Genetic Loci of Plant Pathogenic Dickeya solani IPO 2222 Expressed in Contact with Weed-Host Bittersweet Nightshade (Solanum dulcamara L.) Plants
Abstract
1. Introduction
2. Results
2.1. Identification of the Tn5 Insertion Sites in D. solani Mutants
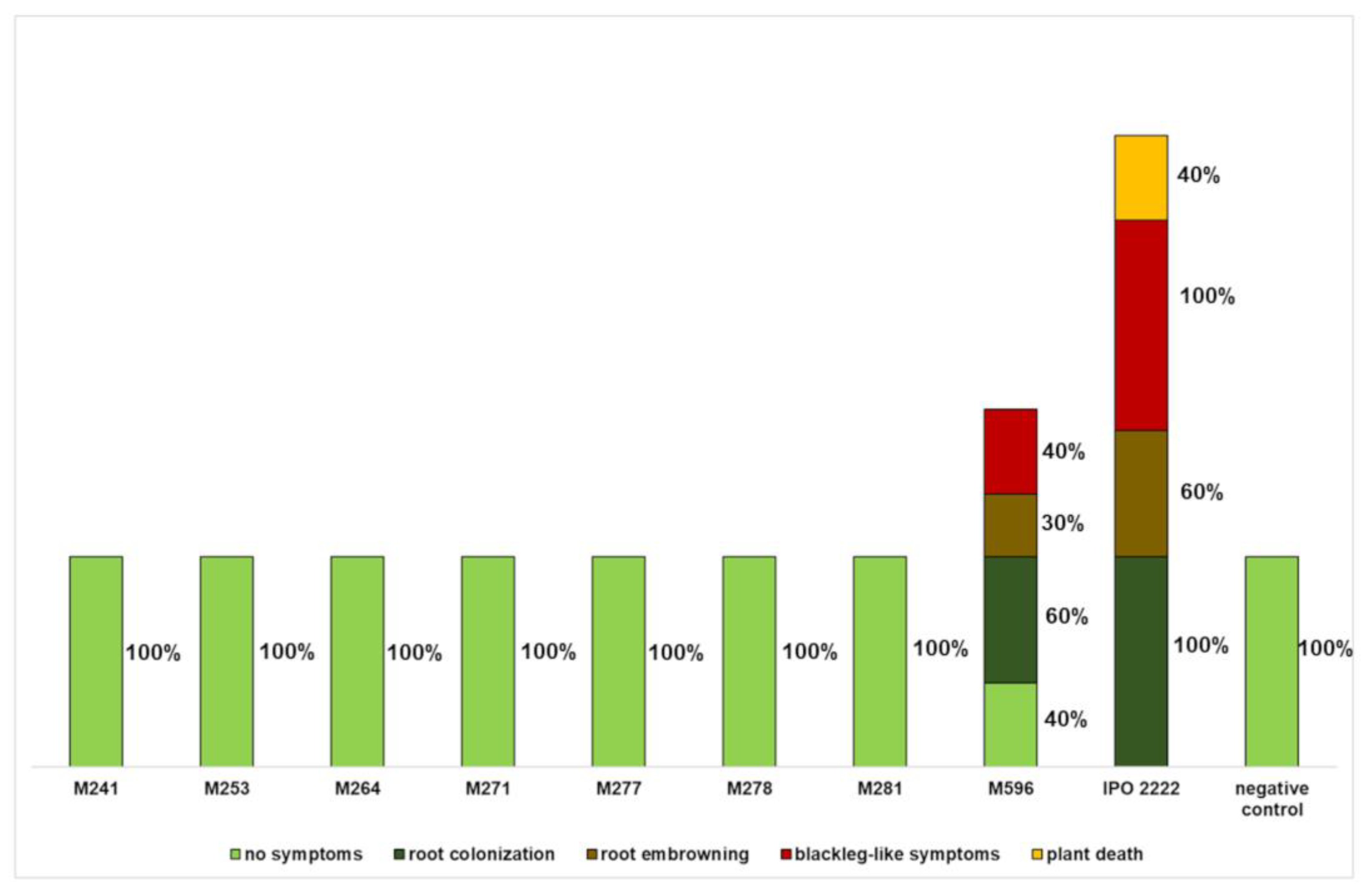
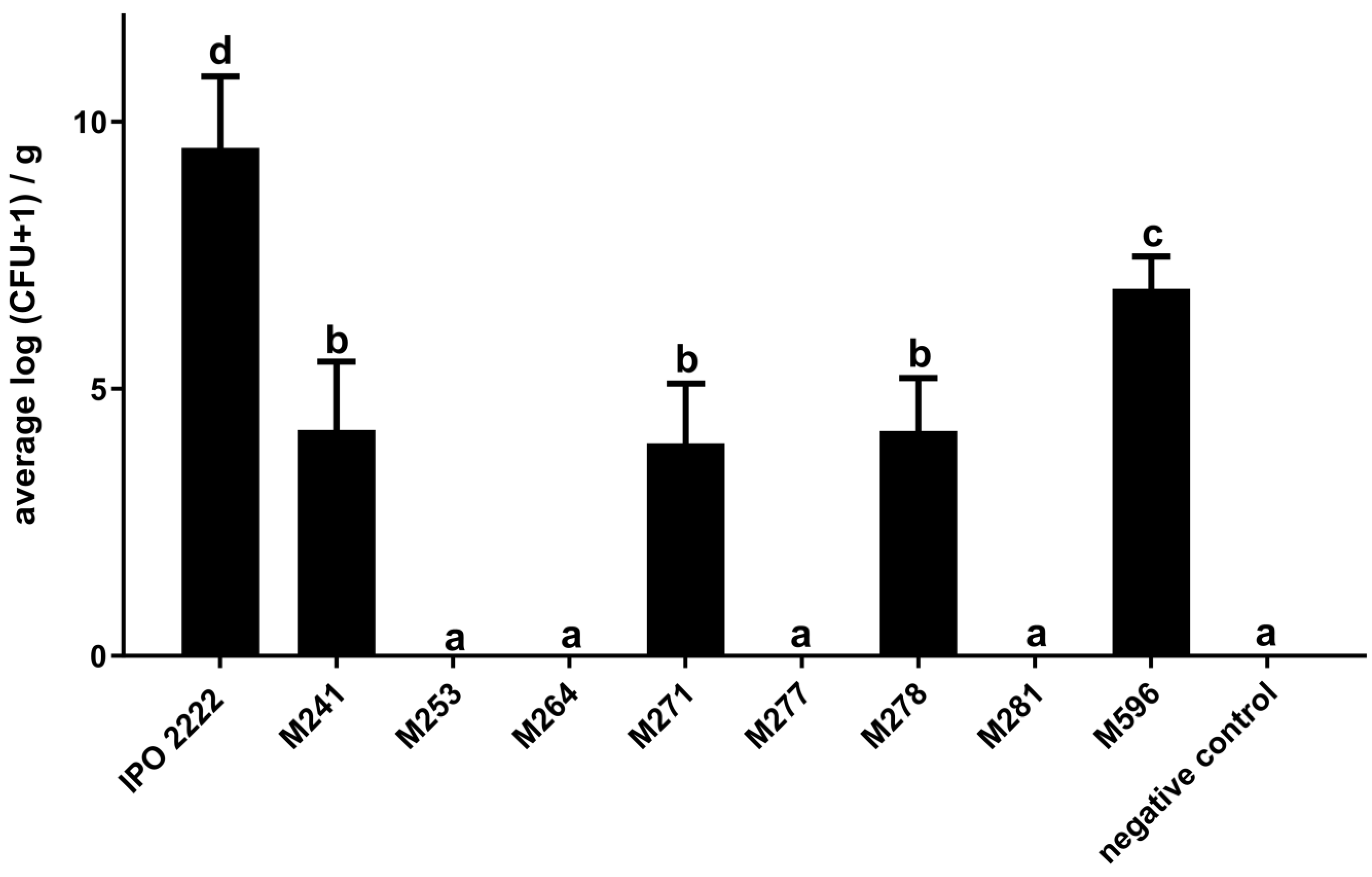
2.2. The Capacity of D. solani Tn5 Mutants to Induce Symptoms in S. dulcamara Plants Cultivated in Culture Tubes
2.3. Phenotypes of D. solani Tn5 Mutants
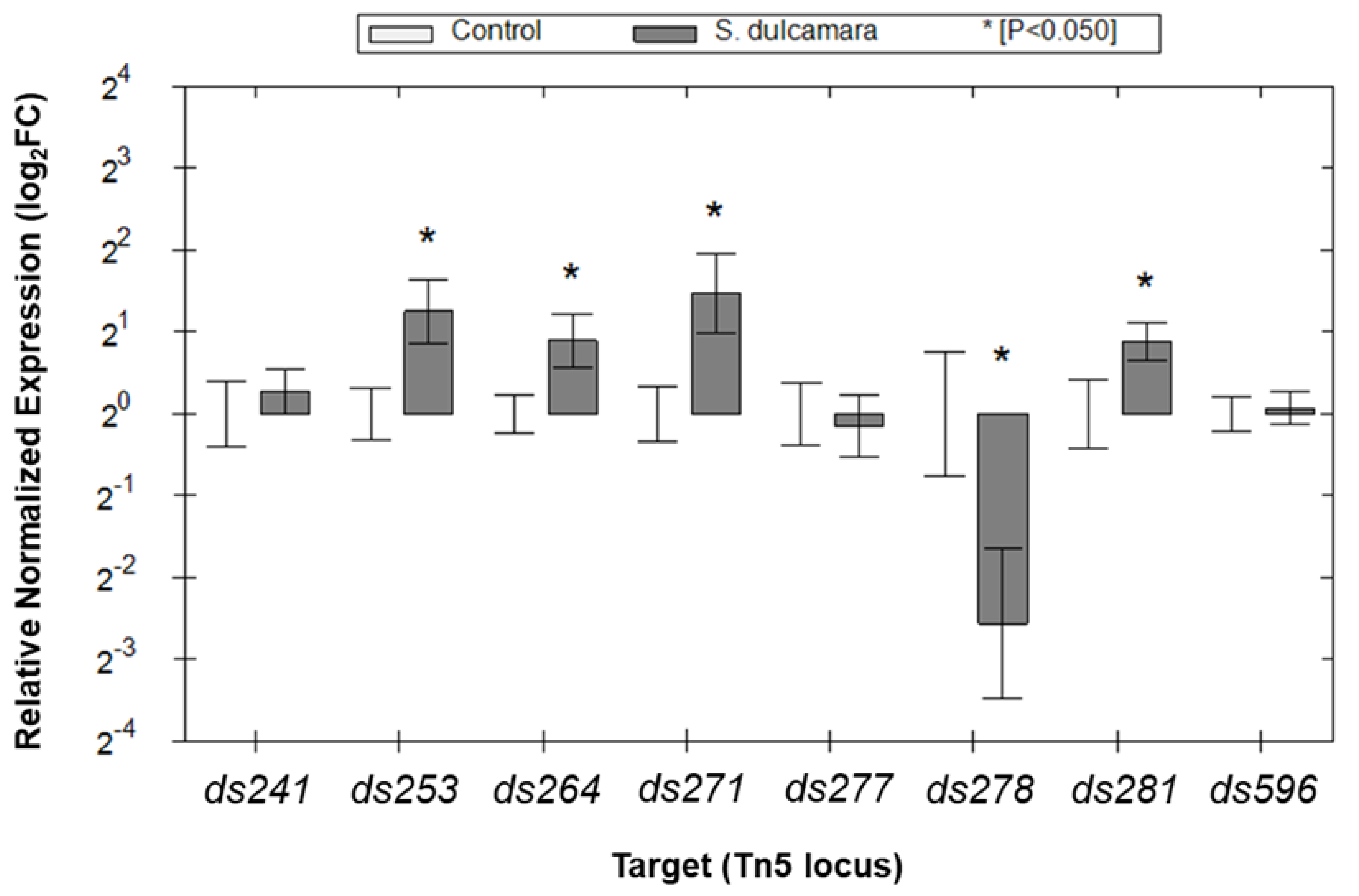
2.4. Expression of the Selected D. solani Genes in the Presence of S. dulcamara Analyzed with qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plants, and Growth Media
4.2. Determination of the Tn5 Insertion Sites through Genome Sequencing and Assessing the Functionality of the Disrupted Genes
4.3. Prediction of Transcriptional Organization and Analysis of Biochemical Pathways and Cellular Enzymatic Networks for D. solani Genes with Tn5 Insertions
4.4. Phenotypic Characterization of D. solani Tn5 Mutants
4.5. Host Colonization and Virulence of D. solani Tn5 Mutants on S. dulcamara Plants Grown in Culture Tubes
4.6. Expression of Selected D. solani Genes Quantified Using qRT-PCR
4.7. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tyler, H.L.; Triplett, E.W. Plants as a habitat for beneficial and/or human pathogenic bacteria. Annu. Rev. Phytopathol. 2008, 46, 53–73. [Google Scholar] [CrossRef]
- Haichar, F.Z.; Marol, C.; Berge, O.; Rangel-Castro, J.I.; Prosser, J.I.; Balesdent, J.; Heulin, T.; Achouak, W. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008, 2, 1221–1230. [Google Scholar] [CrossRef]
- Alfano, J.R.; Collmer, A. Bacterial Pathogens in Plants: Life up against the Wall. Plant Cell 1996, 8, 1683–1698. [Google Scholar] [CrossRef]
- Buonaurio, R. Infection and plant defense responses during plant-bacterial interaction. Plant-Microbe Interact. 2008, 169–197. [Google Scholar]
- Fatima, U.; Senthil-Kumar, M. Plant and pathogen nutrient acquisition strategies. Front. Plant Sci. 2015, 6, 750. [Google Scholar] [CrossRef] [PubMed]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J. How plants recognize pathogens and defend themselves. Cell. Mol. Life Sci. 2007, 64, 2726–2732. [Google Scholar] [CrossRef] [PubMed]
- Mengiste, T. Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 2012, 50, 267–294. [Google Scholar] [CrossRef] [PubMed]
