MiR-290 Family Maintains Pluripotency and Self-Renewal by Regulating MAPK Signaling Pathway in Intermediate Pluripotent Stem Cells
Abstract
1. Introduction
2. Results
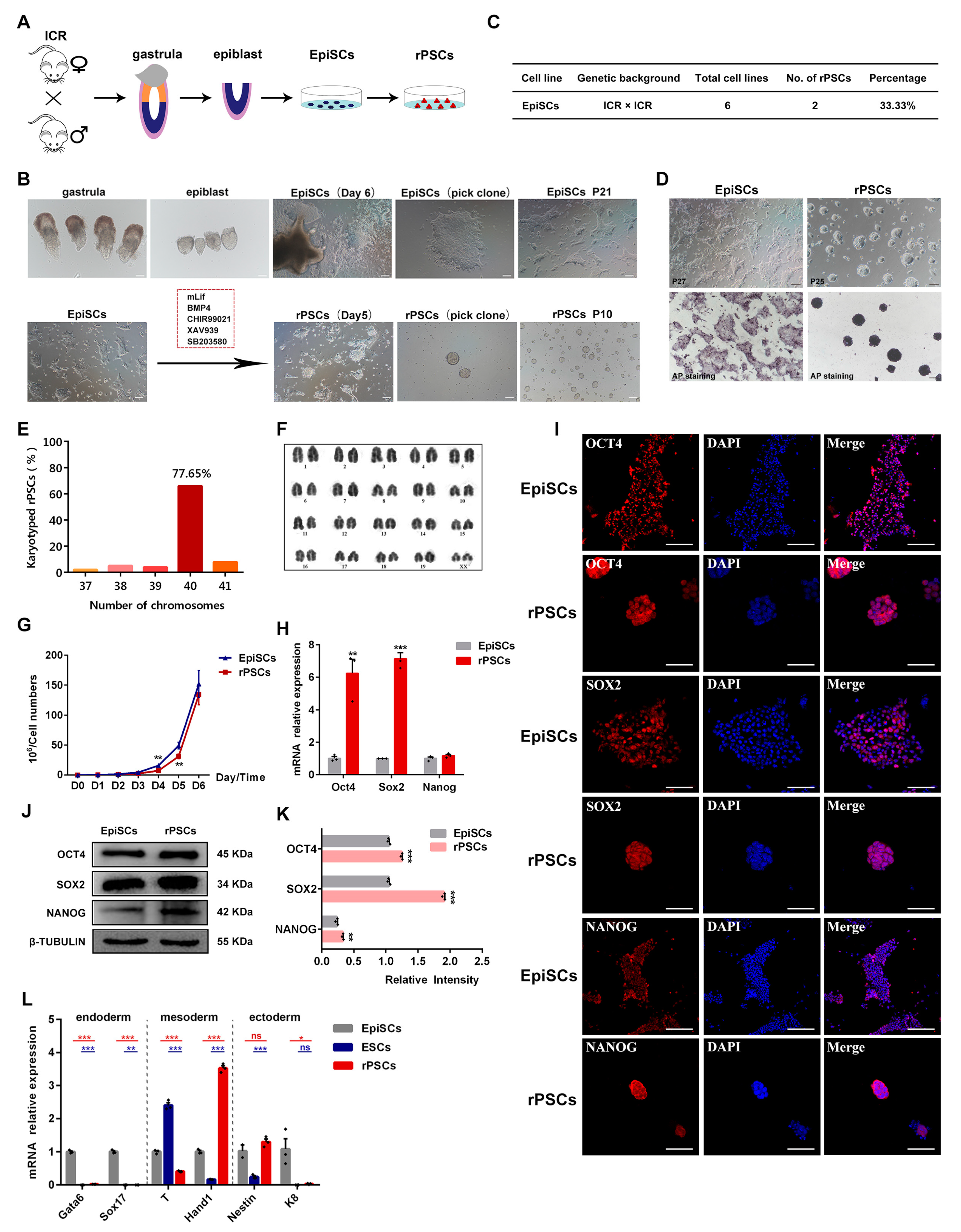
2.1. Conversion of EpiSCs to rPSCs
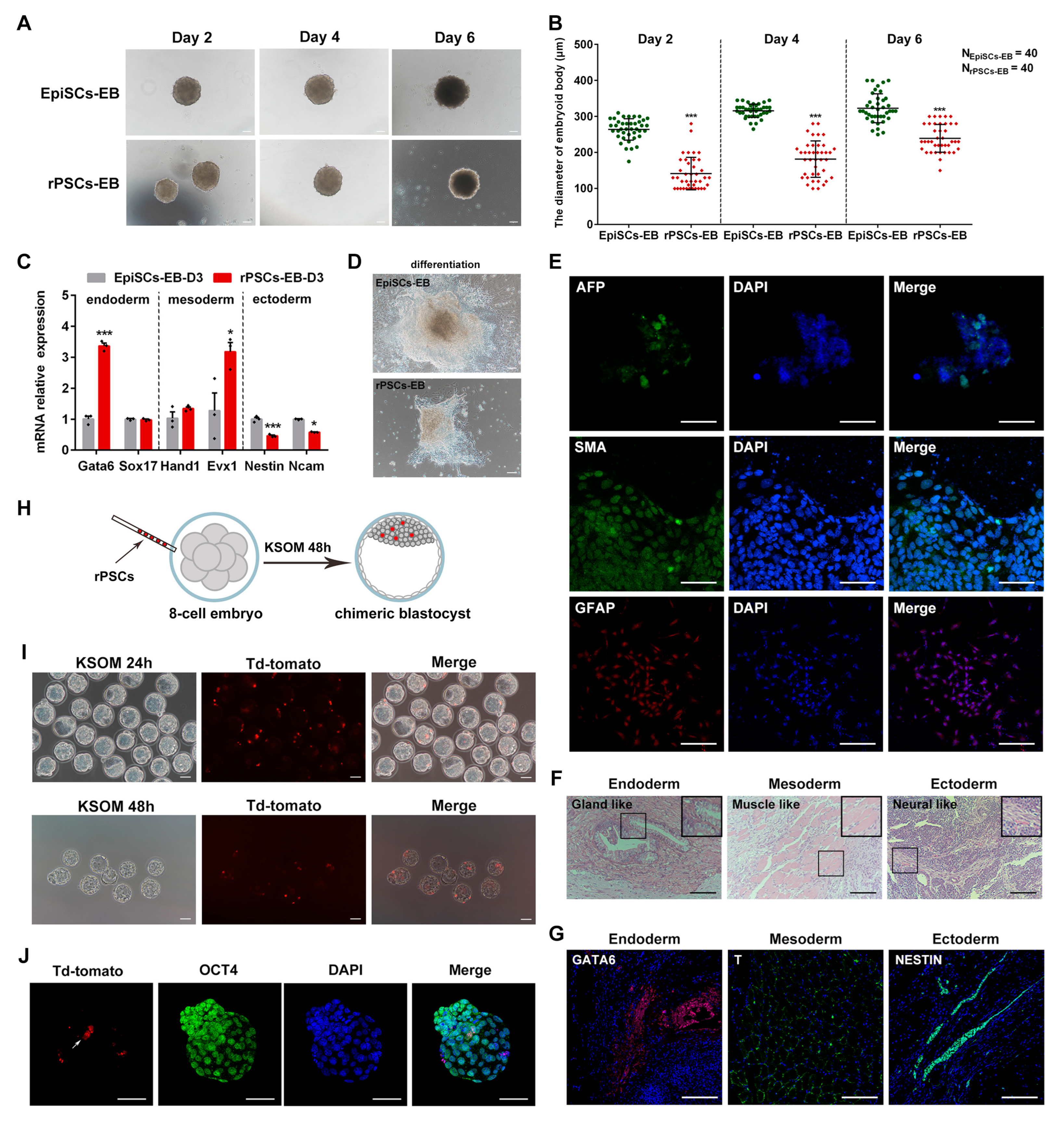
2.2. Developmental and Differentiated Potency of rPSCs
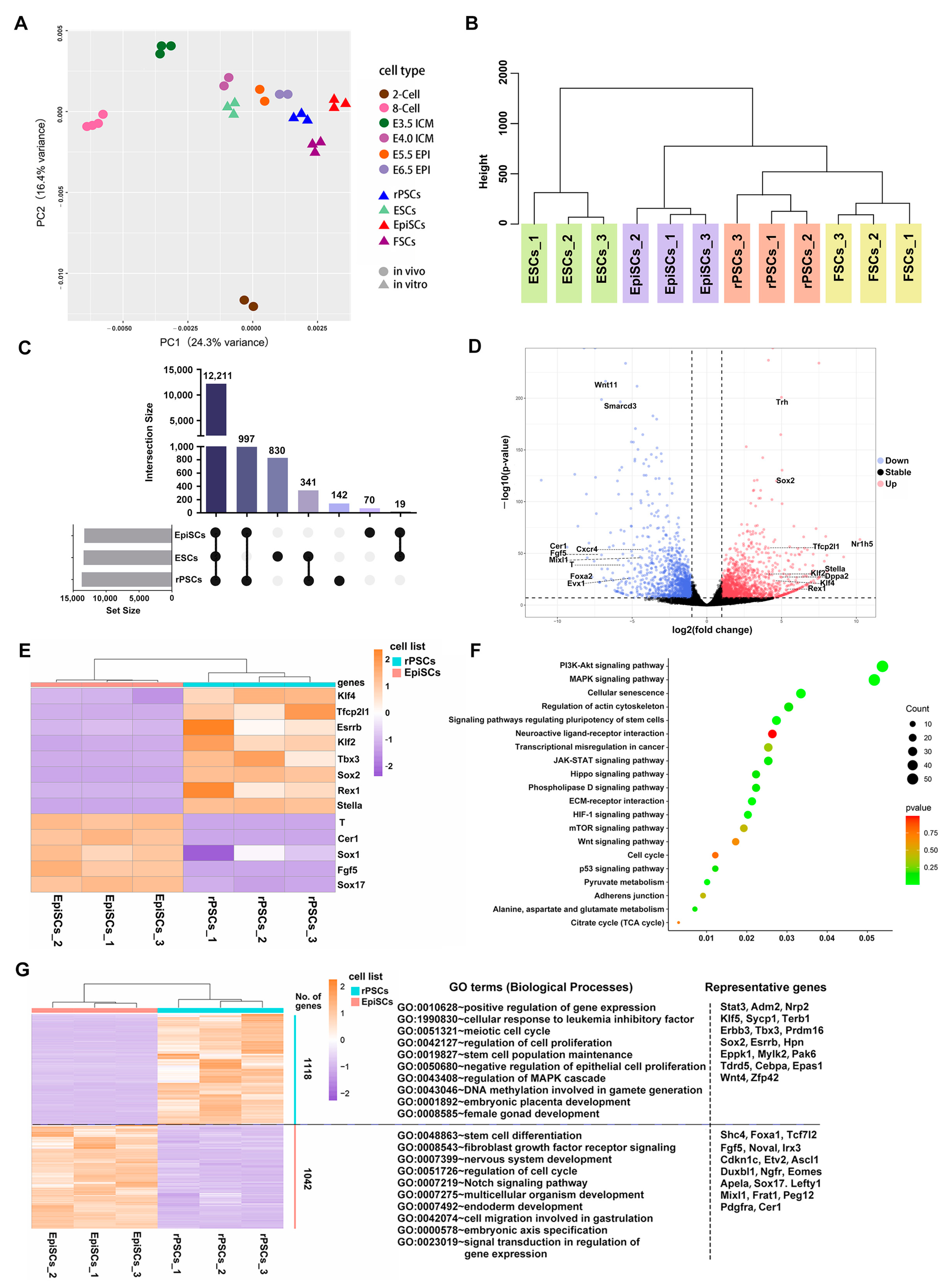
2.3. Molecular Features of rPSCs
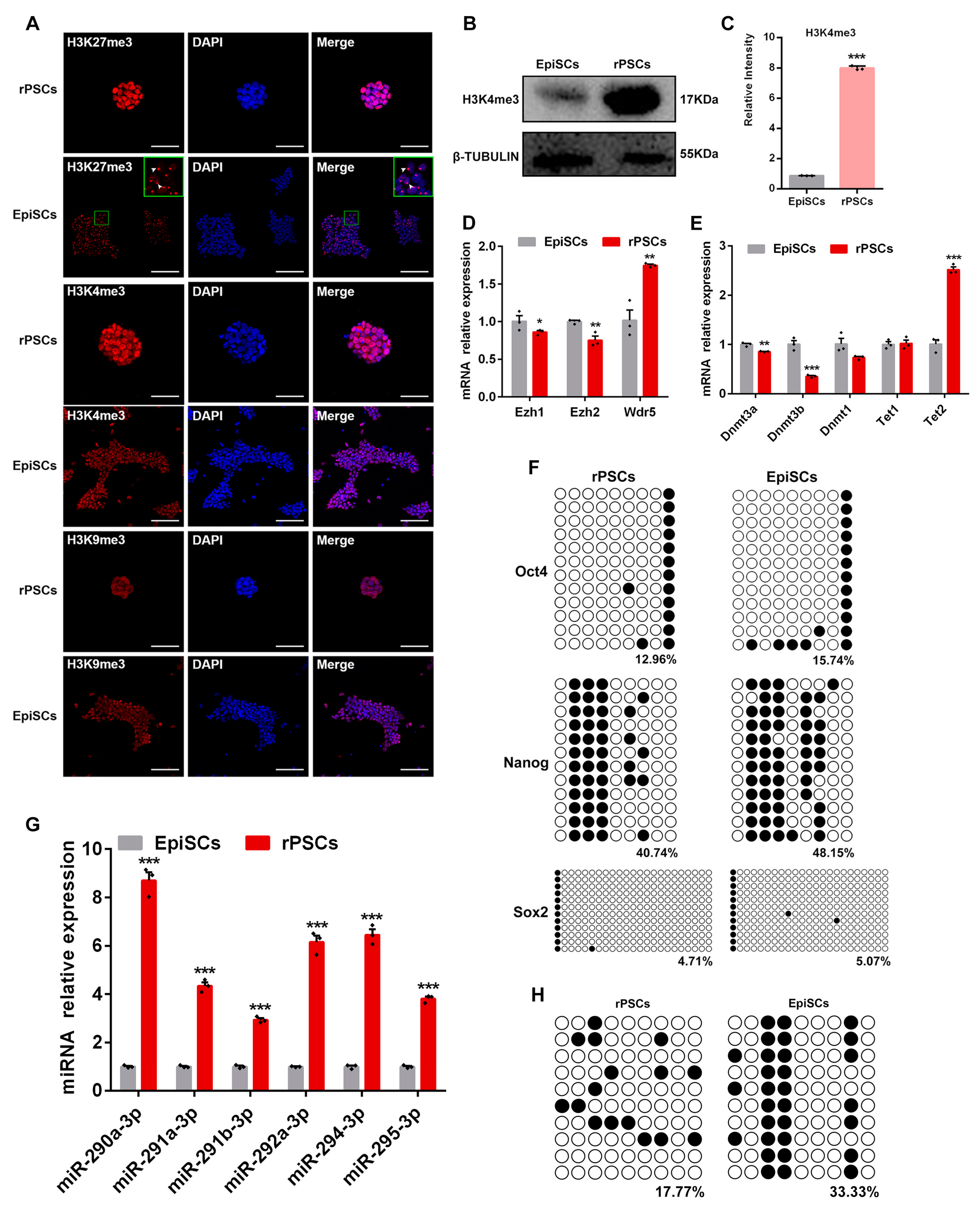
2.4. Epigenetic Changes in rPSCs
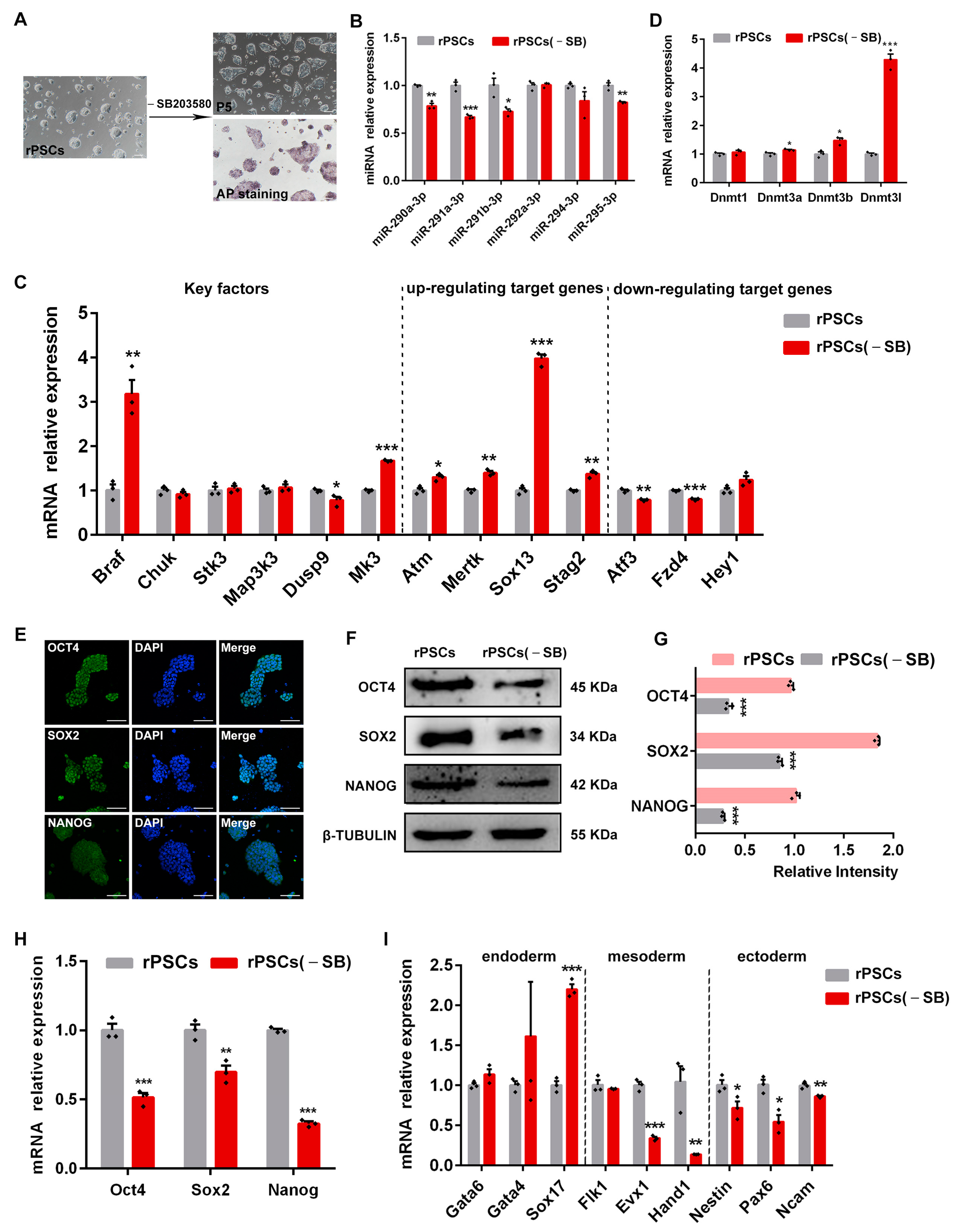
2.5. Removal of SB203580 Reduces the Pluripotency of rPSCs
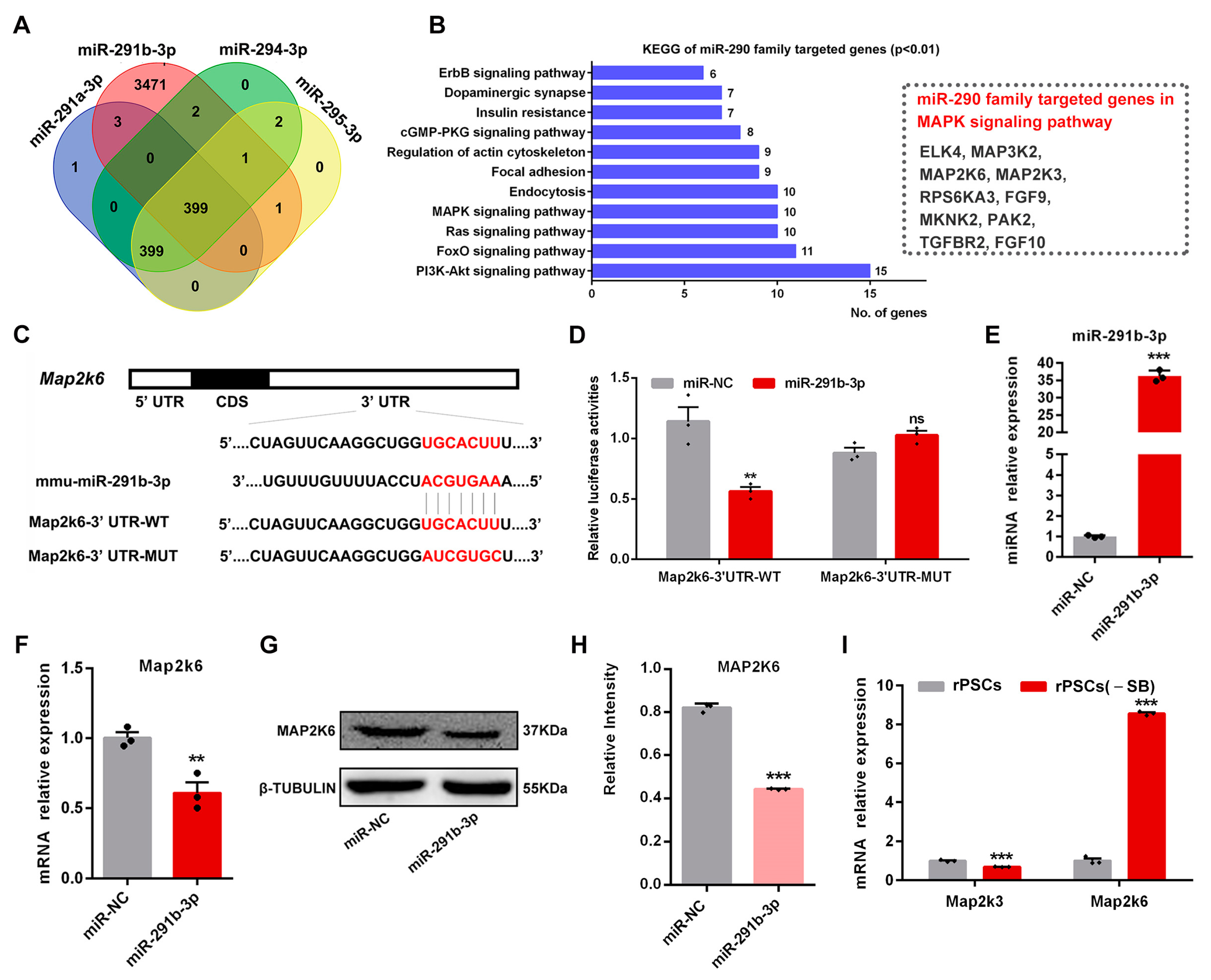
2.6. Map2k6 Is a Direct Target of miR-291b-3p
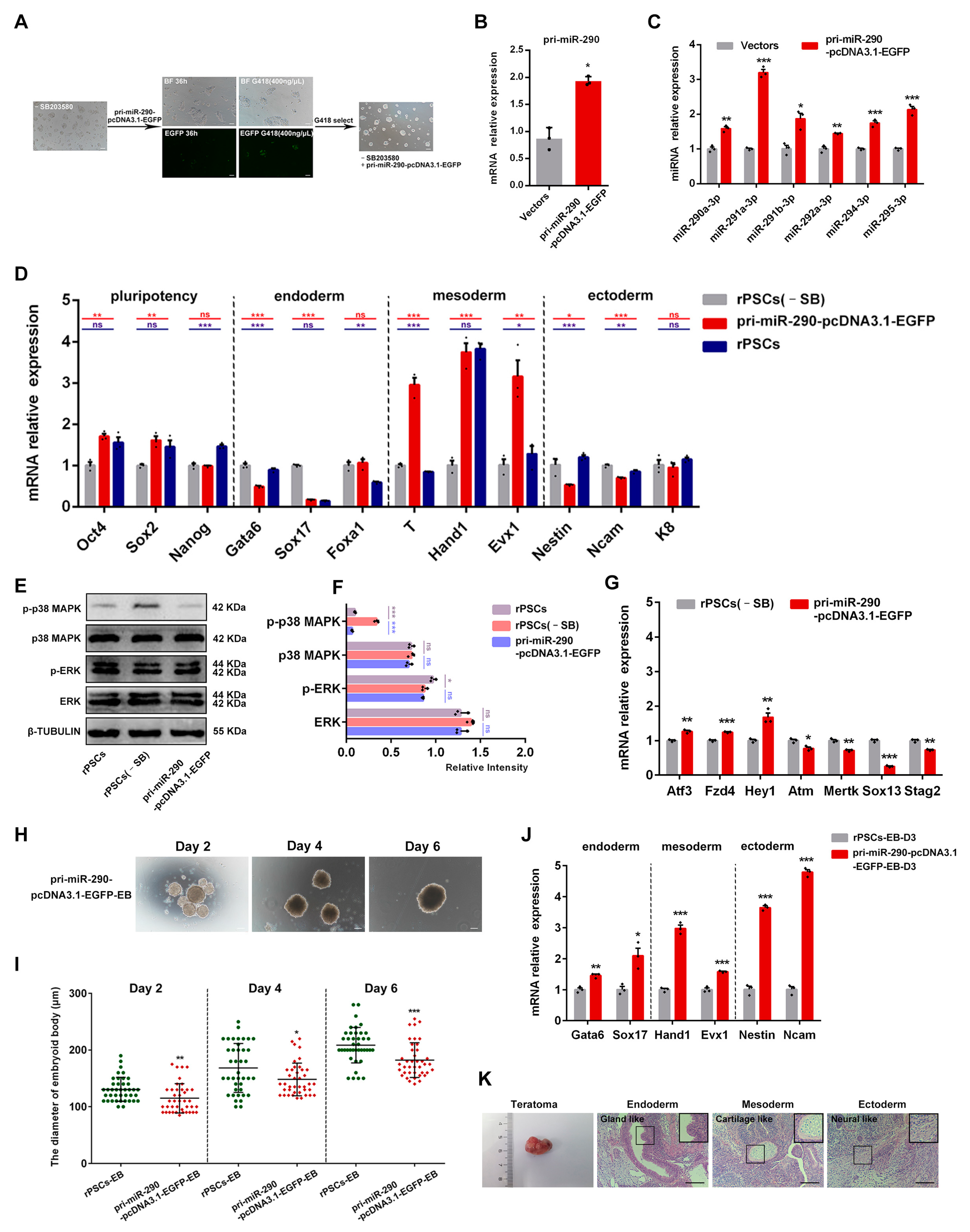
2.7. MiR-290 Family Can Replace MAPK Pathway Inhibitor SB203580 to Maintain the Pluripotency of rPSCs
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Derivation of EpiSCs
4.3. Conversion of EpiSCs to rPSCs
4.4. Culture of Mouse ESCs
4.5. AP Staining
4.6. Karyotype Analysis
4.7. Immunofluorescence
4.8. Total RNA, miRNA Extraction, and RT-qPCR
4.9. Western Blot
4.10. Generation of Teratoma
4.11. Formation of an Embryonic Body
4.12. Generation of Chimera
4.13. DNA Methylation Analysis
4.14. Transcriptome Analysis
4.15. Construction of Expression Plasmid Vectors
4.16. Dual-Iuciferase Reporter Assay
4.17. miR-290 Family Target Genes Analysis
4.18. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hackett, J.A.; Surani, M.A. Regulatory principles of pluripotency: From the ground state up. Cell Stem Cell 2014, 15, 416–430. [Google Scholar] [CrossRef]
- Weinberger, L.; Ayyash, M.; Novershtern, N.; Hanna, J.H. Dynamic stem cell states: Naive to primed pluripotency in rodents and humans. Nat. Rev. Mol. Cell Biol. 2016, 17, 155–169. [Google Scholar] [CrossRef] [PubMed]
- EVANS, M.J. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Brons, I.G.; Smithers, L.E.; Trotter, M.W.; Rugg-Gunn, P.; Sun, B.; Chuva de Sousa Lopes, S.M.; Howlett, S.K.; Clarkson, A.; Ahrlund-Richter, L.; Pedersen, R.A.; et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007, 448, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.; Smith, A. Naive and Primed Pluripotent States. Cell Stem Cell 2009, 4, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Burdon, T.; Chambers, I.; Smith, A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes. Dev. 1998, 12, 2048–2060. [Google Scholar] [CrossRef] [PubMed]
