquitination in Malignant Gastric Carcinoma. World J. Gastroenterol. 2013, 19, 8099–8107. [Google Scholar] [CrossRef]
- van Mierlo, G.; Veenstra, G.J.C.; Vermeulen, M.; Marks, H. The complexity of PRC2 subcomplexes. Trends Cell Biol. 2019, 29, 660–671. [Google Scholar] [CrossRef]
- Hu, H.; Yang, Y.; Ji, Q.; Zhao, W.; Jiang, B.; Liu, R.; Yuan, J.; Liu, Q.; Li, X.; Zou, Y.; et al. CRL4B catalyzes H2AK119 monoubiquitination and coordinates with PRC2 to promote tumorigenesis. Cancer Cell 2012, 22, 781–795. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Quilez, J.; Roig-Soucase, S.; Rodriguez-Navarro, S. Sharing marks: H3K4 methylation and H2B ubiquitination as features of meiotic recombination and transcription. Int. J. Mol. Sci. 2020, 21, 4510. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, M.; Wang, Y. The roles of histone modifications in tumorigenesis and associated inhibitors in cancer therapy. J. Natl. Cancer Cent. 2022, 2, 277–290. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Chan, J.C.; Maze, I. Nothing is yet set in (hi)stone: Novel post-translational modifications regulating chromatin function. Trends Biochem. Sci. 2020, 45, 829–844. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Xu, X.; Ding, J.; Yang, L.; Doan, M.T.; Karmaus, P.W.F.; Snyder, N.W.; Zhao, Y.; Li, J.-L.; Li, X. Histone Crotonylation Promotes Mesoendodermal Commitment of Human Embryonic Stem Cells. Cell Stem Cell 2021, 28, 748–763.e7. [Google Scholar] [CrossRef]
- Zhu, Z.; Han, Z.; Halabelian, L.; Yang, X.; Ding, J.; Zhang, N.; Ngo, L.; Song, J.; Zeng, H.; He, M.; et al. Identification of lysine isobutyrylation as a new histone modification mark. Nucleic Acids Res. 2021, 49, 177–189. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. Histone Lactylation: A New Role for Glucose Metabolism. Trends Biochem. Sci. 2020, 45, 179–182. [Google Scholar] [CrossRef]
- Zhang, D.; Tang, Z.; Huang, H.; Zhou, G.; Cui, C.; Weng, Y.; Liu, W.; Kim, S.; Lee, S.; Perez-Neut, M.; et al. Metabolic Regulation of Gene Expression by Histone Lactylation. Nature 2019, 574, 575–580. [Google Scholar] [CrossRef]
- Leonen, C.J.A.; Shimada, M.; Weller, C.E.; Nakadai, T.; Hsu, P.L.; Tyson, E.L.; Mishra, A.; Shelton, P.M.; Sadilek, M.; Hawkins, R.D.; et al. Sumoylation of the Human Histone H4 Tail Inhibits P300-Mediated Transcription by RNA Polymerase II in Cellular Extracts. Elife 2021, 10, e67952. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg Robert, A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Wellen, K.E.; Thompson, C.B. A two-way street: Reciprocal regulation of metabolism and signaling. Nat. Rev. Mol. Cell Biol. 2012, 13, 270–276. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Sayed, N.; Ditsworth, D.; Thompson, C.B. Brick by brick: Metabolism and tumor cell growth. Curr. Opin. Genet. Dev. 2008, 18, 54–61. [Google Scholar] [CrossRef]
- Ulanovskaya, O.A.; Zuhl, A.M.; Cravatt, B.F. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat. Chem. Biol. 2013, 9, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Zeng, M.; Pan, H.; Liu, H.; He, Y. Nicotinamide N-methyltransferase promotes epithelial-mesenchymal transition in gastric cancer cells by activating transforming growth factor-β1 expression. Oncol. Lett. 2018, 15, 4592–4598. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, C.; Wang, T.; Deng, H. Complex roles of nicotinamide N-methyltransferase in cancer progression. Cell Death Dis. 2022, 13, 267. [Google Scholar] [CrossRef]
- Chen, C.; Wang, X.; Huang, X.; Yong, H.; Shen, J.; Tang, Q.; Zhu, J.; Ni, J.; Feng, Z. Nicotinamide N-methyltransferase: A potential biomarker for worse prognosis in gastric carcinoma. Am. J. Cancer Res. 2016, 6, 649–663. [Google Scholar] [PubMed]
- Wu, M.; Hu, W.; Wang, G.; Yao, Y.; Yu, X.F. Nicotinamide N-Methyltransferase Is a Prognostic Biomarker and Correlated with Immune Infiltrates in Gastric Cancer. Front. Genet. 2020, 11, 580299. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Song, M.; Zhang, F.; Yuan, H.; Chang, W.; Yu, G.; Niu, Y. Accumulation of Nicotinamide N-Methyltransferase (NNMT) in Cancer-associated Fibroblasts: A Potential Prognostic and Predictive Biomarker for Gastric Carcinoma. J. Histochem. Cytochem. 2021, 69, 165–176. [Google Scholar] [CrossRef]
