Transcriptome Analysis by RNA Sequencing of Mouse Embryonic Stem Cells Stocked on International Space Station for 1584 Days in Frozen State after Culture on the Ground
Abstract
1. Introduction
2. Results
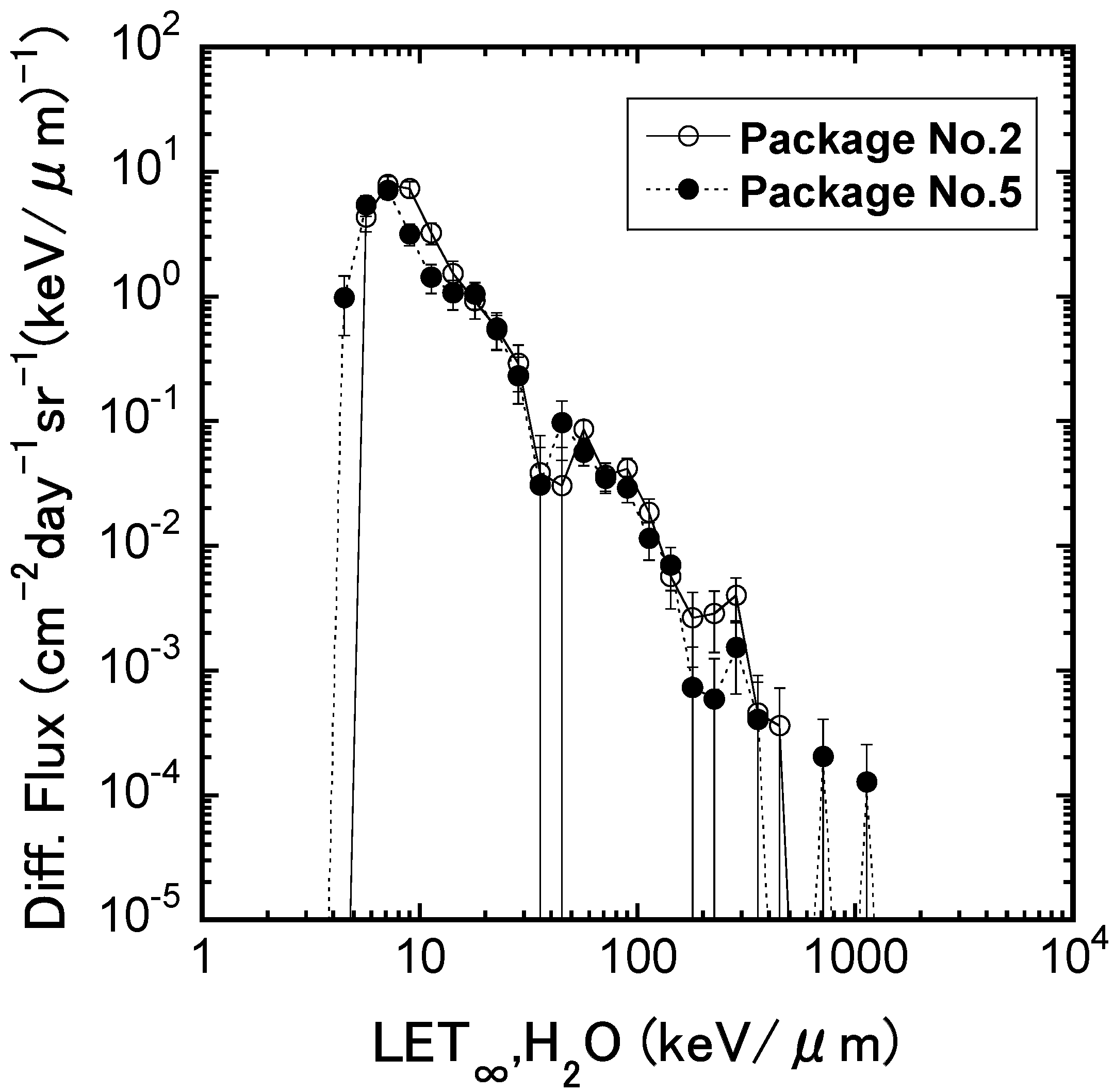
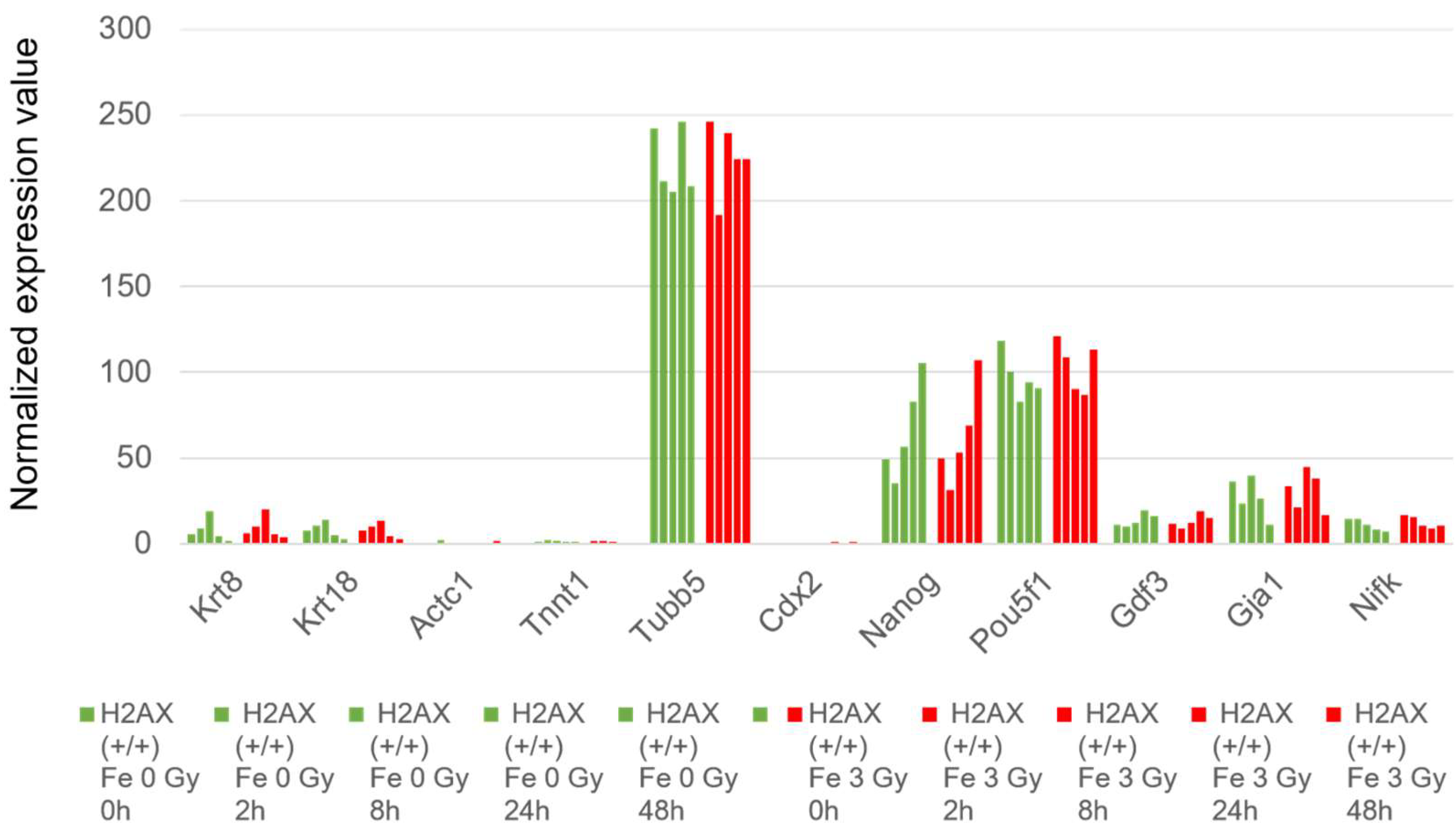
2.1. The Irradiation Effects of Fe Ions on the Mouse ES Cells in terms of Transcriptome Profile
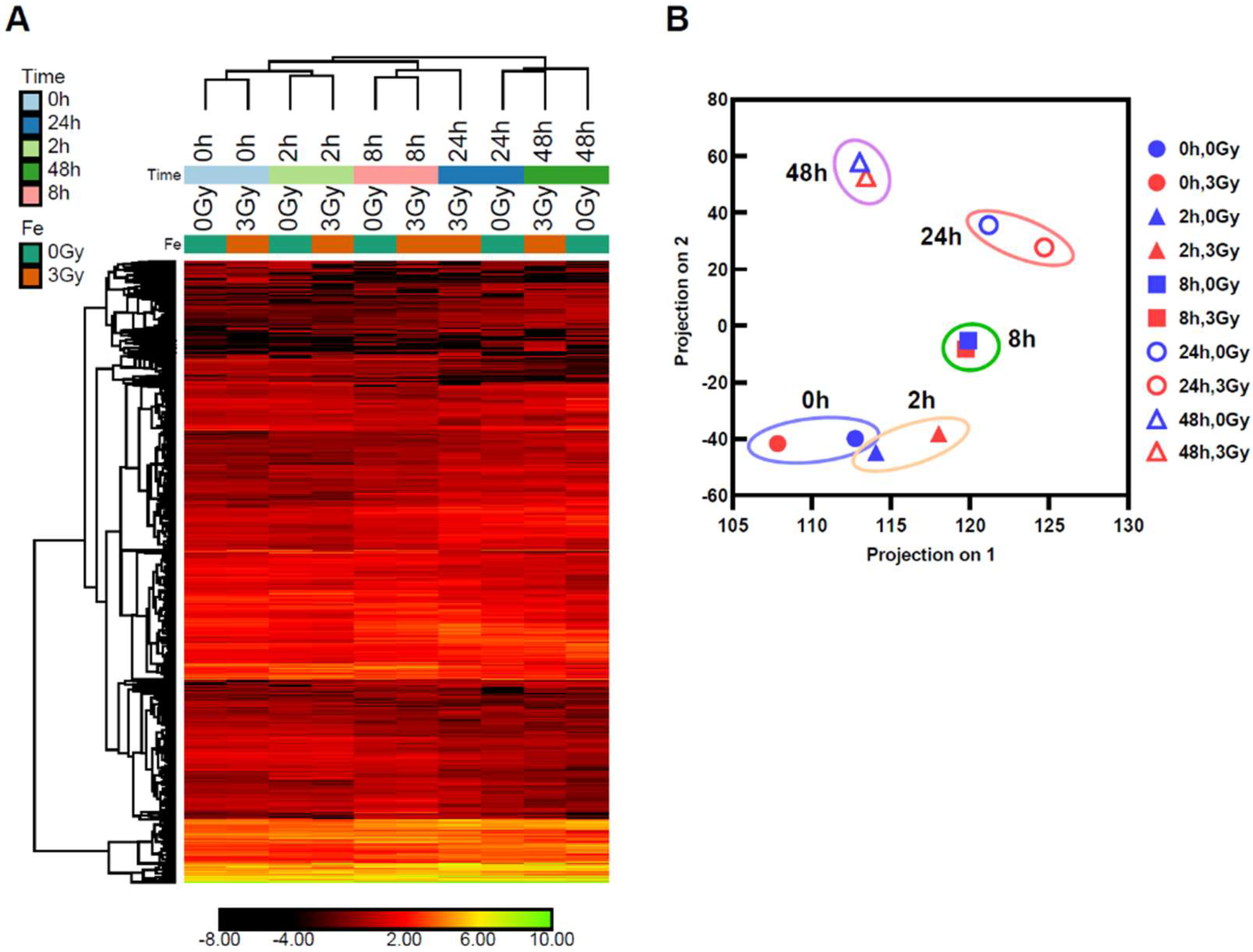
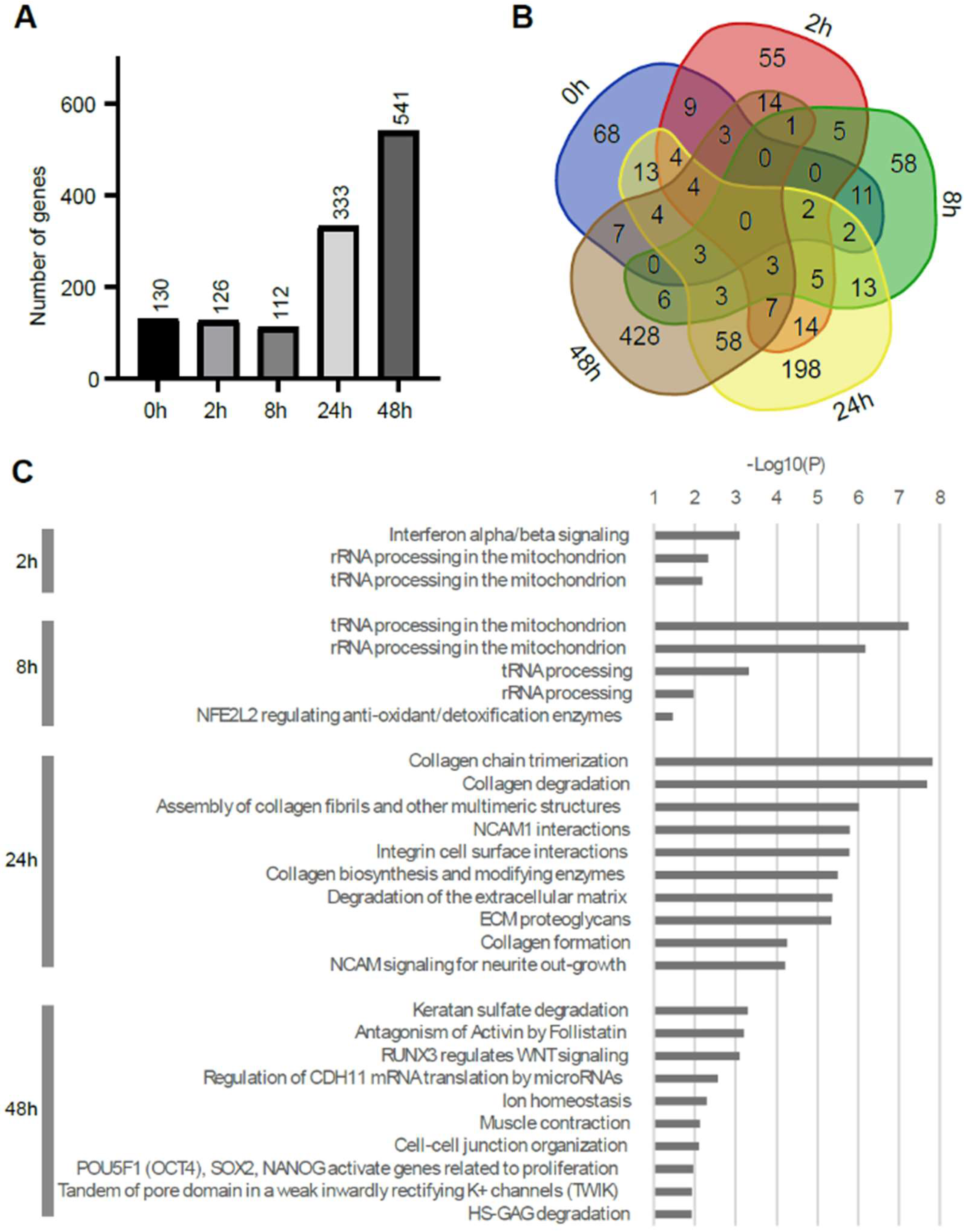
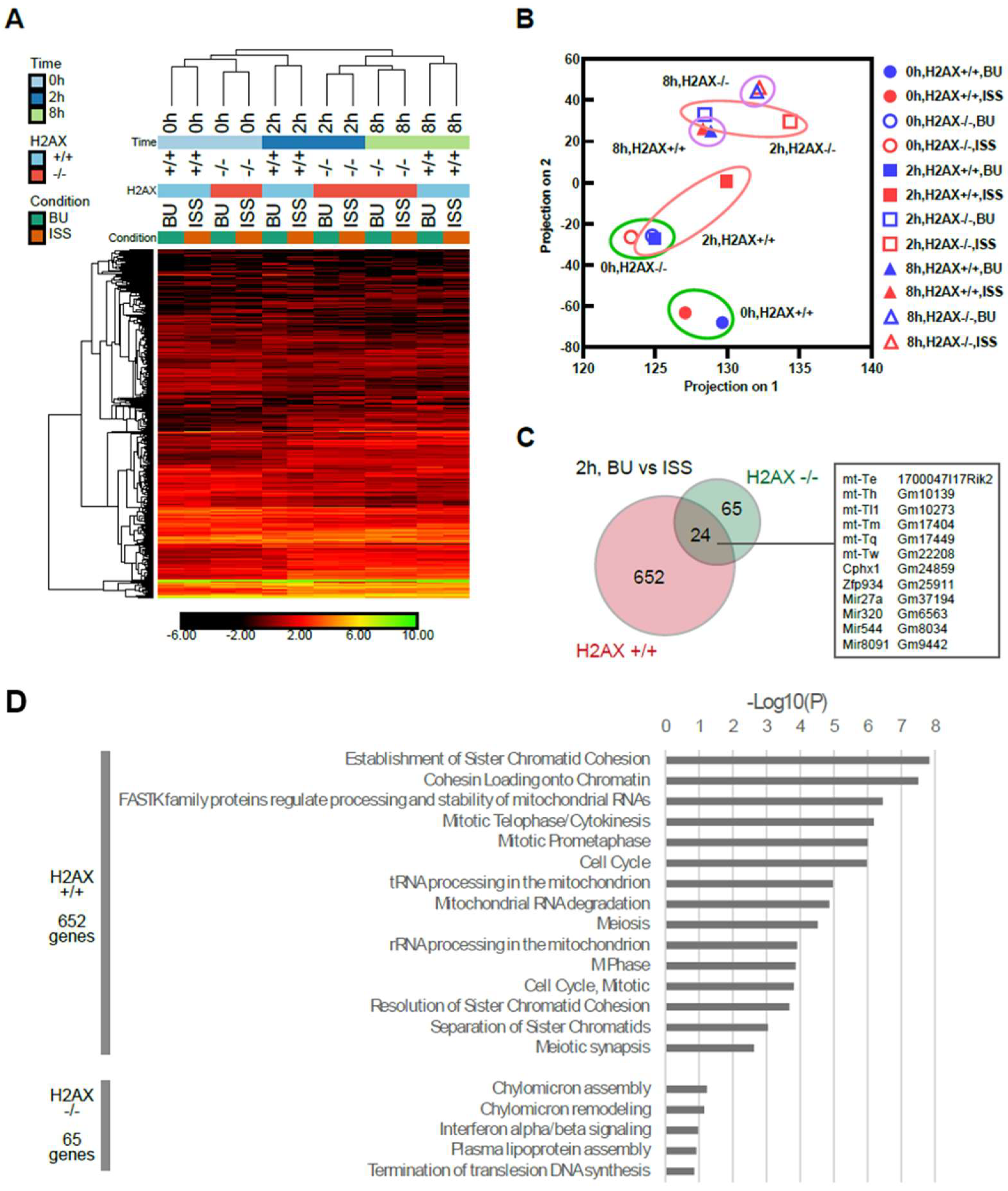
2.2. The Comparison of the Expression Profile of the Transcriptome between Mouse ES Cells of Ground Backup Control and ISS-Stocked Cells