- Abramovitch, R.B.; Martin, G.B. Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 2004, 7, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Ghozlan, M.H.; Eman, E.-A.; Tokgöz, S.; Lakshman, D.K.; Mitra, A. Plant defense against necrotrophic pathogens. Am. J. Plant Sci. 2020, 11, 2122–2138. [Google Scholar] [CrossRef]
- Laluk, K.; Mengiste, T. Necrotroph attacks on plants: Wanton destruction or covert extortion? Arab. Book 2010, 8, e0136. [Google Scholar] [CrossRef] [PubMed]
- Salmond, G.P.C. Secretion of Extracellular Virulence Factors by Plant-Pathogenic Bacteria. Annu. Rev. Phytopathol. 1994, 32, 181–200. [Google Scholar] [CrossRef]
- Pontes, J.G.M.; Fernandes, L.S.; Dos Santos, R.V.; Tasic, L.; Fill, T.P. Virulence Factors in the Phytopathogen-Host Interactions: An Overview. J. Agric. Food Chem. 2020, 68, 7555–7570. [Google Scholar] [CrossRef] [PubMed]
- Toth, I.K.; Barny, M.-A.; Brurberg, M.B.; Condemine, G.; Czajkowski, R.; Elphinstone, J.G.; Helias, V.; Johnson, S.B.; Moleleki, L.N.; Pirhonen, M.; et al. Pectobacterium and Dickeya: Environment to Disease Development. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Van Gijsegem, F., Van Der Wolf, J.M., Toth, I.K., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 39–84. [Google Scholar]
- Charkowski, A.O. The Soft Rot Erwinia. Plant-Assoc. Bact. 2007, 3, 423–505. [Google Scholar]
- Ma, B.; Hibbing, M.E.; Kim, H.S.; Reedy, R.M.; Yedidia, I.; Breuer, J.; Breuer, J.; Glasner, J.D.; Perna, N.T.; Kelman, A.; et al. Host range and molecular phylogenies of the soft rot enterobacterial genera pectobacterium and dickeya. Phytopathology 2007, 97, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Van Gijsegem, F.; Toth, I.K.; Van Der Wolf, J.M. Soft Rot Pectobacteriaceae: A Brief Overview. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Van Gijsegem, F., Van Der Wolf, J.M., Toth, I.K., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 1–11. [Google Scholar]
- Charkowski, A.O. The Changing Face of Bacterial Soft-Rot Diseases. Annu. Rev. Phytopathol. 2018, 56, 269–288. [Google Scholar] [CrossRef]
- Van Gijsegem, F.; Van Der Wolf, J.M.; Toth, I.K. Plant Diseases Caused by Dickeya and Pectobacterium Species; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar] [CrossRef]
- Elphinstone, J. Revised investigation of Erwinia chrysanthemi (Dickeya dianthicola and other Dickeya spp.) able to infect potatoes. Proj. Rep. 2008, 7. [Google Scholar]