- Pozzi, V.; Campagna, R.; Sartini, D.; Emanuelli, M. Nicotinamide N-Methyltransferase as Promising Tool for Management of Gastrointestinal Neoplasms. Biomolecules 2022, 12, 1173. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Q.; Chen, C.; Zhao, X.; Wang, H.; Xu, L.; Fu, Y.; Huang, G.; Li, M.; Xu, J.; et al. NNMT enriches for AQP5+ cancer stem cells to drive malignant progression in early gastric cardia adenocarcinoma. Gut 2024, 73, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, J.; Wang, J.; Song, Y.; Gao, P.; Shi, J.; Chen, P.; Wang, Z. Long Noncoding RNAs in Gastric Cancer: Functions and Clinical Applications. Onco Targets Ther. 2016, 9, 681–697. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wu, D.; Gao, H.; Balic, J.J.; Tsykin, A.; Han, T.-S.; Liu, Y.D.; Kennedy, C.L.; Li, J.K.; Mao, J.Q.; et al. Clinical Utility of a STAT3-Regulated miRNA-200 Family Signature with Prognostic Potential in Early Gastric Cancer. Clin. Cancer Res. 2018, 24, 1459–1472. [Google Scholar] [CrossRef] [PubMed]
- Marchese, F.P.; Huarte, M. Long Non-Coding RNAs and Chromatin Modifiers: Their Place in the Epigenetic Code. Epigenetics 2014, 9, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gao, S.; Zhao, Z.; Liang, G.; Kong, J.; Feng, X. MicroRNA-320d Regulates Tumor Growth and Invasion by Promoting FoxM1 and Predicts Poor Outcome in Gastric Cardiac Adenocarcinoma. Cell Biosci. 2020, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Yadegar, N.; Dadashi, Z.; Shams, K.; Mohammadi, M.; Abyar, M.; Rafat, M. The Prominent Role of miR-942 in Carcinogenesis of Tumors. Adv. Biomed. Res. 2022, 11, 63. [Google Scholar] [CrossRef]
- Abdi, E.; Latifi-Navid, S.; Zahri, S.; Yazdanbod, A.; Pourfarzi, F. Risk Factors Predisposing to Cardia Gastric Adenocarcinoma: Insights and New Perspectives. Cancer Med. 2019, 8, 6114–6126. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Liu, J.; Yang, T.; Zhang, L.; Gong, P.; Li, B.; Zhou, X. Construction and Investigation of MicroRNA-mRNA Regulatory Network of Gastric Cancer with Helicobacter Pylori Infection. Biochem. Res. Int. 2020, 2020, 6285987. [Google Scholar] [CrossRef]
- Gilani, N.; Arabi Belaghi, R.; Aftabi, Y.; Faramarzi, E.; Edgünlü, T.; Somi, M.H. Identifying Potential miRNA Biomarkers for Gastric Cancer Diagnosis Using Machine Learning Variable Selection Approach. Front. Genet. 2021, 12, 779455. [Google Scholar] [CrossRef]
- Qu, X.; Zhao, L.; Zhang, R.; Wei, Q.; Wang, M. Differential microRNA Expression Profiles Associated with Microsatellite Status Reveal Possible Epigenetic Regulation of Microsatellite Instability in Gastric Adenocarcinoma. Ann. Transl. Med. 2020, 8, 484. [Google Scholar] [CrossRef]
- Jia, L.; Chen, J.; Xie, C.; Shao, L.; Xu, Z.; Zhang, L. microRNA-1228⁎ impairs the pro-angiogenic activity of gastric cancer cells by targeting macrophage migration inhibitory factor. Life Sci. 2017, 180, 9–16. [Google Scholar] [CrossRef]
- Motoyama, K.; Inoue, H.; Nakamura, Y.; Uetake, H.; Sugihara, K.; Mori, M. Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin. Cancer Res. 2008, 14, 2334–2340. [Google Scholar] [CrossRef]
- Bie, L.; Luo, S.; Li, D.; Wei, Y.; Mu, Y.; Chen, X.; Wang, S.; Guo, P.; Lu, X. HOTAIR Competitively Binds MiRNA330 as a Molecular Sponge to Increase the Resistance of Gastric Cancer to Trastuzumab. Curr. Cancer Drug Targets 2020, 20, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Shiroki, T.; Nakagawa, T.; Yokoyama, M.; Tamai, K.; Yamanami, H.; Fujiya, T.; Sato, I.; Yamaguchi, K.; Tanaka, N.; et al. Enhanced Expression of Long Non-Coding RNA HOTAIR Is Associated with the Development of Gastric Cancer. PLoS ONE 2013, 8, e77070. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Qin, Y.; Zhi, Q.; Wang, J.; Qin, C. Knockdown of Long Non-Coding RNA HOTAIR Inhibits Cisplatin Resistance of Gastric Cancer Cells through Inhibiting the PI3K/Akt and Wnt/β-Catenin Signaling Pathways by up-Regulating miR-34a. Int. J. Biol. Macromol. 2018, 107, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, J.; Yan, W.; Yao, Z.; Wang, R.; Zhou, X.; Wu, H.; Zhang, G.; Shi, T.; Chen, W. H19 Promotes Aerobic Glycolysis, Proliferation, and Immune Escape of Gastric Cancer Cells through the microRNA-519d-3p/Lactate Dehydrogenase A Axis. Cancer Sci. 2021, 112, 2245–2259. [Google Scholar] [CrossRef] [PubMed]
- Xin, L.; Zhou, L.-Q.; Liu, C.; Zeng, F.; Yuan, Y.-W.; Zhou, Q.; Li, S.-H.; Wu, Y.; Wang, J.-L.; Wu, D.-Z.; et al. Transfer of LncRNA CRNDE in TAM-Derived Exosomes Is Linked with Cisplatin Resistance in Gastric Cancer. EMBO Rep. 2021, 22, e52124. [Google Scholar] [CrossRef]
- Fan, H.; Ge, Y.; Ma, X.; Li, Z.; Shi, L.; Lin, L.; Xiao, J.; Chen, W.; Ni, P.; Yang, L.; et al. Long Non-Coding RNA CCDC144NL-AS1 Sponges miR-143-3p and Regulates MAP3K7 by Acting as a Competing Endogenous RNA in Gastric Cancer. Cell Death Dis. 2020, 11, 521. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Z.; Yu, G.; Wang, T.; Qu, G.; Wang, Y. LINC01232 Promotes Gastric Cancer Proliferation through Interacting with EZH2 to Inhibit the Transcription of KLF2. J. Microbiol. Biotechnol. 2021, 31, 1358–1365. [Google Scholar] [CrossRef]