- Marks, H.; Kalkan, T.; Menafra, R.; Denissov, S.; Jones, K.; Hofemeister, H.; Nichols, J.; Kranz, A.; Stewart, A.F.; Smith, A.; et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell 2012, 149, 590–604. [Google Scholar] [CrossRef]
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448, 196–199. [Google Scholar] [CrossRef]
- Rossant, J. Stem cells and early lineage development. Cell 2008, 132, 527–531. [Google Scholar] [CrossRef]
- Schulz, E.G.; Meisig, J.; Nakamura, T.; Okamoto, I.; Sieber, A.; Picard, C.; Borensztein, M.; Saitou, M.; Blüthgen, N.; Heard, E. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell 2014, 14, 203–216. [Google Scholar] [CrossRef]
- Smith, A. Formative pluripotency: The executive phase in a developmental continuum. Development 2017, 144, 365–373. [Google Scholar] [CrossRef]
- Endoh, M.; Niwa, H. Stepwise pluripotency transitions in mouse stem cells. EMBO Rep. 2022, 23, e55010. [Google Scholar] [CrossRef]
- Kinoshita, M.; Smith, A. Pluripotency Deconstructed. Dev. Growth Differ. 2018, 60, 44–52. [Google Scholar] [CrossRef]
- Acampora, D.; Di Giovannantonio, L.G.; Simeone, A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development 2013, 140, 43–55. [Google Scholar] [CrossRef]
- Kinoshita, M.; Barber, M.; Mansfield, W.; Cui, Y.; Spindlow, D.; Stirparo, G.G.; Dietmann, S.; Nichols, J.; Smith, A. Capture of Mouse and Human Stem Cells with Features of Formative Pluripotency. Cell Stem Cell 2021, 28, 453–471.e458. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wei, Y.; Sun, H.X.; Mahdi, A.K.; Pinzon Arteaga, C.A.; Sakurai, M.; Schmitz, D.A.; Zheng, C.; Ballard, E.D.; Li, J.; et al. Derivation of Intermediate Pluripotent Stem Cells Amenable to Primordial Germ Cell Specification. Cell Stem Cell 2021, 28, 550–567.e512. [Google Scholar] [CrossRef] [PubMed]
- Young, R.A. Control of the embryonic stem cell state. Cell 2011, 144, 940–954. [Google Scholar] [CrossRef] [PubMed]
- Macarthur, B.D.; Ma'ayan, A.; Lemischka, I.R. Systems biology of stem cell fate and cellular reprogramming. Nat. Rev. Mol. Cell Biol. 2009, 10, 672–681. [Google Scholar] [CrossRef]
- Chen, X.; Vega, V.B.; Ng, H.H. Transcriptional regulatory networks in embryonic stem cells. Cold Spring Harb. Symp. Quant. Biol. 2008, 73, 203–209. [Google Scholar] [CrossRef][Green Version]
- Ng, R.K.; Dean, W.; Dawson, C.; Lucifero, D.; Madeja, Z.; Reik, W.; Hemberger, M. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat. Cell Biol. 2008, 10, 1280–1290. [Google Scholar] [CrossRef]