- Fikowicz-Krosko, J.; Wszalek-Rozek, K.; Smolarska, A.; Czajkowski, R. First report of isolation of soft rot Pectobacterium carotovorum subsp. carotovorum from symptomless bittersweet nightshade occuing in rural area of Poland. J. Plant Pathol. 2017, 99, 1. [Google Scholar] [CrossRef]
- Czajkowski, R.; Fikowicz-Krosko, J.; Maciag, T.; Rabalski, L.; Czaplewska, P.; Jafra, S.; Richert, M.; Krychowiak-Masnicka, M.; Hugouvieux-Cotte-Pattat, N. Genome-Wide Identification of Dickeya solani Transcriptional Units Up-Regulated in Response to Plant Tissues from a Crop-Host Solanum tuberosum and a Weed-Host Solanum dulcamara. Front. Plant Sci. 2020, 11, 580330. [Google Scholar] [CrossRef]
- Van Der Wolf, J.M.; Nijhuis, E.H.; Kowalewska, M.J.; Saddler, G.S.; Parkinson, N.; Elphinstone, J.G.; Pritchard, L.; Toth, I.K.; Lojkowska, E.; Potrykus, M.; et al. Dickeya solani sp. nov.: A pectinolytic plant-pathogenic bacterium isolated from potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 2014, 64, 768–774. [Google Scholar] [CrossRef]
- Fikowicz-Krosko, J.; Czajkowski, R. Systemic Colonization and Expression of Disease Symptoms on Bittersweet Nightshade (Solanum dulcamara) Infected with a GFP-Tagged Dickeya solani IPO2222 (IPO2254). Plant Dis. 2018, 102, 619–627. [Google Scholar] [CrossRef]
- Olsson, K. Detection of Erwinia spp. in some Swedish streams. In Report of the International Conference on Potato Blackleg Disease; Potato Marketing Board: Lincolnshire, UK, 1985; pp. 45–46. [Google Scholar]
- Van Gijsegem, F.; Hugouvieux-Cotte-Pattat, N.; Kraepiel, Y.; Lojkowska, E.; Moleleki, L.N.; Gorshkov, V.; Yedidia, I. Molecular Interactions of Pectobacterium and Dickeya with Plants. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Van Gijsegem, F., Van Der Wolf, J.M., Toth, I.K., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 85–147. [Google Scholar]
- Somssich, I.E.; Hahlbrock, K. Pathogen defence in plants—A paradigm of biological complexity. Trends Plant Sci. 1998, 3, 86–90. [Google Scholar] [CrossRef]
- Reverchon, S.; Nasser, W. Dickeya ecology, environment sensing and regulation of virulence programme. Environ. Microbiol. Rep. 2013, 5, 622–636. [Google Scholar] [CrossRef]
- Jiang, X.; Zghidi-Abouzid, O.; Oger-Desfeux, C.; Hommais, F.; Greliche, N.; Muskhelishvili, G.; Nasser, W.; Reverchon, S. Global transcriptional response of Dickeya dadantii to environmental stimuli relevant to the plant infection. Environ. Microbiol. 2016, 18, 3651–3672. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, A.; Georgellis, D. Environmental adaptation and diversification of bacterial two-component systems. Curr. Opin. Microbiol. 2023, 76, 102399. [Google Scholar] [CrossRef]