- Peng, L.; Peng, J.-Y.; Cai, D.-K.; Qiu, Y.-T.; Lan, Q.-S.; Luo, J.; Yang, B.; Xie, H.-T.; Du, Z.-P.; Yuan, X.-Q.; et al. Immune Infiltration and Clinical Outcome of Super-Enhancer-Associated lncRNAs in Stomach Adenocarcinoma. Front. Oncol. 2022, 12, 780493. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Wang, Z.; Wang, G.; Hu, Z.; Ding, H.; Li, R.; Sun, J. Long Non-Coding RNA ZFAS1 Promotes the Expression of EPAS1 in Gastric Cardia Adenocarcinoma. J. Adv. Res. 2021, 28, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Zhou, Y.; Luo, X.; Gao, H.; Deng, X.; Jiang, Y. Long non-coding RNA XIST promotes cell growth and invasion through regulating miR-497/MACC1 axis in gastric cancer. Oncotarget 2017, 8, 4125–4135. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Li, J.; Zhou, X.; Cui, L.; Wang, Y. The lncRNA XIST promotes proliferation, migration and invasion of gastric cancer cells by targeting miR-337. Arab. J. Gastroenterol. 2020, 21, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, L.; Li, P.; Hu, F.; Cao, Y.; Tang, D.; Ye, G.; Li, H.; Wang, D. Silencing lncRNA XIST exhibits antiproliferative and proapoptotic effects on gastric cancer cells by up-regulating microRNA-132 and down-regulating PXN. Aging 2020, 13, 14469–14481. [Google Scholar] [CrossRef]
- Guarnerio, J.; Bezzi, M.; Jeong, J.C.; Paffenholz, S.V.; Berry, K.; Naldini, M.M.; Lo-Coco, F.; Tay, Y.; Beck, A.H.; Pandolfi, P.P. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016, 166, 1055–1056. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Li, T.; Jiang, Y.; Pan, C.; Ding, Y.; Huang, Z.; Yu, H.; Kong, D. Overexpression of Circular RNA ciRS-7 Abrogates the Tumor Suppressive Effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT Signaling Pathway. J. Cell. Biochem. 2018, 119, 440–446. [Google Scholar] [CrossRef]
- Lin, J.; Zhang, Y.; Zeng, X.; Xue, C.; Lin, X. CircRNA CircRIMS Acts as a MicroRNA Sponge to Promote Gastric Cancer Metastasis. ACS Omega 2020, 5, 23237–23246. [Google Scholar] [CrossRef]
- Liu, J.; Zhu, X.; Yan, M.; Li, H. Development of a Two-Circular RNA Panel as Potential Prognostic Biomarker for Gastric Cancer. J. Transl. Med. 2021, 19, 412. [Google Scholar] [CrossRef]
- Han, L.; Zhang, X.; Wang, A.; Ji, Y.; Cao, X.; Qin, Q.; Yu, T.; Huang, H.; Yin, L. A Dual-Circular RNA Signature as a Non-Invasive Diagnostic Biomarker for Gastric Cancer. Front. Oncol. 2020, 10, 184. [Google Scholar] [CrossRef] [PubMed]
- Yesharim, L.; Talebi, S.; Mojbafan, M.; Alemrajabi, M.; Teimourian, S. An Evaluation of Gastric Adenocarcinoma-Associated CircRNAs Based on Microarray Meta-Analysis and ceRNA Networks. Transl. Oncol. 2023, 28, 101611. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, X.; Xu, P.; Wang, H.; Wang, S.; Zhang, L.; Li, Z.; Xie, L.; Sun, G.; Xia, Y.; et al. Circular RNA circLMO7 Acts as a microRNA-30a-3p Sponge to Promote Gastric Cancer Progression via the WNT2/β-Catenin Pathway. J. Exp. Clin. Cancer Res. 2021, 40, 6. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Yu, T.; Jing, X.; Ma, L.; Fan, Y.; Yang, F.; Ma, P.; Jiang, H.; Wu, X.; Shu, Y.; et al. Exosomal circSHKBP1 Promotes Gastric Cancer Progression via Regulating the miR-582-3p/HUR/VEGF Axis and Suppressing HSP90 Degradation. Mol. Cancer 2020, 19, 112. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Gao, H.; Xie, X.; Lu, P. Plasma Exosomal Hsa_circ_0015286 as a Potential Diagnostic and Prognostic Biomarker for Gastric Cancer. Pathol. Oncol. Res. 2022, 28, 1610446. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, H.; Hou, L.; Wang, G.; Zhang, R.; Huang, Y.; Chen, X.; Zhu, J. Circular RNA_LARP4 Inhibits Cell Proliferation and Invasion of Gastric Cancer by Sponging miR-424-5p and Regulating LATS1 Expression. Mol. Cancer 2017, 16, 151. [Google Scholar] [CrossRef]
- Zhang, W.; Zheng, M.; Kong, S.; Li, X.; Meng, S.; Wang, X.; Wang, F.; Tang, C.; Ju, S. Circular RNA Hsa_circ_0007507 May Serve as a Biomarker for the Diagnosis and Prognosis of Gastric Cancer. Front. Oncol. 2021, 11, 699625. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Chen, S.; Chen, H.; Mo, X.; Li, T.; Shao, Y.; Xiao, B.; Guo, J. Using Circular RNA as a Novel Type of Biomarker in the Screening of Gastric Cancer. Clin. Chim. Acta 2015, 444, 132–136. [Google Scholar] [CrossRef]
- PubMed. CircRNA Microarray Profiling Identifies a Novel Circulating Biomarker for Detection of Gastric Cancer. Available online: https://pubmed.ncbi.nlm.nih.gov/30236115/ (accessed on 28 November 2023).