- Tsumura, A.; Hayakawa, T.; Kumaki, Y.; Takebayashi, S.; Sakaue, M.; Matsuoka, C.; Shimotohno, K.; Ishikawa, F.; Li, E.; Ueda, H.R.; et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes. Cells 2006, 11, 805–814. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhuang, L.; Lin, C.P. Roles of MicroRNAs in Establishing and Modulating Stem Cell Potential. Int. J. Mol. Sci. 2019, 20, 3643. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef]
- Neagu, A.; van Genderen, E.; Escudero, I.; Verwegen, L.; Kurek, D.; Lehmann, J.; Stel, J.; Dirks, R.A.M.; van Mierlo, G.; Maas, A.; et al. In vitro capture and characterization of embryonic rosette-stage pluripotency between naive and primed states. Nat. Cell Biol. 2020, 22, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.L.; Nichols, J.; Chambers, I.; Smith, A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 2003, 115, 281–292. [Google Scholar] [CrossRef]
- Yu, S.; Zhou, C.; Cao, S.; He, J.; Cai, B.; Wu, K.; Qin, Y.; Huang, X.; Xiao, L.; Ye, J.; et al. BMP4 resets mouse epiblast stem cells to naive pluripotency through ZBTB7A/B-mediated chromatin remodelling. Nat. Cell Biol. 2020, 22, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Rana, T.M. A kinase inhibitor screen identifies small-molecule enhancers of reprogramming and iPS cell generation. Nat. Commun. 2012, 3, 1085. [Google Scholar] [CrossRef]
- Wang, W.; Li, N.; Li, X.; Tran, M.K.; Han, X.; Chen, J. Tankyrase Inhibitors Target YAP by Stabilizing Angiomotin Family Proteins. Cell Rep. 2015, 13, 524–532. [Google Scholar] [CrossRef]
- Boheler, K.R.; Czyz, J.; Tweedie, D.; Yang, H.T.; Anisimov, S.V.; Wobus, A.M. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ. Res. 2002, 91, 189–201. [Google Scholar] [CrossRef]
- Higuchi, C.; Yamamoto, M.; Shin, S.W.; Miyamoto, K.; Matsumoto, K. Perturbation of maternal PIASy abundance disrupts zygotic genome activation and embryonic development via SUMOylation pathway. Biol. Open 2019, 8, bio048652. [Google Scholar] [CrossRef]
- Wu, J.; Huang, B.; Chen, H.; Yin, Q.; Liu, Y.; Xiang, Y.; Zhang, B.; Liu, B.; Wang, Q.; Xia, W.; et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016, 534, 652–657. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiang, Y.; Yin, Q.; Du, Z.; Peng, X.; Wang, Q.; Fidalgo, M.; Xia, W.; Li, Y.; Zhao, Z.A.; et al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 2018, 50, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Takagi, N.; Sugawara, O.; Sasaki, M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma 1982, 85, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Chuva de Sousa Lopes, S.M.; Hayashi, K.; Shovlin, T.C.; Mifsud, W.; Surani, M.A.; McLaren, A. X chromosome activity in mouse XX primordial germ cells. PLoS Genet. 2008, 4, e30. [Google Scholar] [CrossRef] [PubMed]