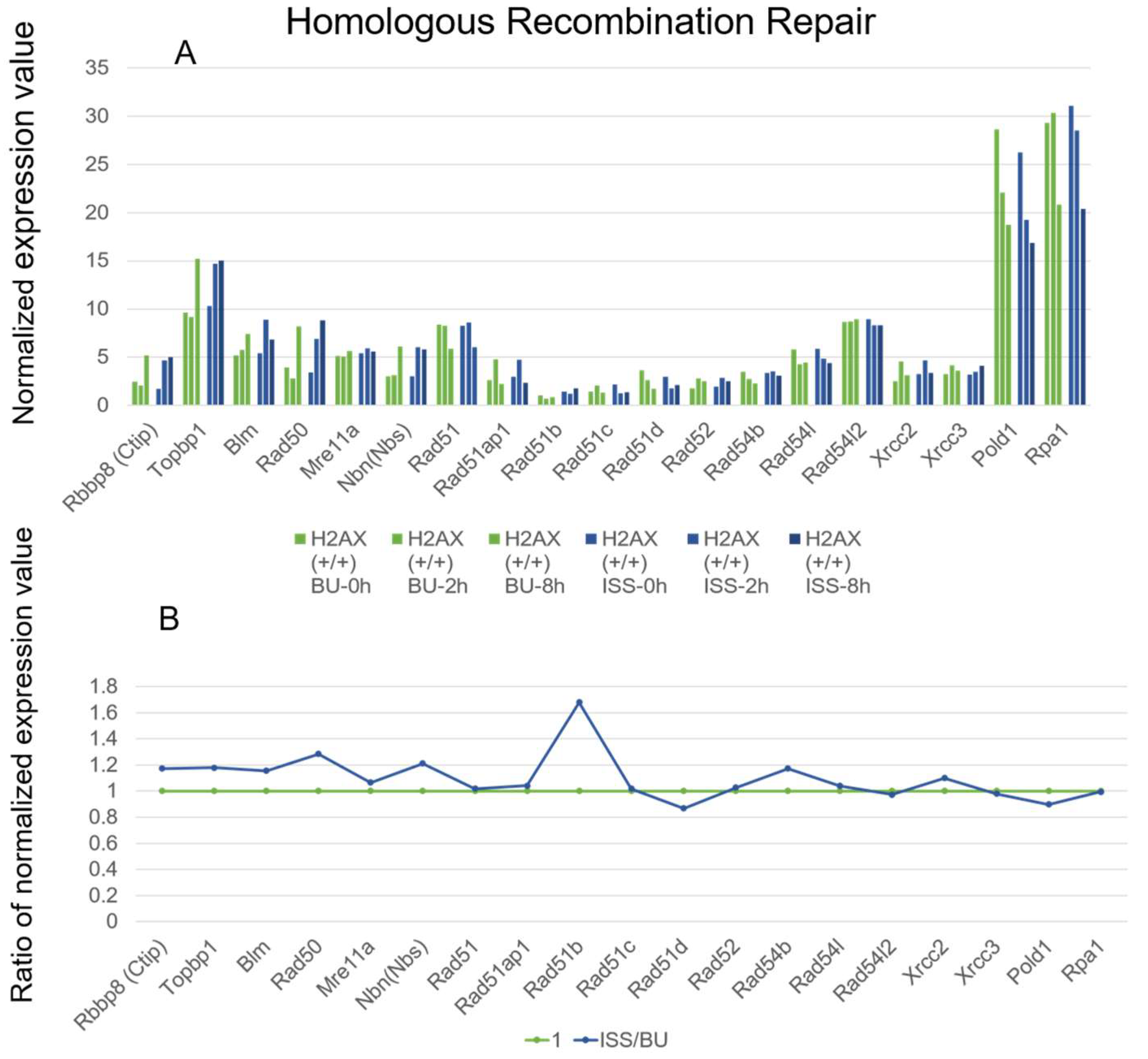
2.3. Homologous Recombination Repair Genes
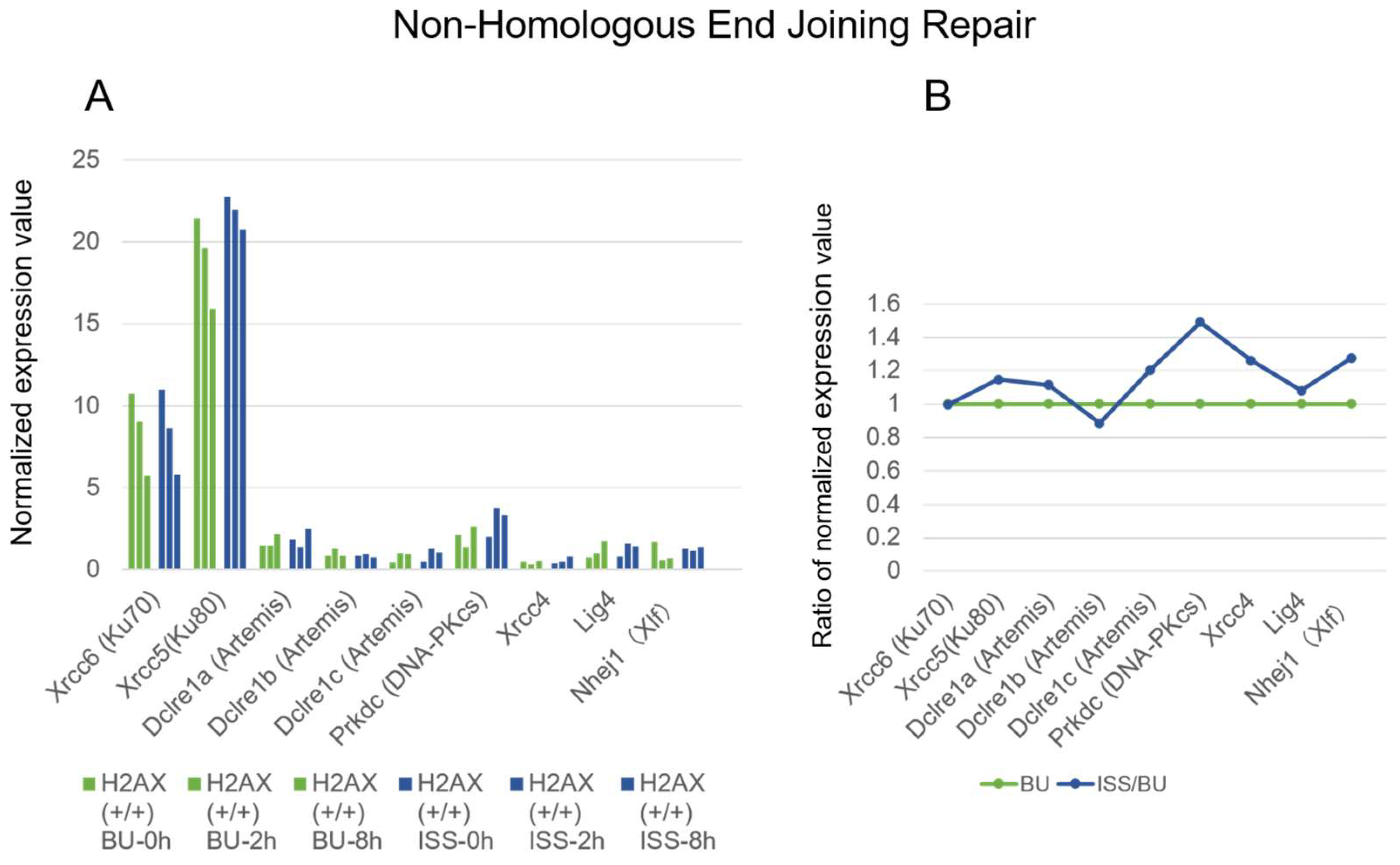
2.4. Non-Homologous End Joining Repair Genes
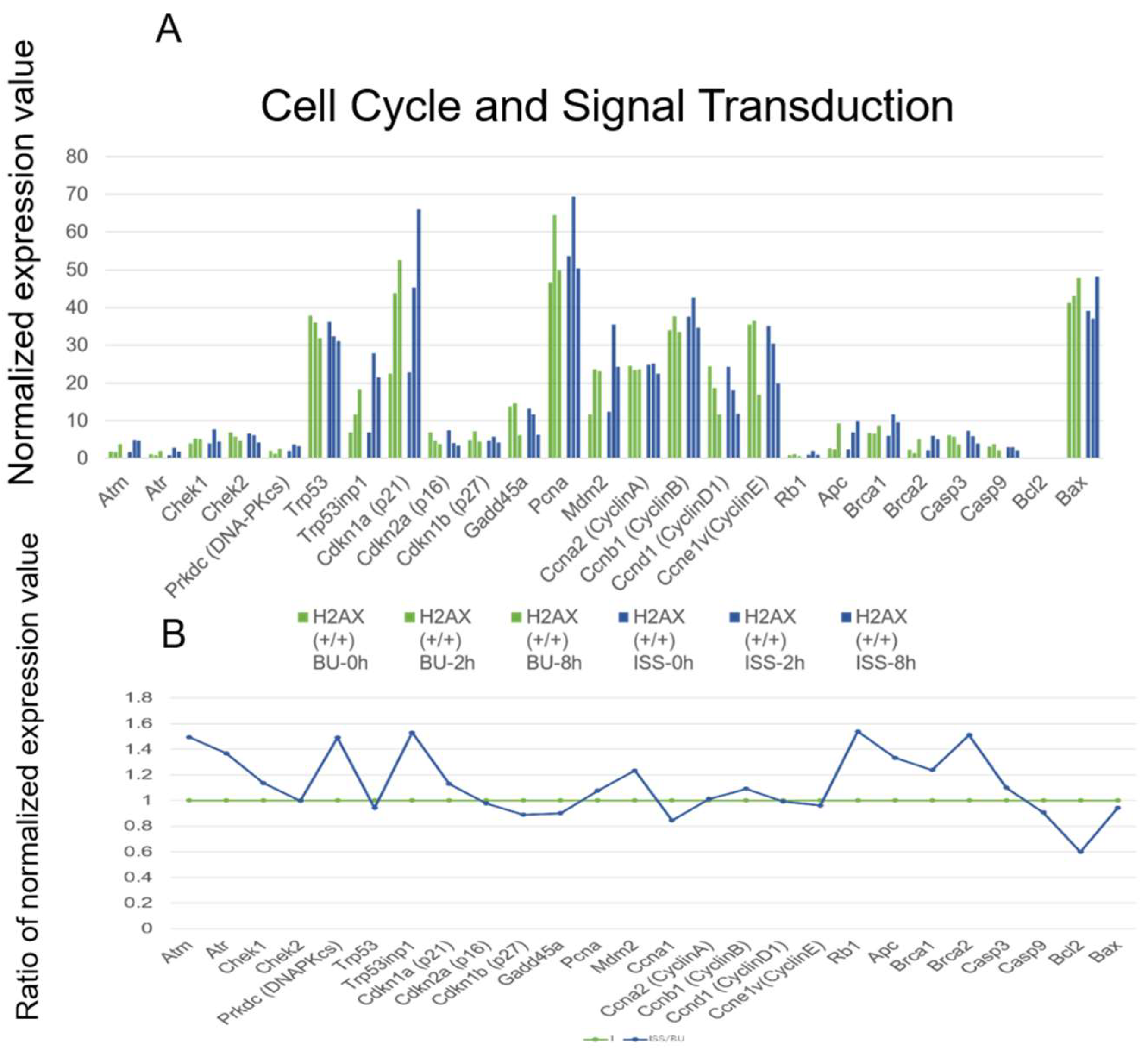
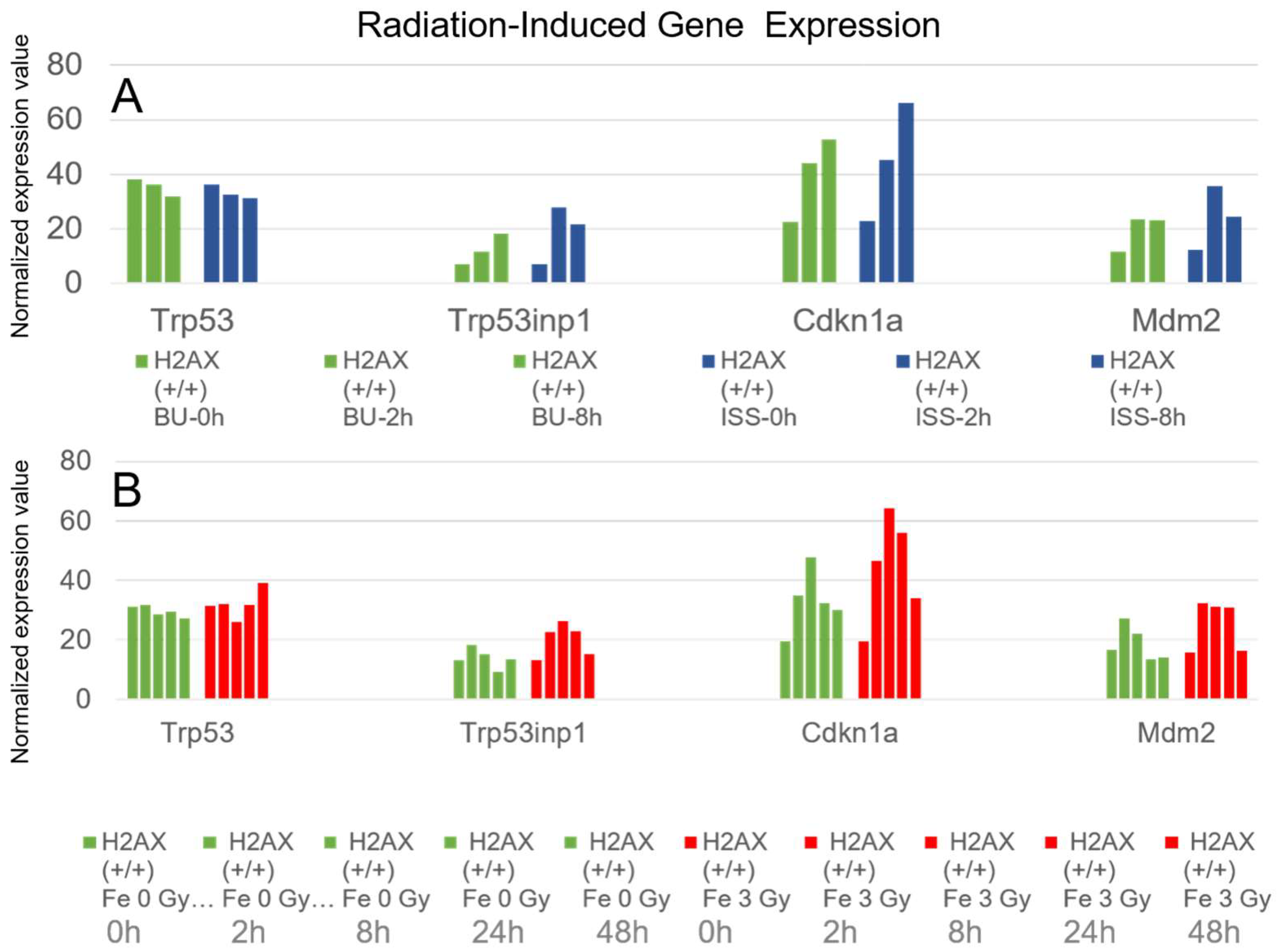
2.5. Cell Cycle Regulation and p53 Pathways Genes
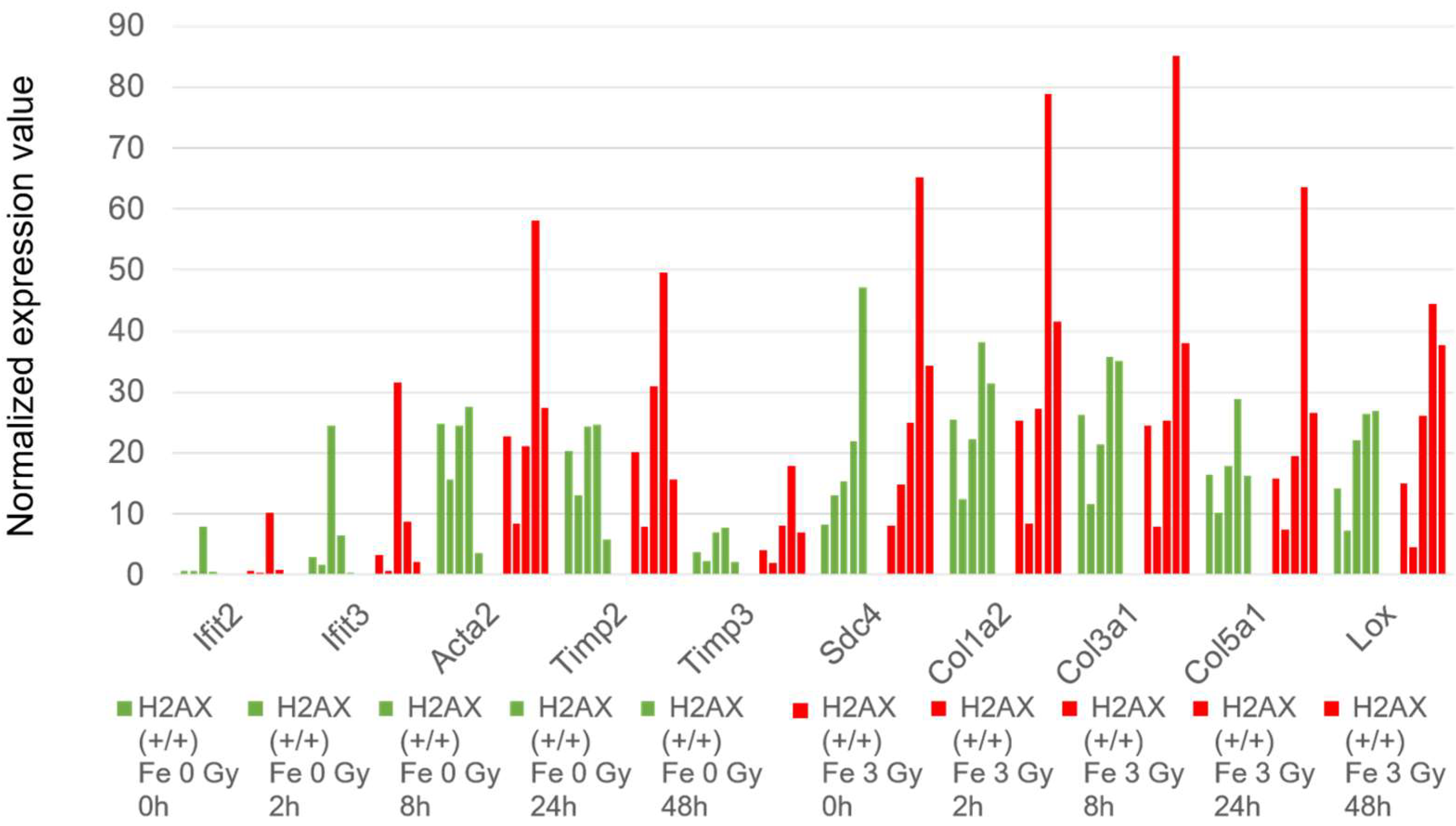
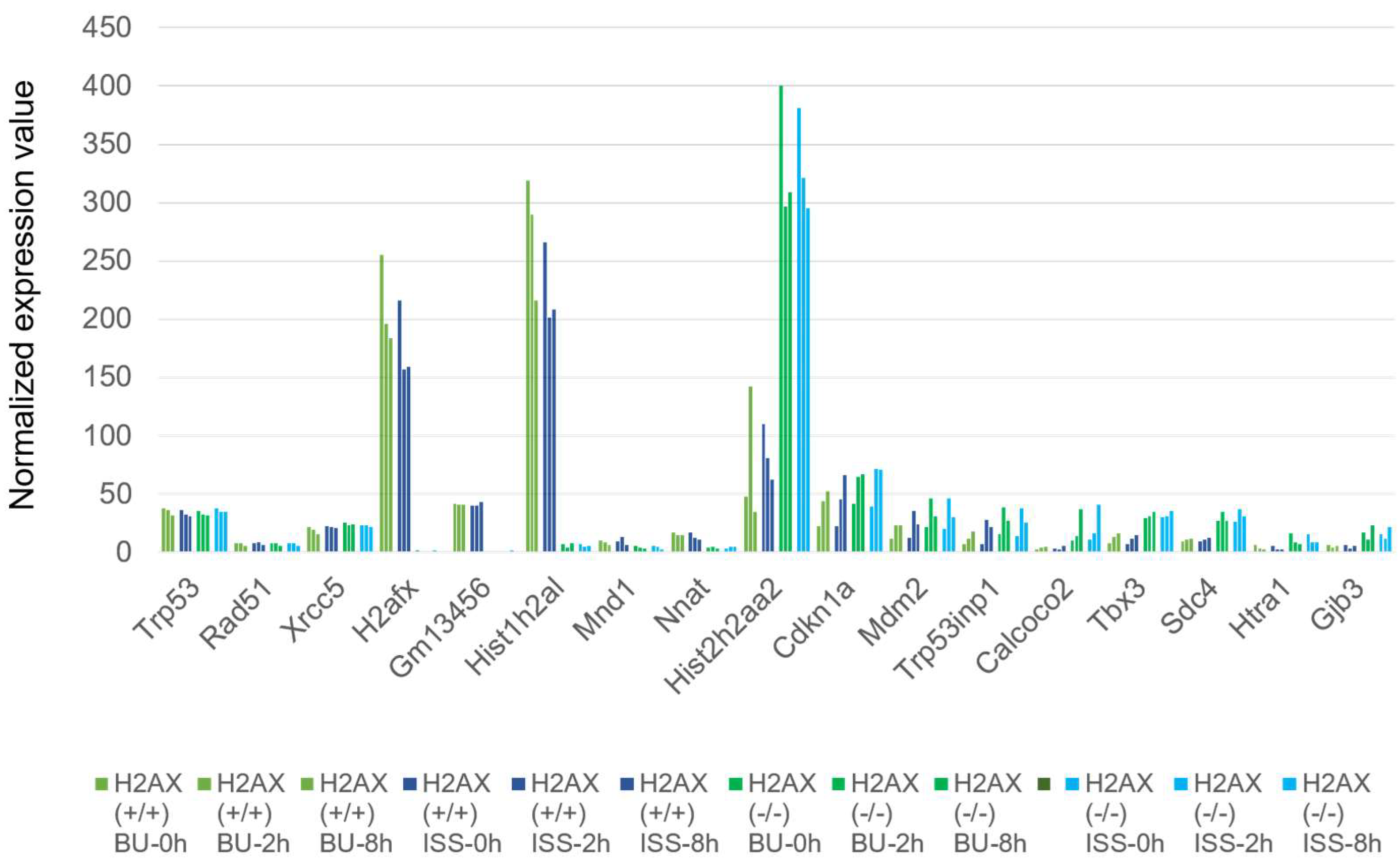
2.6. Histone H2AX-Dependent Gene Expression
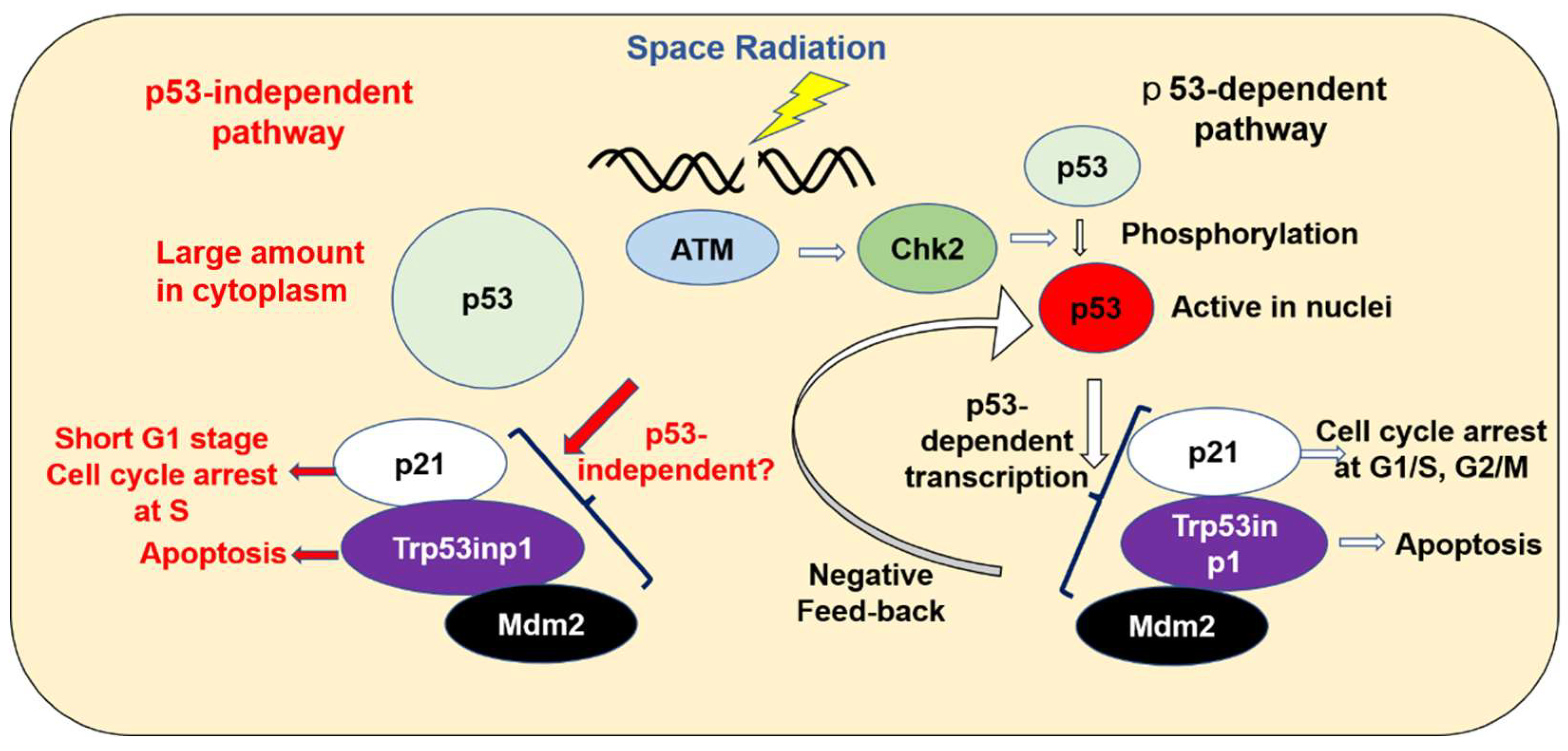
3. Discussion
3.1. The Transcriptome of Mouse ES Cells on the ISS
3.2. The Enhancement of Expression in p53-Related Genes in ES Cells on the ISS