- Zhang, J.; Hou, L.; Liang, R.; Chen, X.; Zhang, R.; Chen, W.; Zhu, J. CircDLST Promotes the Tumorigenesis and Metastasis of Gastric Cancer by Sponging miR-502-5p and Activating the NRAS/MEK1/ERK1/2 Signaling. Mol. Cancer 2019, 18, 80. [Google Scholar] [CrossRef]
- Zhang, L.; Song, X.; Chen, X.; Wang, Q.; Zheng, X.; Wu, C.; Jiang, J. Circular RNA CircCACTIN Promotes Gastric Cancer Progression by Sponging MiR-331-3p and Regulating TGFBR1 Expression. Int. J. Biol. Sci. 2019, 15, 1091–1103. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 Acts as a microRNA-149-5p Sponge to Promote Gastric Cancer Progression via the AKT1/mTOR Pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef]
- Wang, S.; Tang, D.; Wang, W.; Yang, Y.; Wu, X.; Wang, L.; Wang, D. circLMTK2 Acts as a Sponge of miR-150-5p and Promotes Proliferation and Metastasis in Gastric Cancer. Mol. Cancer 2019, 18, 162. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, X.; Li, Z.; Wang, W.; Li, B.; Huang, X.; Sun, G.; Xu, J.; Li, Q.; Xu, Z.; et al. Circular RNA Profile Identifies circOSBPL10 as an Oncogenic Factor and Prognostic Marker in Gastric Cancer. Oncogene 2019, 38, 6985–7001. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Wang, F. circ_AKT3 Knockdown Suppresses Cisplatin Resistance in Gastric Cancer. Open Med. 2022, 17, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.-N.; Chen, Z.-Y.; Chen, X.-Y.; Chen, M.; Yi, Y.-C.; Zhu, J.-S.; Zhang, J. METTL14-Mediated m6A Modification of circORC5 Suppresses Gastric Cancer Progression by Regulating miR-30c-2-3p/AKT1S1 Axis. Mol. Cancer 2022, 21, 51. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.-L.; Sheng, H.; Zhang, D.-S.; Jin, Y.; Zhao, B.-T.; Chen, N.; Song, K.; Xu, R.-H. The Circular RNA circDLG1 Promotes Gastric Cancer Progression and Anti-PD-1 Resistance through the Regulation of CXCL12 by Sponging miR-141-3p. Mol. Cancer 2021, 20, 166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Su, X.; Wang, P.; Lin, W. The Value of Circulating Circular RNA in Cancer Diagnosis, Monitoring, Prognosis, and Guiding Treatment. Front. Oncol. 2021, 11, 736546. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xiu, J.; Baca, Y.; Battaglin, F.; Arai, H.; Kawanishi, N.; Soni, S.; Zhang, W.; Millstein, J.; Salhia, B.; et al. Large-Scale Analysis of KMT2 Mutations Defines a Distinctive Molecular Subset with Treatment Implication in Gastric Cancer. Oncogene 2021, 40, 4894–4905. [Google Scholar] [CrossRef]
- Lordick, F.; Carneiro, F.; Cascinu, S.; Fleitas, T.; Haustermans, K.; Piessen, G.; Vogel, A.; Smyth, E.C.; on behalf of the ESMO Guidelines Committee. Gastric Cancer: ESMO Clinical Practice Guideline for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2022, 33, 1005–1020. [Google Scholar] [CrossRef]
- Wang, F.-H.; Zhang, X.-T.; Li, Y.-F.; Tang, L.; Qu, X.-J.; Ying, J.-E.; Zhang, J.; Sun, L.-Y.; Lin, R.-B.; Qiu, H.; et al. The Chinese Society of Clinical Oncology (CSCO): Clinical Guidelines for the Diagnosis and Treatment of Gastric Cancer, 2021. Cancer Commun. 2021, 41, 747–795. [Google Scholar] [CrossRef]
- Ajani, J.A.; D’Amico, T.A.; Bentrem, D.J.; Chao, J.; Cooke, D.; Corvera, C.; Das, P.; Enzinger, P.C.; Enzler, T.; Fanta, P.; et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2022, 20, 167–192. [Google Scholar] [CrossRef]
- Guan, W.-L.; He, Y.; Xu, R.-H. Gastric Cancer Treatment: Recent Progress and Future Perspectives. J. Hematol. Oncol. 2023, 16, 57. [Google Scholar] [CrossRef]
- Leal, A.; van Grieken, N.C.T.; Palsgrove, D.N.; Phallen, J.; Medina, J.E.; Hruban, C.; Broeckaert, M.A.M.; Anagnostou, V.; Adleff, V.; Bruhm, D.C.; et al. White Blood Cell and Cell-Free DNA Analyses for Detection of Residual Disease in Gastric Cancer. Nat. Commun. 2020, 11, 525. [Google Scholar] [CrossRef]
- Maron, S.B.; Chase, L.M.; Lomnicki, S.; Kochanny, S.; Moore, K.L.; Joshi, S.S.; Landron, S.; Johnson, J.; Kiedrowski, L.A.; Nagy, R.J.; et al. Circulating Tumor DNA Sequencing Analysis of Gastroesophageal Adenocarcinoma. Clin. Cancer Res. 2019, 25, 7098–7112. [Google Scholar] [CrossRef]
- Chiba, T.; Marusawa, H.; Ushijima, T. Inflammation-associated cancer development in digestive organs: Mechanisms and roles for genetic and epigenetic modulation. Gastroenterology 2012, 143, 550–563. [Google Scholar] [CrossRef]
- Hattori, N.; Ushijima, T. Epigenetic impact of infection on carcinogenesis: Mechanisms and applications. Genome Med. 2016, 8, 10. [Google Scholar] [CrossRef]