- Plath, K.; Fang, J.; Mlynarczyk-Evans, S.K.; Cao, R.; Worringer, K.A.; Wang, H.; de la Cruz, C.C.; Otte, A.P.; Panning, B.; Zhang, Y. Role of histone H3 lysine 27 methylation in X inactivation. Science 2003, 300, 131–135. [Google Scholar] [CrossRef]
- Thorvaldsen, J.L.; Verona, R.I.; Bartolomei, M.S. X-tra! X-tra! News from the mouse X chromosome. Dev. Biol. 2006, 298, 344–353. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pasque, V.; Karnik, R.; Chronis, C.; Petrella, P.; Langerman, J.; Bonora, G.; Song, J.; Vanheer, L.; Sadhu Dimashkie, A.; Meissner, A.; et al. X Chromosome Dosage Influences DNA Methylation Dynamics during Reprogramming to Mouse iPSCs. Stem Cell Reports 2018, 10, 1537–1550. [Google Scholar] [CrossRef]
- Cirio, M.C.; Ratnam, S.; Ding, F.; Reinhart, B.; Navara, C.; Chaillet, J.R. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC Dev. Biol. 2008, 8, 9. [Google Scholar] [CrossRef]
- Bourc'his, D.; Xu, G.L.; Lin, C.S.; Bollman, B.; Bestor, T.H. Dnmt3L and the establishment of maternal genomic imprints. Science 2001, 294, 2536–2539. [Google Scholar] [CrossRef]
- Koh, K.P.; Yabuuchi, A.; Rao, S.; Huang, Y.; Cunniff, K.; Nardone, J.; Laiho, A.; Tahiliani, M.; Sommer, C.A.; Mostoslavsky, G.; et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011, 8, 200–213. [Google Scholar] [CrossRef]
- Wang, Y.; Medvid, R.; Melton, C.; Jaenisch, R.; Blelloch, R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007, 39, 380–385. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.K.; Young, A.G.; Bhutkar, A.; Zheng, G.X.; Bosson, A.D.; Nielsen, C.B.; Sharp, P.A. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat. Struct. Mol. Biol. 2011, 18, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.; Ravi, A.; Calabrese, J.M.; Medeiros, L.A.; Kirak, O.; Dennis, L.M.; Jaenisch, R.; Burge, C.B.; Sharp, P.A. A latent pro-survival function for the mir-290-295 cluster in mouse embryonic stem cells. PLoS Genet. 2011, 7, e1002054. [Google Scholar] [CrossRef]
- Ciaudo, C.; Servant, N.; Cognat, V.; Sarazin, A.; Kieffer, E.; Viville, S.; Colot, V.; Barillot, E.; Heard, E.; Voinnet, O. Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet. 2009, 5, e1000620. [Google Scholar] [CrossRef] [PubMed]