3.3. The Effects of Transcriptome Caused by the Lack of the Histone H2AX Gene
4. Materials and Methods
4.1. Mouse ES Cells
4.2. Preparation of Space Samples
4.3. Irradiation by Accelerator
4.4. RNA Sequencing Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durante, M.; Cucinotta, F.A. Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer 2008, 8, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, Y.; Niimi, A.; Isono, M.; Yamauchi, M.; Yasuhara, T.; Limsirichaikul, S.; Oike, T.; Sato, H.; Held, K.D.; Nakano, T.; et al. 3D-structured illumination microscopy reveals clustered DNA double-strand break formation in widespread γH2AX foci after high LET heavy-ion particle radiation. Oncotarget 2017, 8, 109370–109381. [Google Scholar] [CrossRef]
- Blakely, E.A. The 20th Gray lecture 2019: Health and heavy ions. Br. J. Radiol. 2020, 93, 20200172. [Google Scholar] [CrossRef] [PubMed]
- Sihver, L.; Mortazavi, S.M.J. Biological Protection in Deep Space Missions. J. Biomed. Phys. Eng. 2021, 11, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Akamatsu, K.; Tsuda, M.; Tujimoto, A.; Hirayama, R.; Hiromoto, T.; Tamada, T.; Ide, H.; Shikazono, N. Formation of clustered DNA damage in vivo upon irradiation with ionizing radiation: Visualization and analysis with atomic force microscopy. Proc. Natl. Acad. Sci. USA 2022, 119, e2119132119. [Google Scholar] [CrossRef]
- Cacao, E.; Hada, M.; Saganti, P.B.; George, K.A.; Cucinotta, F.A. Relative Biological Effectiveness of HZE Particles for Chromosomal Exchanges and Other Surrogate Cancer Risk Endpoints. PLoS ONE 2016, 11, e0153998. [Google Scholar] [CrossRef]
- Goodhead, D.T. Track Structure and the Quality Factor for Space Radiation Cancer Risk (REID). 2018. Available online: https://three.jsc.nasa.gov/articles/Track_QF_Goodhead.pdf (accessed on 28 September 2018).