- Hyytiainen, H.; Montesano, M.; Palva, E.T. Global regulators ExpA (GacA) and KdgR modulate extracellular enzyme gene expression through the RsmA-rsmB system in Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 2001, 14, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Lebeau, A.; Reverchon, S.; Gaubert, S.; Kraepiel, Y.; Simond-Cote, E.; Nasser, W.; Van Gijsegem, F. The GacA global regulator is required for the appropriate expression of Erwinia chrysanthemi 3937 pathogenicity genes during plant infection. Environ. Microbiol. 2008, 10, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Sepulchre, J.A.; Reverchon, S.; Nasser, W. Modeling the onset of virulence in a pectinolytic bacterium. J. Theor. Biol. 2007, 244, 239–257. [Google Scholar] [CrossRef] [PubMed]
- Garcia, G.A.; Kittendorf, J.D. Transglycosylation: A mechanism for RNA modification (and editing?). Bioorganic Chem. 2005, 33, 229–251. [Google Scholar] [CrossRef]
- Kersten, H.; Kersten, W. Chapter 2 Biosynthesis and Function of Queuine and Queuosine tRNAs. In Chromatography and Modification of Nucleosides—Biological Roles and Function of Modification; Gehrke, C.W., Kuo, K.C.T., Eds.; Elsevier: Amsterdam, The Netherlands, 1990; pp. B69–B108. [Google Scholar]
- Antoine, L.; Bahena-Ceron, R.; Devi Bunwaree, H.; Gobry, M.; Loegler, V.; Romby, P.; Marzi, S. RNA Modifications in Pathogenic Bacteria: Impact on Host Adaptation and Virulence. Genes 2021, 12, 1125. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Rullo, J.; Gonzalez-Pastor, J.E. tRNA queuosine modification is involved in biofilm formation and virulence in bacteria. Nucleic Acids Res. 2023, 51, 9821–9837. [Google Scholar] [CrossRef]
- Koh, C.S.; Sarin, L.P. Transfer RNA modification and infection—Implications for pathogenicity and host responses. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 419–432. [Google Scholar] [CrossRef]
- Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, M.; Khokhani, D.; Nayaka, S.; Srivastava, S.; Keyser, Z.P.; Ranjan, A. Genomic diversity and organization of complex polysaccharide biosynthesis clusters in the genus Dickeya. PLoS ONE 2021, 16, e0245727. [Google Scholar] [CrossRef] [PubMed]
- Szulta, S.; Kornicka, A. Pectobacterium and Dickeya genus-A review on structural variations of polysaccharides and their role in the pathogenic process of plants. Plant Pathol. 2023, 72, 998–1010. [Google Scholar] [CrossRef]