- Houbaviy, H.B.; Murray, M.F.; Sharp, P.A. Embryonic stem cell-specific MicroRNAs. Dev. Cell 2003, 5, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Spruce, T.; Pernaute, B.; Di-Gregorio, A.; Cobb, B.S.; Merkenschlager, M.; Manzanares, M.; Rodriguez, T.A. An early developmental role for miRNAs in the maintenance of extraembryonic stem cells in the mouse embryo. Dev. Cell 2010, 19, 207–219. [Google Scholar] [CrossRef]
- Stelzer, Y.; Shivalila, C.S.; Soldner, F.; Markoulaki, S.; Jaenisch, R. Tracing dynamic changes of DNA methylation at single-cell resolution. Cell 2015, 163, 218–229. [Google Scholar] [CrossRef]
- Yang, J.; Ryan, D.J.; Wang, W.; Tsang, C.H.; Lan, G.; Masaki, H.; Gao, X.; Antunes, L.; Yu, Y.; Zhu, Z. Establishment of mouse expanded potential stem cells. Nature 2017, 550, 393. [Google Scholar] [CrossRef]
- Leitch, H.G.; McEwen, K.R.; Turp, A.; Encheva, V.; Carroll, T.; Grabole, N.; Mansfield, W.; Nashun, B.; Knezovich, J.G.; Smith, A.; et al. Naive pluripotency is associated with global DNA hypomethylation. Nat. Struct. Mol. Biol. 2013, 20, 311–316. [Google Scholar] [CrossRef]
- Habibi, E.; Brinkman, A.B.; Arand, J.; Kroeze, L.I.; Kerstens, H.H.; Matarese, F.; Lepikhov, K.; Gut, M.; Brun-Heath, I.; Hubner, N.C.; et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 2013, 13, 360–369. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Ai, W.B.; Wan, L.Y.; Tan, X.; Wu, J.F. The miR-290-295 cluster as multi-faceted players in mouse embryonic stem cells. Cell Biosci. 2017, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.L.; Chang, D.C.; Lin, C.H.; Ying, S.Y.; Leu, D.; Wu, D.T. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res. 2011, 39, 1054–1065. [Google Scholar] [CrossRef] [PubMed]
- Houbaviy, H.B.; Dennis, L.; Jaenisch, R.; Sharp, P.A. Characterization of a highly variable eutherian microRNA gene. RNA 2005, 11, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Chen, W.; Kang, J. The functions of microRNAs and long non-coding RNAs in embryonic and induced pluripotent stem cells. Genom. Proteom. Bioinform. 2013, 11, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Li, M.A.; He, L. microRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. Bioessays 2012, 34, 670–680. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.; Smith, A. Pluripotency in the embryo and in culture. Cold Spring Harb. Perspect. Biol. 2012, 4, a008128. [Google Scholar] [CrossRef]