- Baba, Y.; Ishimoto, T.; Kurashige, J.; Iwatsuki, M.; Sakamoto, Y.; Yoshida, N.; Watanabe, M.; Baba, H. Epigenetic field cancerization in gastrointestinal cancers. Cancer Lett. 2016, 375, 360–366. [Google Scholar] [CrossRef]
- Kashyap, D.; Baral, B.; Jakhmola, S.; Singh, A.K.; Jha, H.C. Helicobacter pylori and Epstein-Barr virus coinfection stimulates aggressiveness in gastric cancer through the regulation of gankyrin. mSphere 2021, 6, e00751-21. [Google Scholar] [CrossRef]
- Jiang, C.; Pugh, B.F. Nucleosome Positioning and Gene Regulation: Advances through Genomics. Nat. Rev. Genet. 2009, 10, 161–172. [Google Scholar] [CrossRef]
- Matsusaka, K.; Funata, S.; Fukayama, M.; Kaneda, A. DNA methylation in gastric cancer, related to Helicobacter pylori and Epstein-Barr virus. World J. Gastroenterol. 2014, 20, 3916–3926. [Google Scholar] [CrossRef]
- Tan, P.; Yeoh, K.-G. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology 2015, 149, 1153–1162.e3. [Google Scholar] [CrossRef]
- Saletore, Y.; Meyer, K.; Korlach, J.; Vilfan, I.D.; Jaffrey, S.; Mason, C.E. The Birth of the Epitranscriptome: Deciphering the Function of RNA Modifications. Genome Biol. 2012, 13, 175. [Google Scholar] [CrossRef]
- Padmanabhan, N.; Ushijima, T.; Tan, P. How to stomach an epigenetic insult: The gastric cancer epigenome. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 467–478. [Google Scholar] [CrossRef]
- Kaneda, A.; Matsusaka, K.; Aburatani, H.; Fukayama, M. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012, 72, 3445–3450. [Google Scholar] [CrossRef]
- Jácome, A.A.D.A.; Lima, E.M.; Kazzi, A.I.; Chaves, G.F.; de Mendonça, D.C.; Maciel, M.M.; Santos, J.S.D. Epstein-Barr Virus-Positive Gastric Cancer: A Distinct Molecular Subtype of the Disease? Rev. Soc. Bras. Med. Trop. 2016, 49, 150–157. [Google Scholar] [CrossRef]
- Ignatova, E.; Seriak, D.; Fedyanin, M.; Tryakin, A.; Pokataev, I.; Menshikova, S.; Vakhabova, Y.; Smirnova, K.; Tjulandin, S.; Ajani, J.A. Epstein-Barr Virus-Associated Gastric Cancer: Disease That Requires Special Approach. Gastric. Cancer 2020, 23, 951–960. [Google Scholar] [CrossRef]
- Samadani, A.A.; Nikbakhsh, N.; Taheri, H.; Shafaee, S.; Fattahi, S.; Pilehchian Langroudi, M.; Hajian, K.; Akhavan-Niaki, H. CDX1/2 and KLF5 Expression and Epigenetic Modulation of Sonic Hedgehog Signaling in Gastric Adenocarcinoma. Pathol. Oncol. Res. 2019, 25, 1215–1222. [Google Scholar] [CrossRef]
- Heo, H.; Kim, H.-J.; Haam, K.; Sohn, H.A.; Shin, Y.-J.; Go, H.; Jung, H.-J.; Kim, J.-H.; Lee, S.-I.; Song, K.-S.; et al. Epigenetic Activation of Tensin 4 Promotes Gastric Cancer Progression. Mol. Cells 2023, 46, 298–308. [Google Scholar] [CrossRef]
- Moutinho, C.; Martinez-Cardús, A.; Santos, C.; Navarro-Pérez, V.; Martínez-Balibrea, E.; Musulen, E.; Carmona, F.J.; Sartore-Bianchi, A.; Cassingena, A.; Siena, S.; et al. Epigenetic Inactivation of the BRCA1 Interactor SRBC and Resistance to Oxaliplatin in Colorectal Cancer. J. Natl. Cancer Inst. 2014, 106, djt322. [Google Scholar] [CrossRef]
- Rezaei, S.; Hosseinpourfeizi, M.A.; Moaddab, Y.; Safaralizadeh, R. Contribution of DNA Methylation and EZH2 in SRBC Down-Regulation in Gastric Cancer. Mol. Biol. Rep. 2020, 47, 5721–5727. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.-Y.; Ma, H.-X.; Shang, Y.-H.; Jin, M.-S.; Kong, F.; Jia, Z.-F.; Cao, D.-H.; Wang, Y.-P.; Suo, J.; Jiang, J. DNA Methyltransferase3a Expression Is an Independent Poor Prognostic Indicator in Gastric Cancer. World J. Gastroenterol. 2014, 20, 8201–8208. [Google Scholar] [CrossRef]
- Purkait, S.; Patra, S.; Mitra, S.; Behera, M.M.; Panigrahi, M.K.; Kumar, P.; Kar, M.; Hallur, V.; Chandra Samal, S. Elevated Expression of DNA Methyltransferases and Enhancer of Zeste Homolog 2 in Helicobacter Pylori—Gastritis and Gastric Carcinoma. Dig. Dis. 2022, 40, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi Hedayati, M.; Ahmadi, A.; Khatooni, Z. DNMT1 Gene Expression in Patients with Helicobacter Pylori Infection. Sci. World J. 2022, 2022, 2386891. [Google Scholar] [CrossRef]
- Song, W.; Ren, J.; Wang, W.-J.; Wang, C.-T.; Fu, T. Genome-Wide Methylation and Expression Profiling Identify a Novel Epigenetic Signature in Gastrointestinal Pan-Adenocarcinomas. Epigenomics 2020, 12, 907–920. [Google Scholar] [CrossRef]
- Han, L.C.; Chen, Y. Targeting EZH2 for Cancer Therapy: Progress and Perspective. Curr. Protein Pept. Sci. 2015, 16, 559–570. [Google Scholar] [CrossRef]