- Reverchon, S.; Muskhelisvili, G.; Nasser, W. Chapter Three—Virulence Program of a Bacterial Plant Pathogen: The Dickeya Model. In Progress in Molecular Biology and Translational Science; San Francisco, M., San Francisco, B., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 51–92. [Google Scholar]
- Bartnik, P.; Lewtak, K.; Fiolka, M.; Czaplewska, P.; Narajczyk, M.; Czajkowski, R. Resistance of Dickeya solani strain IPO 2222 to lytic bacteriophage PhiD5 results in fitness tradeoffs for the bacterium during infection. Sci. Rep. 2022, 12, 10725. [Google Scholar] [CrossRef] [PubMed]
- Schwabe, J.; Perez-Burgos, M.; Herfurth, M.; Glatter, T.; Sogaard-Andersen, L. Evidence for a Widespread Third System for Bacterial Polysaccharide Export across the Outer Membrane Comprising a Composite OPX/beta-Barrel Translocon. mBio 2022, 13, e0203222. [Google Scholar] [CrossRef] [PubMed]
- Coplin, D.L.; Cook, D. Molecular genetics of extracellular polysaccharide biosynthesis in vascular phytopathogenic bacteria. Mol. Plant Microbe Interact. 1990, 3, 271–279. [Google Scholar] [CrossRef]
- Taylor, C.M.; Roberts, I.S. Capsular Polysaccharides and Their Role in Virulence. In Concepts in Bacterial Virulence; Russell, W., Herwald, H., Eds.; S. Karger AG: Basel, Switzerland, 2004. [Google Scholar]
- Pasquina, L.W.; Santa Maria, J.P.; Walker, S. Teichoic acid biosynthesis as an antibiotic target. Curr. Opin. Microbiol. 2013, 16, 531–537. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P., Jr.; Walker, S. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Kiriukhin, M.Y.; Neuhaus, F.C. D-alanylation of lipoteichoic acid: Role of the D-alanyl carrier protein in acylation. J. Bacteriol. 2001, 183, 2051–2058. [Google Scholar] [CrossRef]
- Von Aulock, C.D.; Marois, C.; Harel, M.J.; De La Cruz Domínguez-Punaro, M.; Fittipaldi, S.N.; Sekizaki, T.; Takamatsu, D. d-Alanylation of Lipoteichoic Acid. Infect. Immun. 2008, 76, 3587. [Google Scholar]
- Hankins, J.V.; Madsen, J.A.; Giles, D.K.; Brodbelt, J.S.; Trent, M.S. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in gram-positive and gram-negative bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 8722–8727. [Google Scholar] [CrossRef]
- Koprivnjak, T.; Peschel, A. Bacterial resistance mechanisms against host defense peptides. Cell. Mol. Life Sci. 2011, 68, 2243–2254. [Google Scholar] [CrossRef] [PubMed]