- Cucinotta, F.A.; Cacao, E. Risks of cognitive detriments after low dose heavy ion and proton exposures. Int. J. Radiat. Biol. 2019, 95, 985–998. [Google Scholar] [CrossRef]
- Huang, E.G.; Wang, R.-Y.; Xie, L.; Chang, P.; Yao, G.; Zhang, B.; Ham, D.W.; Lin, Y.; Blakely, E.A.; Sachs, R.K. Simulating galactic cosmic ray effects: Synergy modeling of murine tumor prevalence after exposure to two one-ion beams in rapid sequence. Life Sci. Space Res. 2020, 25, 107–118. [Google Scholar] [CrossRef]
- Cucinotta, F.A.; Pak, S. Cancer and circulatory disease risks for the largest solar particle events in the space age. Life Sci. Space Res. 2024, 40, 1–7. [Google Scholar] [CrossRef]
- Cucinotta, F.A.; Kim, M.-H.Y.; Willingham, V.; George, K.A. Physical and Biological Organ Dosimetry Analysis for International Space Station Astronauts. Radiat. Res. 2008, 170, 127–138. [Google Scholar] [CrossRef]
- Nagamatsu, A.; Murakami, K.; Kitajo, K.; Shimada, K.; Kumagai, H.; Tawara, H. Area radiation monitoring on ISS Increments 17 to 22 using PADLES in the Japanese Experiment Module Kibo. Radiat. Meas. 2013, 59, 84–93. [Google Scholar] [CrossRef]
- Naito, M.; Hasebe, N.; Shikishima, M.; Amano, Y.; Haruyama, J.; A Matias-Lopes, J.; Kim, K.J.; Kodaira, S. Radiation dose and its protection in the Moon from galactic cosmic rays and solar energetic particles: At the lunar surface and in a lava tube. J. Radiol. Prot. 2020, 40, 947–961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wimmer-Schweingruber, R.F.; Yu, J.; Wang, C.; Fu, Q.; Zou, Y.; Sun, Y.; Wang, C.; Hou, D.; Böttcher, S.I.; et al. First measurements of the radiation dose on the lunar surface. Sci. Adv. 2020, 6, eaaz1334. [Google Scholar] [CrossRef]
- Kim, M.Y.; Cucinotta, F.A.; Nounu, H.; Zeitlin, C.; Hassler, D.M.; Rafkin, S.C.R.; Wimmer-Schweingruber, B.; Ehresmann, D.E.; Brinza, S.; Böttcher, E.; et al. Comparison of Martian Surface Ionizing Radiation Measurements from MSL-RAD with Badhwar-O’Neill 2011/HZETRN Model Calculations. J. Geophys. Res. 2014, 119, 1311–1321. [Google Scholar] [CrossRef]
- A Cucinotta, F.; Wu, H.; Shavers, M.R.; George, K. Radiation dosimetry and biophysical models of space radiation effects. Gravitational Space Biol. 2003, 16, 11–18. [Google Scholar]
- Zeitlin, C.; Hassler, D.M.; Cucinotta, F.A.; Ehresmann, B.; Wimmer-Schweingruber, R.F.; Brinza, D.E.; Kang, S.; Weigle, G.; Böttcher, S.; Böhm, E.; et al. Measurements of Energetic Particle Radiation in Transit to Mars on the Mars Science Laboratory. Science 2013, 340, 1080–1084. [Google Scholar] [CrossRef] [PubMed]
- Zeitlin, C.; Hassler, D.; Ehresmann, B.; Rafkin, S.; Guo, J.; Wimmer-Schweingruber, R.; Berger, T.; Matthiä, D. Measurements of radiation quality factor on Mars with the Mars Science Laboratory Radiation Assessment Detector. Life Sci. Space Res. 2019, 22, 89–97. [Google Scholar] [CrossRef]
- Yoshida, K.; Hada, M.; Kizu, A.; Kitada, K.; Eguchi-Kasai, K.; Kokubo, T.; Teramura, T.; Yano, S.; Suzuki, H.H.; Watanabe, H.; et al. Comparison of biological measurement and physical estimates of space radiation in the International Space Station. Heliyon 2022, 8, e10266. [Google Scholar] [CrossRef] [PubMed]
- Cucinotta, F.A.; To, K.; Cacao, E. Predictions of space radiation fatality risk for exploration missions. Life Sci. Space Res. 2017, 13, 1–11. [Google Scholar] [CrossRef]
- Fensterl, V.; Sen, G.C. Interferon-Induced Ifit Proteins: Their Role in Viral Pathogenesis. J. Virol. 2015, 89, 2462–2468. [Google Scholar] [CrossRef]
- Midwood, K.S.; Valenick, L.V.; Hsia, H.C.; Schwarzbauer, J.E. Coregulation of Fibronectin Signaling and Matrix Contraction by Tenascin-C and Syndecan-4. Mol. Biol. Cell 2004, 15, 5670–5677. [Google Scholar] [CrossRef]
- Schenke-Layland, K.; Angelis, E.; Rhodes, K.E.; Heydarkhan-Hagvall, S.; Mikkola, H.K.; MacLellan, W.R.; MacLellan, W.R. Collagen IV Induces Trophoectoderm Differentiation of Mouse Embryonic Stem Cells. Stem Cells 2007, 25, 1529–1538. [Google Scholar] [CrossRef]
- Ando, Y.; Okeyo, K.O.; Adachi, T. Pluripotency state of mouse ES cells determines their contribution to self-organized layer formation by mesh closure on microstructured adhesion-limiting substrates. Biochem. Biophys. Res. Commun. 2021, 590, 97–102. [Google Scholar] [CrossRef]
- Ho, M.; Thompson, B.; Fisk, J.N.; Nebert, D.W.; Bruford, E.A.; Vasiliou, V.; Bunick, C.G. Update of the keratin gene family: Evolution, tissue-specific expression patterns, and relevance to clinical disorders. Hum. Genom. 2022, 16, 1–21. [Google Scholar] [CrossRef]
- Ralston, A.; Rossant, J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev. Biol. 2008, 313, 614–629. [Google Scholar] [CrossRef]
- Bhattacharya, B.; Miura, T.; Brandenberger, R.; Mejido, J.; Luo, Y.; Yang, A.X.; Joshi, B.H.; Ginis, I.; Thies, R.S.; Amit, M.; et al. Gene expression in human embryonic stem cell lines: Unique molecular signature. Blood 2004, 103, 2956–2964. [Google Scholar] [CrossRef]
- Abujarour, R.; Efe, J.; Ding, S. Genome-wide gain-of-function screen identifies novel regulators of pluripotency. Stem Cells 2010, 28, 1487–1497. [Google Scholar] [CrossRef]
- Saha, J.; Wang, M.; Cucinotta, F.A. Investigation of switch from ATM to ATR signaling at the sites of DNA damage induced by low and high LET radiation. DNA Repair. 2013, 12, 1143–1151. [Google Scholar] [CrossRef]
- 1990 Recommendations of the International Commission on Radiological Protection (ICRP Publication 60). Ann. ICRP 1991, 21, 1–3.