- Fattahi, S.; Golpour, M.; Amjadi-Moheb, F.; Sharifi-Pasandi, M.; Khodadadi, P.; Pilehchian-Langroudi, M.; Ashrafi, G.H.; Akhavan-Niaki, H. DNA Methyltransferases and Gastric Cancer: Insight into Targeted Therapy. Epigenomics 2018, 10, 1477–1497. [Google Scholar] [CrossRef]
- Takeshima, H.; Niwa, T.; Takahashi, T.; Wakabayashi, M.; Yamashita, S.; Ando, T.; Inagawa, Y.; Taniguchi, H.; Katai, H.; Sugiyama, T.; et al. Frequent involvement of chromatin remodeler alterations in gastric field cancerization. Cancer Lett. 2015, 357, 328–338. [Google Scholar] [CrossRef]
- Santos, J.C.; Gambeloni, R.Z.; Roque, A.T.; Oeck, S.; Ribeiro, M.L. Epigenetic Mechanisms of ATM Activation after Helicobacter Pylori Infection. Am. J. Pathol. 2018, 188, 329–335. [Google Scholar] [CrossRef]
- Okabe, A.; Huang, K.K.; Matsusaka, K.; Fukuyo, M.; Xing, M.; Ong, X.; Hoshii, T.; Usui, G.; Seki, M.; Mano, Y.; et al. Cross-Species Chromatin Interactions Drive Transcriptional Rewiring in Epstein-Barr Virus-Positive Gastric Adenocarcinoma. Nat. Genet. 2020, 52, 919–930. [Google Scholar] [CrossRef]
- Abd El Fattah, Y.K.; Abulsoud, A.I.; AbdelHamid, S.G.; Hamdy, N.M. Interactome Battling of lncRNA CCDC144NL-AS1: Its Role in the Emergence and Ferocity of Cancer and Beyond. Int. J. Biol. Macromol. 2022, 222, 1676–1687. [Google Scholar] [CrossRef]
- Liu, X.-H.; Sun, M.; Nie, F.-Q.; Ge, Y.-B.; Zhang, E.-B.; Yin, D.-D.; Kong, R.; Xia, R.; Lu, K.-H.; Li, J.-H.; et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 2014, 13, 92. [Google Scholar] [CrossRef]
- Jin, Y.; Ma, J.; Wang, L. Comprehensive Analysis of Immune-Related lncRNAs and Their Clinical Relevance in Gastric Adenocarcinoma. Funct. Integr. Genom. 2023, 23, 28. [Google Scholar] [CrossRef]
- Kipkeeva, F.; Muzaffarova, T.; Korotaeva, A.; Nikulin, M.; Grishina, K.; Mansorunov, D.; Apanovich, P.; Karpukhin, A. MicroRNA in Gastric Cancer Development: Mechanisms and Biomarkers. Diagnostics 2020, 10, 891. [Google Scholar] [CrossRef]
- Kang, J.; Huang, X.; Dong, W.; Zhu, X.; Li, M.; Cui, N. MicroRNA-1269b Inhibits Gastric Cancer Development through Regulating Methyltransferase-like 3 (METTL3). Bioengineered 2021, 12, 1150–1160. [Google Scholar] [CrossRef]
- Ueda, T.; Volinia, S.; Okumura, H.; Shimizu, M.; Taccioli, C.; Rossi, S.; Alder, H.; Liu, C.G.; Oue, N.; Yasui, W.; et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010, 11, 136–146. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA Is Enriched and Stable in Exosomes: A Promising Biomarker for Cancer Diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef]
- Reis-das-Mercês, L.; Vinasco-Sandoval, T.; Pompeu, R.; Ramos, A.C.; Anaissi, A.K.M.; Demachki, S.; de Assumpção, P.P.; Vidal, A.F.; Ribeiro-Dos-Santos, Â.; Magalhães, L. CircRNAs as Potential Blood Biomarkers and Key Elements in Regulatory Networks in Gastric Cancer. Int. J. Mol. Sci. 2022, 23, 650. [Google Scholar] [CrossRef]
- Li, T.; Shao, Y.; Fu, L.; Xie, Y.; Zhu, L.; Sun, W.; Yu, R.; Xiao, B.; Guo, J. Plasma Circular RNA Profiling of Patients with Gastric Cancer and Their Droplet Digital RT-PCR Detection. J. Mol. Med. 2018, 96, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Lv, J.; Xuan, Z.; Li, B.; Li, Z.; He, Z.; Li, F.; Xu, J.; Wang, S.; Xia, Y.; et al. Circular CPM Promotes Chemoresistance of Gastric Cancer via Activating PRKAA2-Mediated Autophagy. Clin. Transl. Med. 2022, 12, e708. [Google Scholar] [CrossRef] [PubMed]
- Saviana, M.; Le, P.; Micalo, L.; Del Valle-Morales, D.; Romano, G.; Acunzo, M.; Li, H.; Nana-Sinkam, P. Crosstalk between miRNAs and DNA Methylation in Cancer. Genes 2023, 14, 1075. [Google Scholar] [CrossRef]
- Yuan, R.; Wang, G.; Xu, Z.; Zhao, H.; Chen, H.; Han, Y.; Wang, B.; Zhou, J.; Hu, H.; Guo, Z.; et al. Up-regulated Circulating miR-106a by DNA Methylation Promised a Potential Diagnostic and Prognostic Marker for Gastric Cancer. Anticancer. Agents Med. Chem. 2016, 16, 1093–1100. [Google Scholar] [CrossRef]
- Szczepanek, J.; Tretyn, A. MicroRNA-Mediated Regulation of Histone-Modifying Enzymes in Cancer: Mechanisms and Therapeutic Implications. Biomolecules 2023, 13, 1590. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, D.; Wang, D.; Jin, T.; Yang, L.; Wu, H.; Li, Y.; Zhao, J.; Du, F.; Song, M.; et al. HEDD: The human epigenetic drug database. Database 2016, 2016, baw159. [Google Scholar] [CrossRef]
- Sahafnejad, Z.; Ramazi, S.; Allahverdi, A. An Update of Epigenetic Drugs for the Treatment of Cancers and Brain Diseases: A Comprehensive Review. Genes 2023, 14, 873. [Google Scholar] [CrossRef]