- Costechareyre, D.; Chich, J.F.; Strub, J.M.; Rahbe, Y.; Condemine, G. Transcriptome of Dickeya dadantii infecting Acyrthosiphon pisum reveals a strong defense against antimicrobial peptides. PLoS ONE 2013, 8, e54118. [Google Scholar] [CrossRef] [PubMed]
- Balleza, E.; Lopez-Bojorquez, L.N.; Martinez-Antonio, A.; Resendis-Antonio, O.; Lozada-Chavez, I.; Balderas-Martinez, Y.I.; Encarnacion, S.; Collado-Vides, J. Regulation by transcription factors in bacteria: Beyond description. FEMS Microbiol. Rev. 2009, 33, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Cases, I.; De Lorenzo, V.; Ouzounis, C.A. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 2003, 11, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Van Gijsegem, F.; Wlodarczyk, A.; Cornu, A.; Reverchon, S.; Hugouvieux-Cotte-Pattat, N. Analysis of the LacI family regulators of Erwinia chrysanthemi 3937, involvement in the bacterial phytopathogenicity. Mol. Plant-Microbe Interact. 2008, 21, 1471–1481. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Li, S.; Shao, F. Sweet Talk: Protein Glycosylation in Bacterial Interaction with the Host. Trends Microbiol. 2015, 23, 630–641. [Google Scholar] [CrossRef]
- Li, Y.; Powell, D.A.; Shaffer, S.A.; Rasko, D.A.; Pelletier, M.R.; Leszyk, J.D.; Scott, A.J.; Masoudi, A.; Goodlett, D.R.; Wang, X.; et al. LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 8716–8721. [Google Scholar] [CrossRef]
- Li, J.Y.; Wang, N. The gene encoding a glycosyltransferase is important for polysaccharide production and required for full virulence in Xanthomonas citri subsp. citri. BMC Microbiol. 2012, 12, 31. [Google Scholar] [CrossRef]
- Santaella, C.; Schue, M.; Berge, O.; Heulin, T.; Achouak, W. The exopolysaccharide of Rhizobium sp. YAS34 is not necessary for biofilm formation on Arabidopsis thaliana and Brassica napus roots but contributes to root colonization. Environ. Microbiol. 2008, 10, 2150–2163. [Google Scholar] [CrossRef]
- Chatterjee, A.K.; Thurn, K.K.; Feese, D.A. Tn5-Induced Mutations in the Enterobacterial Phytopathogen Erwinia chrysanthemi. Appl. Environ. Microbiol. 1983, 45, 644–650. [Google Scholar] [CrossRef]
- Coutinho, B.G.; Licastro, D.; Mendonca-Previato, L.; Camara, M.; Venturi, V. Plant-Influenced Gene Expression in the Rice Endophyte Burkholderia kururiensis M130. Mol. Plant Microbe Interact. 2015, 28, 10–21. [Google Scholar] [CrossRef]
- Heroven, A.K.; Nuss, A.M.; Dersch, P. RNA-based mechanisms of virulence control in Enterobacteriaceae. RNA Biol. 2017, 14, 471–487. [Google Scholar] [CrossRef]