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef]
- Smith, J.; Tho, L.M.; Xu, N.; Gillespie, D.A. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010, 108, 73–112. [Google Scholar] [CrossRef]
- Mozaffari, N.L.; Pagliarulo, F.; Sartori, A.A. Human CtIP: A ‘double agent’ in DNA repair and tumorigenesis. Semin. Cell Dev. Biol. 2020, 113, 47–56. [Google Scholar] [CrossRef]
- Li, F.; Mladenov, E.; Sun, Y.; Soni, A.; Stuschke, M.; Timmermann, B.; Iliakis, G. Low CDK Activity and Enhanced Degradation by APC/CCDH1 Abolishes CtIP Activity and Alt-EJ in Quiescent Cells. Cells 2023, 12, 1530. [Google Scholar] [CrossRef]
- Xu, Z.-X.; Timanova-Atanasova, A.; Zhao, R.-X.; Chang, K.-S. PML Colocalizes with and Stabilizes the DNA Damage Response Protein TopBP1. Mol. Cell. Biol. 2003, 23, 4247–4256. [Google Scholar] [CrossRef][Green Version]
- Nickoloff, J.A.; Jaiswal, A.S.; Sharma, N.; Williamson, E.A.; Tran, M.T.; Arris, D.; Yang, M.; Hromas, R. Cellular Responses to Widespread DNA Replication Stress. Int. J. Mol. Sci. 2023, 24, 16903. [Google Scholar] [CrossRef]
- Yamane, K.; Wu, X.; Chen, J. A DNA Damage-Regulated BRCT-Containing Protein, TopBP1, Is Required for Cell Survival. Mol. Cell. Biol. 2002, 22, 555–566. [Google Scholar] [CrossRef]
- Mladenova, V.; Mladenov, E.; Stuschke, M.; Iliakis, G. DNA Damage Clustering after Ionizing Radiation and Consequences in the Processing of Chromatin Breaks. Molecules 2022, 27, 1540. [Google Scholar] [CrossRef] [PubMed]
- Morita, T.; Yoshimura, Y.; Yamamoto, A.; Murata, K.; Mori, M.; Yamamoto, H.; Matsushiro, A. A mouse homolog of the Escherichia coli recA and Saccharomyces cervisiae RAD51 genes. Proc. Natl. Acad. Sci. USA 1993, 90, 6577–6580. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, A.; Yagi, H.; Habu, T.; Yoshimura, Y.; Matsushiro, A.; Nishimune, Y.; Morita, T.; Taki, T.; Yoshida, K.; Yamamoto, K. Cell cycle-dependent expression of the mouseRad51 gene in proliferating cells. Mol. Genet. Genom. 1996, 251, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tsuzuki, T.; Fujii, Y.; Sakumi, K.; Tominaga, Y.; Nakao, K.; Sekiguchi, M.; Matsushiro, A.; Yoshimura, Y.; Morita, T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 1996, 93, 6236–6240. [Google Scholar] [CrossRef] [PubMed]
- Inagawa, T.; Wennink, T.; Lebbink, J.H.G.; Keijzers, G.; Florea, B.I.; Verkaik, N.S.; van Gent, D.C. C-Terminal Extensions of Ku70 and Ku80 Differentially Influence DNA End Binding Properties. Int. J. Mol. Sci. 2020, 21, 6725. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pannicke, U.; Schwarz, K.; Lieber, M.R. Hairpin Opening and Overhang Processing by an Artemis/DNA-Dependent Protein Kinase Complex in Nonhomologous End Joining and V(D)J Recombination. Cell 2002, 108, 781–794. [Google Scholar] [CrossRef]
- Zhang, X.; Succi, J.; Feng, Z.; Prithivirajsingh, S.; Story, M.D.; Legerski, R.J. Artemis is a phosphorylation target of ATM and ATR and is involved in the G2/M DNA damage checkpoint response. Mol. Cell Biol. 2004, 24, 9207–9220. [Google Scholar] [CrossRef]
- Shahbazi, J.; Lock, R.; Liu, T. Tumor Protein 53-Induced Nuclear Protein 1 Enhances p53 Function and Represses Tumorigenesis. Front. Genet. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Sándor, N.; Schilling-Tóth, B.; Kis, E.; Fodor, L.; Mucsányi, F.; Sáfrány, G.; Hegyesi, H. TP53inp1 Gene Is Implicated in Early Radiation Response in Human Fibroblast Cells. Int. J. Mol. Sci. 2015, 16, 25450–25465. [Google Scholar] [CrossRef] [PubMed]
- Lagunas-Martínez, A.; García-Villa, E.; Arellano-Gaytán, M.; Contreras-Ochoa, C.O.; Dimas-González, J.; López-Arellano, M.E.; Madrid-Marina, V.; Gariglio, P. MG132 plus apoptosis antigen-1 (APO-1) antibody cooperate to restore p53 activity inducing autophagy and p53-dependent apoptosis in HPV16 E6-expressing keratinocytes. Apoptosis 2016, 22, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Galluzzi, L. BAX and BAK dynamics control mitochondrial DNA release during apoptosis. Cell Death Differ. 2022, 29, 1296–1298. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.; Emi, M.; Tanabe, K. Caspase-dependent and -independent cell death pathways after DNA damage (Review). Oncol. Rep. 2005, 14, 595–599. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Stead, E.; Faast, R.; Conn, S.; Cartwright, P.; Dalton, S. Developmental Activation of the Rb–E2F Pathway and Establishment of Cell Cycle-regulated Cyclin-dependent Kinase Activity during Embryonic Stem Cell Differentiation. Mol. Biol. Cell 2005, 16, 2018–2027. [Google Scholar] [CrossRef]
- Conklin, J.F.; Baker, J.; Sage, J. The RB family is required for the self-renewal and survival of human embryonic stem cells. Nat. Commun. 2012, 3, 1244. [Google Scholar] [CrossRef]
- Vélez-Cruz, R.; Manickavinayaham, S.; Biswas, A.K.; Clary, R.W.; Premkumar, T.; Cole, F.; Johnson, D.G. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Minerva Anestesiol. 2016, 30, 2500–2512. [Google Scholar] [CrossRef]
- Pouyet, L.; Carrier, A. Mutant mouse models of oxidative stress. Transgenic. Res. 2009, 19, 155–164. [Google Scholar] [CrossRef]
- Ohshige, T.; Iwata, M.; Omori, S.; Tanaka, Y.; Hirose, H.; Kaku, K.; Maegawa, H.; Watada, H.; Kashiwagi, A.; Kawamori, R.; et al. Association of New Loci Identified in European Genome-Wide Association Studies with Susceptibility to Type 2 Diabetes in the Japanese. PLoS ONE 2011, 6, e26911. [Google Scholar] [CrossRef] [PubMed]
- Bassing, C.H.; Chua, K.F.; Sekiguchi, J.; Suh, H.; Whitlow, S.R.; Fleming, J.C.; Monroe, B.C.; Ciccone, D.N.; Yan, C.; Vlasakova, K.; et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA 2002, 99, 8173–8178. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Morita, T. Control of Radiosensitivity of F9 Mouse Teratocarcinoma Cells by Regulation of Histone H2AX Gene Expression using a Tetracycline Turn-Off System. Cancer Res. 2004, 64, 4131–4136. [Google Scholar] [CrossRef] [PubMed]
- Franco, S.; Gostissa, M.; Zha, S.; Lombard, D.B.; Murphy, M.M.; Zarrin, A.A.; Yan, C.; Tepsuporn, S.; Morales, J.C.; Adams, M.M.; et al. H2AX Prevents DNA Breaks from Progressing to Chromosome Breaks and Translocations. Mol. Cell 2006, 21, 201–214. [Google Scholar] [CrossRef]
- Nishida, H.; Tomaru, Y.; Oho, Y.; Hayashizaki, Y. Naturally occurring antisense RNA of histone H2a in mouse cultured cell lines. BMC Genet. 2005, 6, 23. [Google Scholar] [CrossRef]
- Beheshti, A.; Sachs, R.K.; Peluso, M.; Rietman, E.; Hahnfeldt, P.; Hlatky, L. Age and Space Irradiation Modulate Tumor Progression: Implications for Carcinogenesis Risk. Radiat. Res. 2013, 179, 208–220. [Google Scholar] [CrossRef]
- Wage, J.; Ma, L.; Peluso, M.; Lamont, C.; Evens, A.M.; Hahnfeldt, P.; Hlatky, L.; Beheshti, A. Proton irradiation impacts age-driven modulations of cancer progression influenced by immune system transcriptome modifications from splenic tissue. J. Radiat. Res. 2015, 56, 792–803. [Google Scholar] [CrossRef]
- Overbey, E.G.; Paul, A.M.; da Silveira, W.A.; Tahimic, C.G.T.; Reinsch, S.S.; Szewczyk, N.; Stanbouly, S.; Wang, C.; Galazka, J.M.; Mao, X.W. Mice Exposed to Combined Chronic Low-Dose Irradiation and Modeled Microgravity Develop Long-Term Neurological Sequelae. Int. J. Mol. Sci. 2019, 20, 4094. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.M.; Overbey, E.G.; da Silveira, W.A.; Szewczyk, N.; Nishiyama, N.C.; Pecaut, M.J.; Anand, S.; Galazka, J.M.; Mao, X.W. Immunological and hematological outcomes following protracted low dose/low dose rate ionizing radiation and simulated microgravity. Sci. Rep. 2021, 11, 1–11. [Google Scholar] [CrossRef]
- Schenten, V.; Gueguinou, N.; Baatout, S.; Frippiat, J.-P. Modulation of Pleurodeles waltl DNA Polymerase mu Expression by Extreme Conditions Encountered during Spaceflight. PLoS ONE 2013, 8, e69647. [Google Scholar] [CrossRef]
- Beheshti, A.; Ray, S.; Fogle, H.; Berrios, D.; Costes, S.V. A microRNA signature and TGF-β1 response were identified as the key master regulators for spaceflight response. PLoS ONE 2018, 13, e0199621. [Google Scholar] [CrossRef]
- Fujita, S.-I.; Rutter, L.; Ong, Q.; Muratani, M. Integrated RNA-seq Analysis Indicates Asynchrony in Clock Genes between Tissues under Spaceflight. Life 2020, 10, 196. [Google Scholar] [CrossRef]
- Veliz, A.L.; Mamoun, L.; Hughes, L.; Vega, R.; Holmes, B.; Monteon, A.; Bray, J.; Pecaut, M.J.; Kearns-Jonker, M. Transcriptomic Effects on the Mouse Heart Following 30 Days on the International Space Station. Biomolecules 2023, 13, 371. [Google Scholar] [CrossRef]