- Abdelfatah, E.; Kerner, Z.; Nanda, N.; Ahuja, N. Epigenetic therapy in gastrointestinal cancer: The right combination. Ther. Adv. Gastroenterol. 2016, 9, 560–579. [Google Scholar] [CrossRef]
- Gao, Y.; van Haren, M.J.; Buijs, N.; Innocenti, P.; Zhang, Y.; Sartini, D.; Campagna, R.; Emanuelli, M.; Parsons, R.B.; Jespers, W.; et al. Potent Inhibition of Nicotinamide N-Methyltransferase by Alkene-Linked Bisubstrate Mimics Bearing Electron Deficient Aromatics. J. Med. Chem. 2021, 64, 12938–12963. [Google Scholar] [CrossRef]
- van Haren, M.J.; Zhang, Y.; Thijssen, V.; Buijs, N.; Gao, Y.; Mateuszuk, L.; Fedak, F.A.; Kij, A.; Campagna, R.; Sartini, D.; et al. Macrocyclic peptides as allosteric inhibitors of nicotinamide N-methyltransferase (NNMT). RSC Chem. Biol. 2021, 2, 1546–1555. [Google Scholar] [CrossRef] [PubMed]
- van Haren, M.J.; Gao, Y.; Buijs, N.; Campagna, R.; Sartini, D.; Emanuelli, M.; Mateuszuk, L.; Kij, A.; Chlopicki, S.; Escudé Martinez de Castilla, P.; et al. Esterase-Sensitive Prodrugs of a Potent Bisubstrate Inhibitor of Nicotinamide N-Methyltransferase (NNMT) Display Cellular Activity. Biomolecules 2021, 11, 1357. [Google Scholar] [CrossRef] [PubMed]
- Moro, H.; Hattori, N.; Nakamura, Y.; Kimura, K.; Imai, T.; Maeda, M.; Yashiro, M.; Ushijima, T. Epigenetic priming sensitizes gastric cancer cells to irinotecan and cisplatin by restoring multiple pathways. Gastric. Cancer 2020, 23, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Hu, N.; Li, W.; Hong, Y.; Zeng, Z.; Zhang, J.; Wu, X.; Zhou, K.; Wu, F. A PD1 targeted nano-delivery system based on epigenetic alterations of T cell responses in the treatment of gastric cancer. Mol. Ther.-Oncolytics 2022, 24, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Turcan, S. Epigenetic Drugs and Their Immune Modulating Potential in Cancers. Biomedicines 2022, 10, 211. [Google Scholar] [CrossRef]
- Miranda Furtado, C.L.; Dos Santos Luciano, M.C.; da Silva Santos, R.; Furtado, G.P.; Moraes, M.O.; Pessoa, C. Epidrugs: Targeting epigenetic marks in cancer treatment. Epigenetics 2019, 14, 1164–1176. [Google Scholar] [CrossRef]
| Gene | Role | Expression | Role in Gastric Carcinomas | References |
|---|---|---|---|---|
| CDH1 | Tumor-suppressor gene | Silenced |
| [18] |
| MLH1 | Mismatch-repair mechanism | Silenced |
| [6,19] |
| CDKN2A | Cell-cycle arrest | Silenced | GC development through silencing mediated by Hp and EBV infections. Potential therapeutic target. | [20,21] |
| LAMA4 | Encodes laminin subunit alpha 4, a member of extracellular matrix glycoproteins | Overexpressed |
| [22] |
| FBX032 | Tumor-suppressor gene mediates in cell-survival regulation | Downregulated or loss of function | Predicts metastasis and poor prognosis in stage-III and -IV gastric-cancer patients. Potential prognostic and therapeutic biomarker. | [23] |
| CDH11 | Tumor-suppressor gene | Silenced | Potential prognostic biomarker of malignant behavior. | [24,25] |
| IGFBP7 | Regulation of insulin-like growth factors (IGFs)—potential tumor suppressor gene | Silenced | Suppressive effect on gastric-cancer development when it is expressed. Potential prognostic and therapeutic biomarker. | [26] |
| Claudin-3 | Cell-adhesion molecule | Downregulated | Predictor of high metastatic status and LN spread. Potential prognostic and therapeutic biomarker. | [27] |
| hTERT | Part of the telomerase complex—mediates in cellular immortalization | Overexpressed | Poor prognosis and shorter OS in GC patients. Potential diagnostic and prognostic biomarker. | [28,29] |
| PD-L1 | Immunosuppressive molecule—acts as an oncogene | Silenced | Resistance to immunotherapy. Potential therapeutic biomarker. | [30] |
| PD-L2 | Immunosuppressive molecule | Overexpressed | Predictor of response after PD-L1 therapy. Potential predictive biomarker. | [31] |
| SRBC | Tumor-suppressor gene | Downregulated | Chemoresistance (against oxaliplatin). Potential therapeutic target. | [30] |
| PCAF | Loss of function | Poor outcomes. Potential prognostic biomarker. | [32,33] |
| Micro-RNA | Action | Role in Gastric Carcinomas | References |
|---|---|---|---|
| miRNA200 | Promotes oncogenesis | Predictor of OS Prognostic biomarker | [95,96] |
| miR-320d | Tumor suppression | Treatment of GC Prognostic and therapeutic biomarker | [97] |
| miR-942-3p/-5p | Oncogenic/tumor suppression | Potential prognostic biomarker | [98] |
| miR-141 | Inhibits the proliferation of cancerous cells and triggers apoptosis | Predictor of OS Potential therapeutic biomarker | [98,99] |