- Buecker, C.; Srinivasan, R.; Wu, Z.; Calo, E.; Acampora, D.; Faial, T.; Simeone, A.; Tan, M.; Swigut, T.; Wysocka, J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell 2014, 14, 838–853. [Google Scholar] [CrossRef]
- Stuart, H.T.; Stirparo, G.G.; Lohoff, T.; Bates, L.E.; Kinoshita, M.; Lim, C.Y.; Sousa, E.J.; Maskalenka, K.; Radzisheuskaya, A.; Malcolm, A.A.; et al. Distinct Molecular Trajectories Converge to Induce Naive Pluripotency. Cell Stem Cell 2019, 25, 388–406.e388. [Google Scholar] [CrossRef]
- Adachi, K.; Kopp, W.; Wu, G.; Heising, S.; Greber, B.; Stehling, M.; Araúzo-Bravo, M.J.; Boerno, S.T.; Timmermann, B.; Vingron, M.; et al. Esrrb Unlocks Silenced Enhancers for Reprogramming to Naive Pluripotency. Cell Stem Cell 2018, 23, 266–275.e266. [Google Scholar] [CrossRef]
- Okashita, N.; Suwa, Y.; Nishimura, O.; Sakashita, N.; Kadota, M.; Nagamatsu, G.; Kawaguchi, M.; Kashida, H.; Nakajima, A.; Tachibana, M.; et al. PRDM14 Drives OCT3/4 Recruitment via Active Demethylation in the Transition from Primed to Naive Pluripotency. Stem Cell Reports 2016, 7, 1072–1086. [Google Scholar] [CrossRef] [PubMed]
- Dunn, S.J.; Li, M.A.; Carbognin, E.; Smith, A.; Martello, G. A common molecular logic determines embryonic stem cell self-renewal and reprogramming. EMBO J. 2019, 38, e100003. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yamashita, J.K. AMPK activators contribute to maintain naïve pluripotency in mouse embryonic stem cells. Biochem. Biophys. Res. Commun. 2019, 509, 24–31. [Google Scholar] [CrossRef]
- Chung, Y.; Klimanskaya, I.; Becker, S.; Marh, J.; Lu, S.J.; Johnson, J.; Meisner, L.; Lanza, R. Embryonic and extraembryonic stem cell lines derived from single mouse blastomeres. Nature 2006, 439, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, M.; Yamamoto, T.; Tanoue, T.; Yuasa, Y.; Chisaka, O.; Nishida, E. Requirement of the MAP kinase signaling pathways for mouse preimplantation development. Development 2005, 132, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Liu, K.; Zhao, Y.; Li, H.; Zhu, D.; Du, Y.; Xiang, C.; Li, X.; Liu, H.; Miao, Z.; et al. Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell Stem Cell 2014, 15, 488–497. [Google Scholar] [CrossRef]
- Chen, F.; Shi, D.; Zou, L.; Yang, X.; Qiao, S.; Zhang, R.; Yang, S.; Deng, Y. Two Small Molecule Inhibitors Promote Reprogramming of Guangxi Bama Mini-Pig Mesenchymal Stem Cells Into Naive-Like State Induced Pluripotent Stem Cells. Cell Reprogram 2021, 23, 158–167. [Google Scholar] [CrossRef]