- Gridley, D.S.; Mao, X.W.; Tian, J.; Cao, J.D.; Perez, C.; Stodieck, L.S.; Ferguson, V.L.; A Bateman, T.; Pecaut, M.J. Genetic and Apoptotic Changes in Lungs of Mice Flown on the STS-135 Mission in Space. In Vivo 2015, 29, 423–433. [Google Scholar]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA Twins Study: A multidimensional analysis of a year-long human spaceflight. Science 2019, 364, eaau8650. [Google Scholar] [CrossRef]
- da Silveira, W.A.; Fazelinia, H.; Rosenthal, S.B.; Laiakis, E.C.; Kim, M.S.; Meydan, C.; Kidane, Y.; Rathi, K.S.; Smith, S.M.; Steer, B.; et al. Comprehensive Multi-omics Analysis Reveals Mitochondrial Stress as a Central Biological Hub for Spaceflight Impact. Cell 2020, 183, 1185–1201.e20. [Google Scholar] [CrossRef] [PubMed]
- Tilton, S.C.; Markillie, L.M.; Hays, S.; Taylor, R.C.; Stenoien, D.L. Identification of Differential Gene Expression Patterns after Acute Exposure to High and Low Doses of Low-LET Ionizing Radiation in a Reconstituted Human Skin Tissue. Radiat. Res. 2016, 186, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Cano, C.E.; Gommeaux, J.; Pietri, S.; Culcasi, M.; Garcia, S.; Seux, M.; Barelier, S.; Vasseur, S.; Spoto, R.P.; Peébusque, M.-J.; et al. Tumor Protein 53–Induced Nuclear Protein 1 Is a Major Mediator of p53 Antioxidant Function. Cancer Res. 2008, 69, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Sarin, N.; Engel, F.; Kalayda, G.V.; Mannewitz, M.; Cinatl, J.; Rothweiler, F.; Michaelis, M.; Saafan, H.; Ritter, C.A.; Jaehde, U.; et al. Cisplatin resistance in non-small cell lung cancer cells is associated with an abrogation of cisplatin-induced G2/M cell cycle arrest. PLoS ONE 2017, 12, e0181081. [Google Scholar] [CrossRef]
- El Husseini, N.; Schlisser, A.E.; Hales, B.F. Editor’s Highlight: Hydroxyurea Exposure Activates the P53 Signaling Pathway in Murine Organogenesis-Stage Embryos. Toxicol. Sci. 2016, 152, 297–308. [Google Scholar] [CrossRef]
- Hong, Y.; Cervantes, R.B.; Tichy, E.; Tischfield, J.A.; Stambrook, P.J. Protecting genomic integrity in somatic cells and embryonic stem cells. Mutat. Res. Mol. Mech. Mutagen. 2007, 614, 48–55. [Google Scholar] [CrossRef]
- Aladjem, M.I.; Spike, B.T.; Rodewald, L.W.; Hope, T.J.; Klemm, M.; Jaenisch, R.; Wahl, G.M. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr. Biol. 1998, 8, 145–155. [Google Scholar] [CrossRef]
- Solozobova, V.; Rolletschek, A.; Blattner, C. Nuclear accumulation and activation of p53 in embryonic stem cells after DNA damage. BMC Cell Biol. 2009, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Solozobova, V.; Blattner, C. Regulation of p53 in embryonic stem cells. Exp. Cell Res. 2010, 316, 2434–2446. [Google Scholar] [CrossRef] [PubMed]
- Ballabeni, A.; Park, I.-H.; Zhao, R.; Wang, W.; Lerou, P.H.; Daley, G.Q.; Kirschner, M.W. Cell cycle adaptations of embryonic stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 19252–19257. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Kastner, P.K.; Jardine, K.; Cormier, M.; McBurney, M.W. Absence of p53-dependent cell cycle regulation in pluripotent mouse cell lines. Oncogene 1998, 16, 3003–3011. [Google Scholar] [CrossRef] [PubMed]
- Neganova, I.; Lako, M. G1 to S phase cell cycle transition in somatic ad embryonic stem cells. J. Anat. 2008, 213, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Prost, S.; Bellamy, C.O.; Clarke, A.R.; Wyllie, A.H.; Harrison, D.J. p53-independent DNA repair and cell cycle arrest in embryonic stem cells. FEBS Lett. 1998, 425, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Nakao, K.; Nakamura, K.; Saito, I.; Katsuki, M.; Arai, K.; Masai, H. Inactivation of Cdc7 kinase in mouse ES cells results in S-phase arrest and p53-dependent cell death. EMBO J. 2002, 21, 2168–2179. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; He, Y.; Dubois, W.; Wu, X.; Shi, J.; Huang, J. Distinct Regulatory Mechanisms and Functions for p53-Activated and p53-Repressed DNA Damage Response Genes in Embryonic Stem Cells. Mol. Cell 2012, 46, 30–42. [Google Scholar] [CrossRef] [PubMed]
- ter Huurne, M.; Peng, T.; Yi, G.; van Mierlo, G.; Marks, H.; Stunnenberg, H.G. Critical Role for P53 in Regulating the Cell Cycle of ground state embryonic stem cells. Stem Cell Rep. 2020, 14, 175–183. [Google Scholar] [CrossRef]
- Tomasini, R.; Seux, M.; Nowak, J.; Bontemps, C.; Carrier, A.; Dagorn, J.-C.; Pébusque, M.-J.; Iovanna, J.L.; Dusetti, N.J. TP53INP1 is a novel p73 target gene that induces cell cycle arrest and cell death by modulating p73 transcriptional activity. Oncogene 2005, 24, 8093–8104. [Google Scholar] [CrossRef]
- Hong, H.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Kanagawa, O.; Nakagawa, M.; Okita, K.; Yamanaka, S. Suppression of induced pluripotent stem cell generation by the p53–p21 pathway. Nature 2009, 460, 1132–1135. [Google Scholar] [CrossRef] [PubMed]
- Stillman, B. Origin recognition and the chromosome cycle. FEBS Lett. 2005, 579, 877–884. [Google Scholar] [CrossRef]
- O’donnell, M.; Langston, L.; Stillman, B. Principles and Concepts of DNA Replication in Bacteria, Archaea, and Eukarya. Cold Spring Harb. Perspect. Biol. 2013, 5, a010108. [Google Scholar] [CrossRef]
- Hu, Y.; Stillman, B. Origins of DNA replication in eukaryotes. Mol. Cell 2023, 83, 352–372. [Google Scholar] [CrossRef]
- Fishel, R. Mismatch Repair. J. Biol. Chem. 2015, 290, 26395–26403. [Google Scholar] [CrossRef]
- Chaudhuri, A.R.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Apostolou, Z.; Chatzinikolaou, G.; Stratigi, K.; Garinis, G.A. Nucleotide Excision Repair and Transcription-Associated Genome Instability. BioEssays 2019, 41, e1800201. [Google Scholar] [CrossRef] [PubMed]
- Krokan, H.E.; Bjørås, M. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2013, 5, a012583. [Google Scholar] [CrossRef] [PubMed]
- Beard, W.A.; Horton, J.K.; Prasad, R.; Wilson, S.H. Eukaryotic Base Excision Repair: New Approaches Shine Light on Mechanism. Annu. Rev. Biochem. 2019, 88, 137–162. [Google Scholar] [CrossRef] [PubMed]
- Marteijn, J.A.; Lans, H.; Vermeulen, W.; Hoeijmakers, J.H.J. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014, 15, 465–481. [Google Scholar] [CrossRef]
- Basu, U.; Bostwick, A.M.; Das, K.; Dittenhafer-Reed, K.E.; Patel, S.S. Structure, mechanism, and regulation of mitochondrial DNA transcription initiation. J. Biol. Chem. 2020, 295, 18406–18425. [Google Scholar] [CrossRef]
- Kotrys, A.V.; Szczesny, R.J. Mitochondrial Gene Expression and Beyond—Novel Aspects of Cellular Physiology. Cells 2019, 9, 17. [Google Scholar] [CrossRef]
| ES Cell | C57BL6/129 Sv | B6-G | |
|---|---|---|---|
| Genotypes | H2AX (+/+) | H2AX (+/+) | H2AX (+/+) |
| Comparison | ISS/BU | Fe; 3 Gy/0 Gy | Fe; 3 Gy/0 Gy |
| Duration | 0 h–8 h | 0 h–8 h | 0 h–48 h |
| Trp53 (p53) | 0.94 | 0.98 | 1.09 |
| Trp53inp1 | 1.53 | 1.33 | 1.45 |
| Cdkn1a (p21) | 1.13 | 1.28 | 1.34 |
| Mdm2 | 1.23 | 1.20 | 1.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, K.; Hada, M.; Hayashi, M.; Kizu, A.; Kitada, K.; Eguchi-Kasai, K.; Kokubo, T.; Teramura, T.; Hashizume Suzuki, H.; Watanabe, H.; et al. Transcriptome Analysis by RNA Sequencing of Mouse Embryonic Stem Cells Stocked on International Space Station for 1584 Days in Frozen State after Culture on the Ground. Int. J. Mol. Sci. 2024, 25, 3283. https://doi.org/10.3390/ijms25063283
Yoshida K, Hada M, Hayashi M, Kizu A, Kitada K, Eguchi-Kasai K, Kokubo T, Teramura T, Hashizume Suzuki H, Watanabe H, et al. Transcriptome Analysis by RNA Sequencing of Mouse Embryonic Stem Cells Stocked on International Space Station for 1584 Days in Frozen State after Culture on the Ground. International Journal of Molecular Sciences. 2024; 25(6):3283. https://doi.org/10.3390/ijms25063283
Chicago/Turabian StyleYoshida, Kayo, Megumi Hada, Masami Hayashi, Akane Kizu, Kohei Kitada, Kiyomi Eguchi-Kasai, Toshiaki Kokubo, Takeshi Teramura, Hiromi Hashizume Suzuki, Hitomi Watanabe, and et al. 2024. "Transcriptome Analysis by RNA Sequencing of Mouse Embryonic Stem Cells Stocked on International Space Station for 1584 Days in Frozen State after Culture on the Ground" International Journal of Molecular Sciences 25, no. 6: 3283. https://doi.org/10.3390/ijms25063283
APA StyleYoshida, K., Hada, M., Hayashi, M., Kizu, A., Kitada, K., Eguchi-Kasai, K., Kokubo, T., Teramura, T., Hashizume Suzuki, H., Watanabe, H., Kondoh, G., Nagamatsu, A., Saganti, P., Muratani, M., Cucinotta, F. A., & Morita, T. (2024). Transcriptome Analysis by RNA Sequencing of Mouse Embryonic Stem Cells Stocked on International Space Station for 1584 Days in Frozen State after Culture on the Ground. International Journal of Molecular Sciences, 25(6), 3283. https://doi.org/10.3390/ijms25063283