| miR-1269b | Inhibits the development of GC, suppresses migration and invasion | Predictor of OS Potential diagnostic and prognostic biomarker | [98,99] |
| miR-203 a/b | Tumor suppressor | Potential prognostic biomarker | [98,99] |
| miR-196b-3p | Associates with the age of onset | Potential prognostic biomarker | [100,101] |
| miR-30-3p/miR-105-5p | Modifies expression of DNA damage-repair genes in MSI-H tumors | Predictive biomarkers for microsatellite instability | [102] |
| miR-1343-3p | Tumor suppressor Antiangiogenic role | Potential diagnostic biomarker | [101] |
| miR-8073 | Tumor suppressor | Potential diagnostic biomarker | [101] |
| miR-1228-5p | Negative regulator of gastric-cancer growth and angiogenesis | Potential diagnostic biomarker and therapeutic target for anti-angiogenic therapy | [103] |
| miR-let7g | Predictive biomarker of DFS | Potential prognostic biomarker | [104] |
| LncRNA | Action | Role in Gastric Carcinomas | References |
|---|---|---|---|
| HOTAIR | Oncogenic |
| [106,107] |
| H19 | Oncogenic |
| [108] |
| CRNDE | Tumor suppressor |
| [109] |
| CCDC144NL-AS1 | Acts as competing endogenous RNA |
| [110] |
| LINC01232 | Oncogenic |
| [111] |
| TM4SF1-AS1 | Involved in the tumor’s immune microenvironment |
| [112] |
| ZFAS1 | Oncogenic |
| [113] |
| XIST | Acts as competing endogenous RNA |
| [114] |
| Circ-RNA | Effect | Role in Gastric Carcinomas | References |
|---|---|---|---|
| ciRs-7 | GC progression | Prospective prognostic and therapeutic biomarker | [118] |
| circRIMS | Predicts invasive metastasis | Potential diagnostic and therapeutic biomarker | [119] |
| hsa_circ_0005092/hsa_circ_0002647 | Upregulated | Prognostic and predictive for post-operative recurrence biomarker | [120] |
| hsa_circ_0021087/hsa_circ_0005051 | Occurrence and development of GC | Non-invasive diagnostic biomarkers | [121] |
| hsa_circ_0002019/hsa_circ_00074736 | Regulates the expression of genes linked to GC survival | Predictors of OS Potential prognostic biomarker | [122] |
| CircAKT3/circLM07 | GC progression | Predictor of OS Potential prognostic biomarker | [123] |
| CircSHKBP1 | GC progression, poor survival | Potential non-invasive diagnostic and prognostic biomarker | [124] |
| has_circ_0015286 | Non-invasive diagnostic biomarker | Potential diagnostic and prognostic biomarker | [125] |
| circLARP4 | Tumor suppressor | Prognostic factor for OS | [126] |
| hsa_circ_0007507 | Differentially expressed in GC patients, post-operative GC patients, gastritis patients, intestinal metaplasia patients | Potential diagnostic and monitoring biomarker | [127] |
| hsa_circ_002059 | Correlation with TNM stage, distant metastasis, and age of onset | Potential diagnostic biomarker | [128] |
| hsa_circ_0000467/hsa_circ_KIAA1244 | Correlation with TNM and metastasis | Potential prognostic biomarker | [129] |
| circDLST, circCACTIN, circNRIP1 | Promote oncogenesis, migration, and invasion | Potential prognostic biomarker | [130,131,132] |
| CircLMTK2 | Correlation with TNM | Potential prognostic and therapeutic biomarker | [133] |
| CircOSBPL10 | Promotes tumor growth | Potential prognostic biomarker | [134] |
| Circ_AKT3 | Association with cisplatin sensitivity | Potential therapeutic biomarker | [135] |
| CircCPM | Regulates autophagy and 5-Fluro-Uracil resistance | Potential therapeutic biomarker | [136] |
| CircDLG1 | Increases distant metastasis, anti-PD-L1 resistance | Predictor of OS Potential therapeutic biomarker | [137] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christodoulidis, G.; Koumarelas, K.-E.; Kouliou, M.-N.; Thodou, E.; Samara, M. Gastric Cancer in the Era of Epigenetics. Int. J. Mol. Sci. 2024, 25, 3381. https://doi.org/10.3390/ijms25063381
Christodoulidis G, Koumarelas K-E, Kouliou M-N, Thodou E, Samara M. Gastric Cancer in the Era of Epigenetics. International Journal of Molecular Sciences. 2024; 25(6):3381. https://doi.org/10.3390/ijms25063381
Chicago/Turabian StyleChristodoulidis, Grigorios, Konstantinos-Eleftherios Koumarelas, Marina-Nektaria Kouliou, Eleni Thodou, and Maria Samara. 2024. "Gastric Cancer in the Era of Epigenetics" International Journal of Molecular Sciences 25, no. 6: 3381. https://doi.org/10.3390/ijms25063381
APA StyleChristodoulidis, G., Koumarelas, K.-E., Kouliou, M.-N., Thodou, E., & Samara, M. (2024). Gastric Cancer in the Era of Epigenetics. International Journal of Molecular Sciences, 25(6), 3381. https://doi.org/10.3390/ijms25063381