- Van Opijnen, T.; Bodi, K.L.; Camilli, A. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microrganisms. Nat. Methods 2009, 6, 767. [Google Scholar] [CrossRef]
- Royet, K.; Parisot, N.; Rodrigue, A.; Gueguen, E.; Condemine, G. Identification of Tn-seq of Dickeya dadantii genes required for survival in chicory plants. Mol. Plant Pathool. 2019, 20, 287. [Google Scholar] [CrossRef]
- Robic, K.; Munier, E.; Effantin, G.; Lachat, J.; Naquin, D.; Gueguen, E.; Faure, D. Dissimilar gene reportoires of Dickeya solani involved in the colonization of lessions and roots of Solanum tuberosum. Front. Plant Sci. 2023, 14, 1154110. [Google Scholar] [CrossRef] [PubMed]
- Helmann, T.C.; Filiatrault, M.J.; Stodghill, P.V. Genome-wide identification of genes important for growth of Dickeya dadantii and Dickeya dianthicola in potato (Solanum tuberosum) tubers. Front. Microbiol. 2022, 13, 778927. [Google Scholar] [CrossRef] [PubMed]
- Czajkowski, R.; Kaczyńska, N.; Jafra, S.; Narajczyk, M.; Lojkowska, E. Temperature-responsive genetic loci in pectinolytic plant pathogenic Dickeya solani. Plant Pathol. 2016, 66, 584–594. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; Dejongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Khayi, S.; Blin, P.; Chong, T.M.; Chan, K.-G.; Faure, D. Complete genome anatomy of the emerging potato pathogen Dickeya solani type strain IPO 2222. Stand. Genomic Sci. 2016, 11, 2–6. [Google Scholar]
- Lisicka, W.; Fikowicz-Krosko, J.; Jafra, S.; Narajczyk, M.; Czaplewska, P.; Czajkowski, R. Oxygen Availability Influences Expression of Dickeya solani Genes Associated with Virulence in Potato (Solanum tuberosum L.) and Chicory (Cichorium intybus L.). Front. Plant Sci. 2018, 9, 374. [Google Scholar] [CrossRef] [PubMed]
- Kurowski, M.A.; Bujnicki, J.M. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003, 31, 3305–3307. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Koonin, E.V. Iterated profile searches with PSI-BLAST—A tool for discovery in protein databases. Trends Biochem. Sci. 1998, 23, 444–447. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Nykyri, J.; Mattinen, L.; Niemi, O.; Adhikari, S.; Koiv, V.; Somervuo, P.; Fang, X.; Auvinen, P.; Mae, A.; Palva, E.T.; et al. Role and regulation of the Flp/Tad pilus in the virulence of Pectobacterium atrosepticum SCRI1043 and Pectobacterium wasabiae SCC3193. PLoS ONE 2013, 8, e73718. [Google Scholar] [CrossRef] [PubMed]
- Roth, V. Doubling Time Computing. 2006. Available online: http://www.doubling-time.com/compute.php (accessed on 1 July 2023).