- Tan, B.S.; Kwek, J.; Wong, C.K.; Saner, N.J.; Yap, C.; Felquer, F.; Morris, M.B.; Gardner, D.K.; Rathjen, P.D.; Rathjen, J. Src Family Kinases and p38 Mitogen-Activated Protein Kinases Regulate Pluripotent Cell Differentiation in Culture. PLoS ONE 2016, 11, e0163244. [Google Scholar] [CrossRef] [PubMed]
- Yap, C.; Goh, H.N.; Familari, M.; Rathjen, P.D.; Rathjen, J. The formation of proximal and distal definitive endoderm populations in culture requires p38 MAPK activity. J. Cell Sci. 2014, 127, 2204–2216. [Google Scholar]
- Binétruy, B.; Heasley, L.; Bost, F.; Caron, L.; Aouadi, M. Concise review: Regulation of embryonic stem cell lineage commitment by mitogen-activated protein kinases. Stem Cells 2007, 25, 1090–1095. [Google Scholar] [CrossRef]
- Trouillas, M.; Saucourt, C.; Duval, D.; Gauthereau, X.; Thibault, C.; Dembele, D.; Feraud, O.; Menager, J.; Rallu, M.; Pradier, L.; et al. Bcl2, a transcriptional target of p38alpha, is critical for neuronal commitment of mouse embryonic stem cells. Cell Death Differ. 2008, 15, 1450–1459. [Google Scholar] [CrossRef]
- Shirane, K.; Kurimoto, K.; Yabuta, Y.; Yamaji, M.; Satoh, J.; Ito, S.; Watanabe, A.; Hayashi, K.; Saitou, M.; Sasaki, H. Global Landscape and Regulatory Principles of DNA Methylation Reprogramming for Germ Cell Specification by Mouse Pluripotent Stem Cells. Dev. Cell 2016, 39, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Marson, A.; Levine, S.S.; Cole, M.F.; Frampton, G.M.; Brambrink, T.; Johnstone, S.; Guenther, M.G.; Johnston, W.K.; Wernig, M.; Newman, J.; et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 2008, 134, 521–533. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Y.; Mu, Q.; Tian, J.; Yu, H. MiR-290 family maintains developmental potential by targeting p21 in mouse preimplantation embryos‡. Biol. Reprod. 2022, 106, 425–440. [Google Scholar] [CrossRef]
- Jouneau, A.; Ciaudo, C.; Sismeiro, O.; Brochard, V.; Jouneau, L.; Vandormael-Pournin, S.; Coppee, J.Y.; Zhou, Q.; Heard, E.; Antoniewski, C.; et al. Naive and primed murine pluripotent stem cells have distinct miRNA expression profiles. RNA 2012, 18, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Orlando, D.A.; Hnisz, D.; Abraham, B.J.; Lin, C.Y.; Kagey, M.H.; Rahl, P.B.; Lee, T.I.; Young, R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013, 153, 307–319. [Google Scholar] [CrossRef]
- Gu, K.L.; Zhang, Q.; Yan, Y.; Li, T.T.; Duan, F.F.; Hao, J.; Wang, X.W.; Shi, M.; Wu, D.R.; Guo, W.T.; et al. Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res. 2016, 26, 350–366. [Google Scholar] [CrossRef]
- Judson, R.L.; Babiarz, J.E.; Venere, M.; Blelloch, R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 2009, 27, 459–461. [Google Scholar] [CrossRef]
- Melton, C.; Judson, R.L.; Blelloch, R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 2010, 463, 621–626. [Google Scholar] [CrossRef]
- Rosa, A.; Papaioannou, M.D.; Krzyspiak, J.E.; Brivanlou, A.H. miR-373 is regulated by TGFβ signaling and promotes mesendoderm differentiation in human Embryonic Stem Cells. Dev. Biol. 2014, 391, 81–88. [Google Scholar] [CrossRef]
- Ishikura, Y.; Ohta, H.; Sato, T.; Murase, Y.; Yabuta, Y.; Kojima, Y.; Yamashiro, C.; Nakamura, T.; Yamamoto, T.; Ogawa, T.; et al. In vitro reconstitution of the whole male germ-cell development from mouse pluripotent stem cells. Cell Stem Cell 2021, 28, 2167–2179.e2169. [Google Scholar] [CrossRef] [PubMed]
- Tarazi, S.; Aguilera-Castrejon, A.; Joubran, C.; Ghanem, N.; Ashouokhi, S.; Roncato, F.; Wildschutz, E.; Haddad, M.; Oldak, B.; Gomez-Cesar, E.; et al. Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs. Cell 2022, 185, 3290–3306.e3225. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Li, X.; Ma, X.; Du, Q.; Wang, J.; Yu, H. MiR-290 Family Maintains Pluripotency and Self-Renewal by Regulating MAPK Signaling Pathway in Intermediate Pluripotent Stem Cells. Int. J. Mol. Sci. 2024, 25, 2681. https://doi.org/10.3390/ijms25052681
Liu Y, Li X, Ma X, Du Q, Wang J, Yu H. MiR-290 Family Maintains Pluripotency and Self-Renewal by Regulating MAPK Signaling Pathway in Intermediate Pluripotent Stem Cells. International Journal of Molecular Sciences. 2024; 25(5):2681. https://doi.org/10.3390/ijms25052681
Chicago/Turabian StyleLiu, Yueshi, Xiangnan Li, Xiaozhuang Ma, Qiankun Du, Jiemin Wang, and Haiquan Yu. 2024. "MiR-290 Family Maintains Pluripotency and Self-Renewal by Regulating MAPK Signaling Pathway in Intermediate Pluripotent Stem Cells" International Journal of Molecular Sciences 25, no. 5: 2681. https://doi.org/10.3390/ijms25052681
APA StyleLiu, Y., Li, X., Ma, X., Du, Q., Wang, J., & Yu, H. (2024). MiR-290 Family Maintains Pluripotency and Self-Renewal by Regulating MAPK Signaling Pathway in Intermediate Pluripotent Stem Cells. International Journal of Molecular Sciences, 25(5), 2681. https://doi.org/10.3390/ijms25052681