- Czajkowski, R.; De Boer, W.J.; Van Veen, J.A.; Van Der Wolf, J.M. Characterization of bacterial isolates from rotting potato tuber tissue showing antagonism to Dickeya sp. biovar 3 in vitro and in planta. Plant Pathol. 2012, 61, 169–182. [Google Scholar] [CrossRef]
- Dickey, R.S. Erwinia chrysanthemi: A comparative study of phenotypic properties of strains from several hosts and other Erwinia species. Phytopathology 1979, 69, 324–329. [Google Scholar] [CrossRef]
- Krzyzanowska, D.M.; Maciag, T.; Siwinska, J.; Krychowiak, M.; Jafra, S.; Czajkowski, R. Compatible Mixture of Bacterial Antagonists Developed to Protect Potato Tubers from Soft Rot Caused by Pectobacterium spp. and Dickeya spp. Plant Dis. 2019, 103, 1374–1382. [Google Scholar] [CrossRef] [PubMed]
- Perombelon, M.C.M.; Van Der Wolf, J.M. Methods for the detection and quantification of Erwinia carotovora subsp. atroseptica (Pectobacterium carotovorum subsp. atrosepticum) on potatoes: A laboratory manual. Scott. Crop Res. Inst. Annu. Rep. 2002, 10, 1–82. [Google Scholar]
- Czajkowski, R.; Van Der Wolf, J.M.; Krolicka, A.; Ozymko, Z.; Narajczyk, M.; Kaczynska, N.; Lojkowska, E. Salicylic acid can reduce infection symptoms caused by Dickeya solani in tissue culture grown potato (Solanum tuberosum L.) plants. Eur. J. Plant Pathol. 2014, 141, 545–558. [Google Scholar] [CrossRef][Green Version]
- Hommais, F.; Zghidi-Abouzid, O.; Oger-Desfeux, C.; Pineau-Chapelle, E.; Van Gijsegem, F.; Nasser, W.; Reverchon, S. lpxC and yafS are the most suitable internal controls to normalize real time RT-qPCR expression in the phytopathogenic bacteria Dickeya dadantii. PLoS ONE 2011, 6, e20269. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Krzyzanowska, D.M.; Supernat, A.; Maciag, T.; Matuszewska, M.; Jafra, S. Selection of reference genes for measuring the expression of aiiO in Ochrobactrum quorumnocens A44 using RT-qPCR. Sci. Rep. 2019, 9, 13129. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Tukey’s honestly significant difference (HSD) test. Encycl. Res. Des. 2010, 3, 1–5. [Google Scholar]
- Hammer, Ø.; Harper, D.a.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Student. The Probable Error of a Mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Shieh, G.; Jan, S.L. The effectiveness of randomized complete block design. Stat. Neerl. 2004, 58, 111–124. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czajkowski, R.; Krzyżanowska, D.M.; Sokolova, D.; Rąbalski, Ł.; Kosiński, M.; Jafra, S.; Królicka, A. Genetic Loci of Plant Pathogenic Dickeya solani IPO 2222 Expressed in Contact with Weed-Host Bittersweet Nightshade (Solanum dulcamara L.) Plants. Int. J. Mol. Sci. 2024, 25, 2794. https://doi.org/10.3390/ijms25052794
Czajkowski R, Krzyżanowska DM, Sokolova D, Rąbalski Ł, Kosiński M, Jafra S, Królicka A. Genetic Loci of Plant Pathogenic Dickeya solani IPO 2222 Expressed in Contact with Weed-Host Bittersweet Nightshade (Solanum dulcamara L.) Plants. International Journal of Molecular Sciences. 2024; 25(5):2794. https://doi.org/10.3390/ijms25052794
Chicago/Turabian StyleCzajkowski, Robert, Dorota M. Krzyżanowska, Daryna Sokolova, Łukasz Rąbalski, Maciej Kosiński, Sylwia Jafra, and Aleksandra Królicka. 2024. "Genetic Loci of Plant Pathogenic Dickeya solani IPO 2222 Expressed in Contact with Weed-Host Bittersweet Nightshade (Solanum dulcamara L.) Plants" International Journal of Molecular Sciences 25, no. 5: 2794. https://doi.org/10.3390/ijms25052794
APA StyleCzajkowski, R., Krzyżanowska, D. M., Sokolova, D., Rąbalski, Ł., Kosiński, M., Jafra, S., & Królicka, A. (2024). Genetic Loci of Plant Pathogenic Dickeya solani IPO 2222 Expressed in Contact with Weed-Host Bittersweet Nightshade (Solanum dulcamara L.) Plants. International Journal of Molecular Sciences, 25(5), 2794. https://doi.org/10.3390/ijms25052794







