Investigation of the General Molecular Mechanisms of Gallic Acid via Analyses of Its Transcriptome Profile
Abstract
1. Introduction
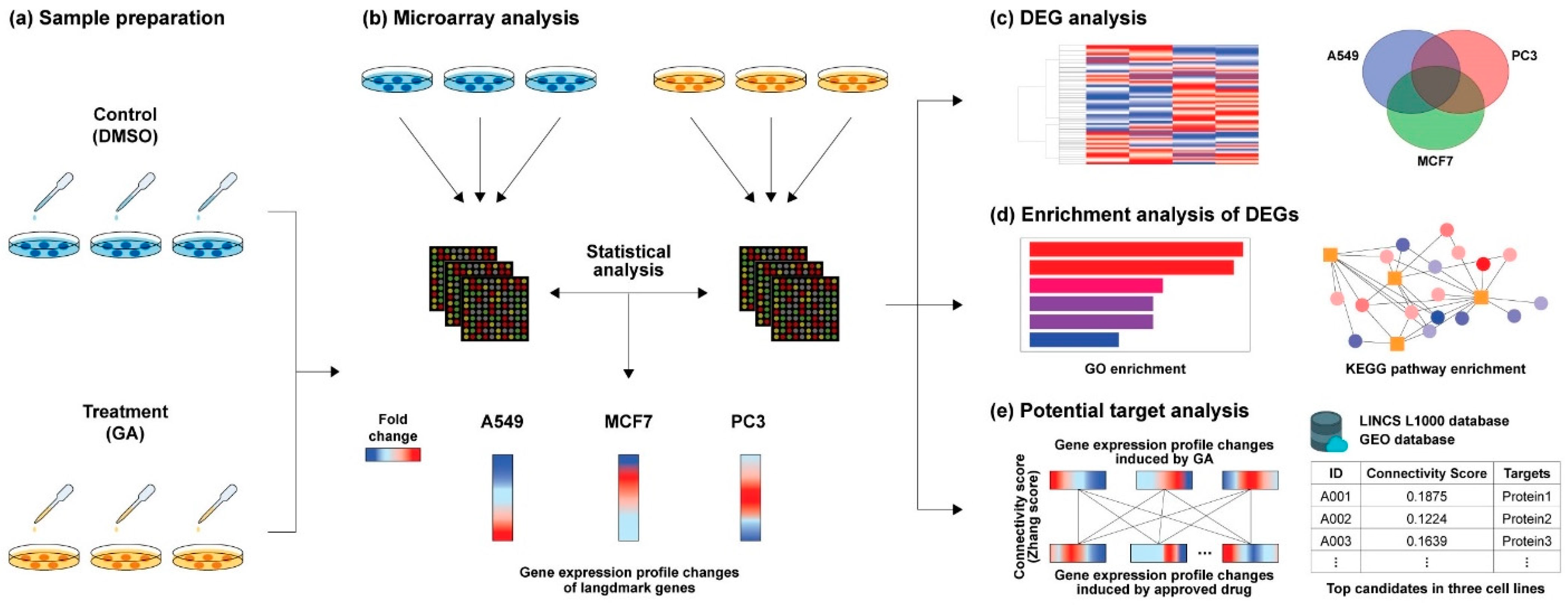
2. Results
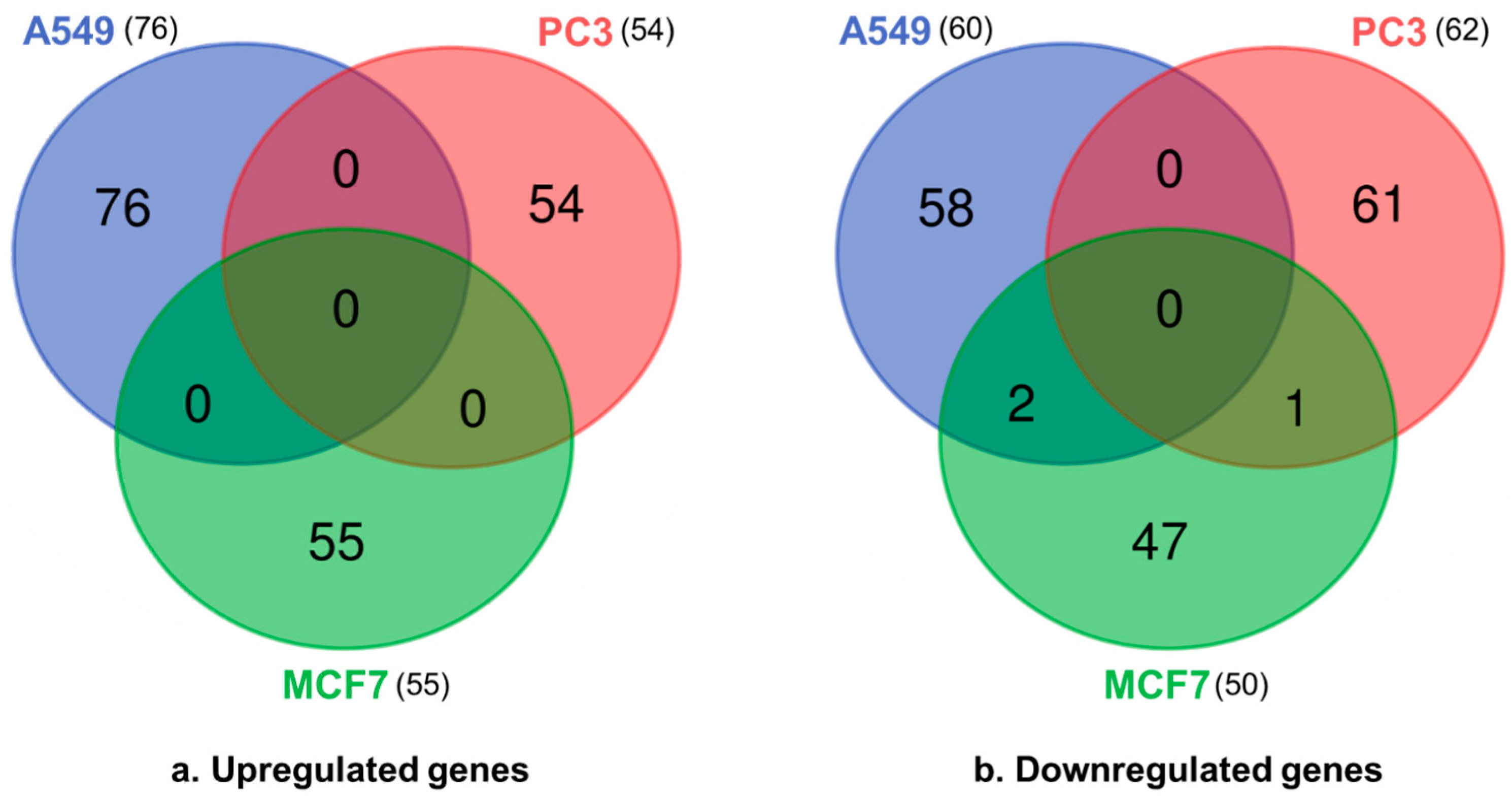
2.1. Data Analysis of GA-Induced GEPs
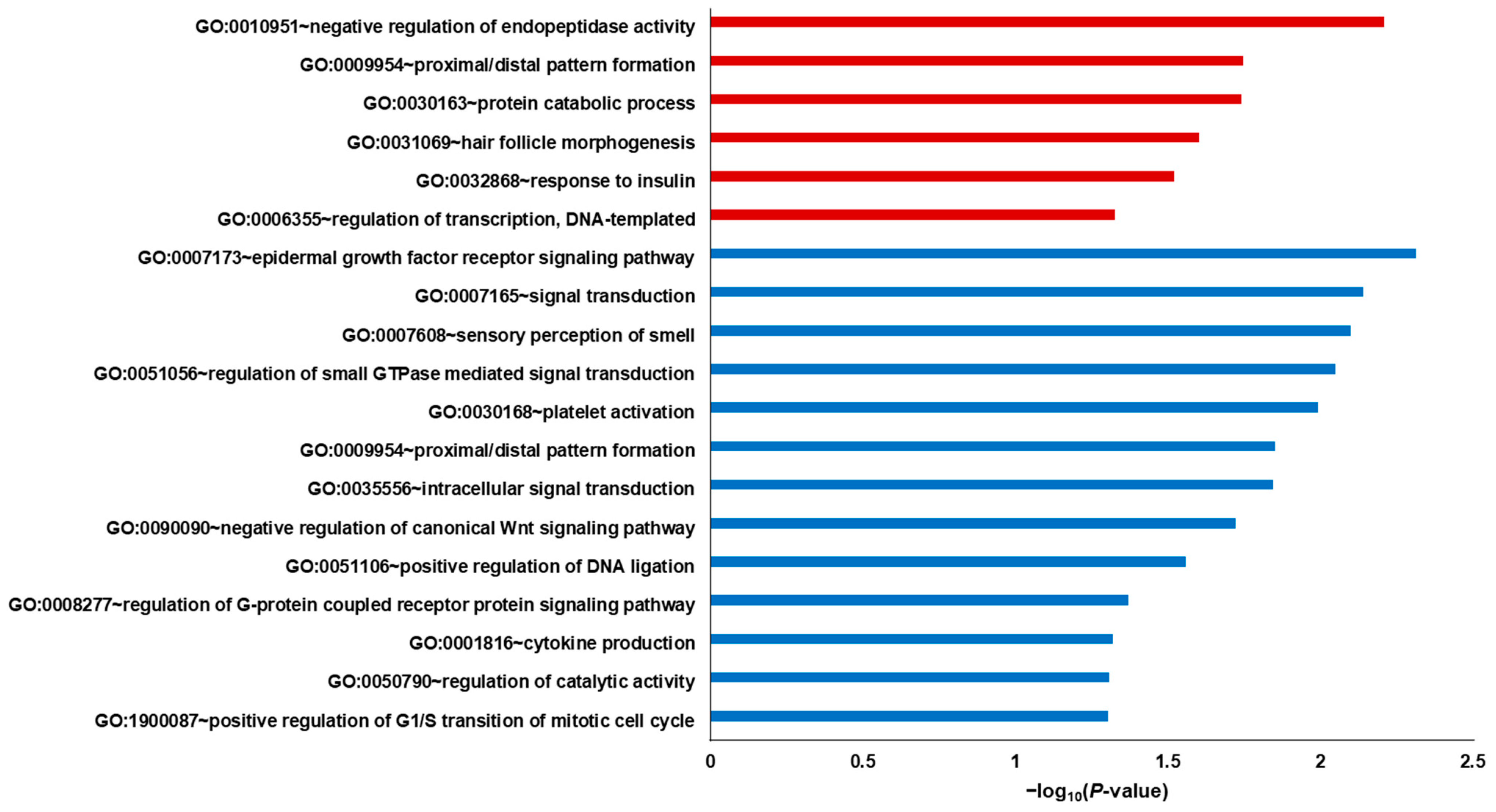
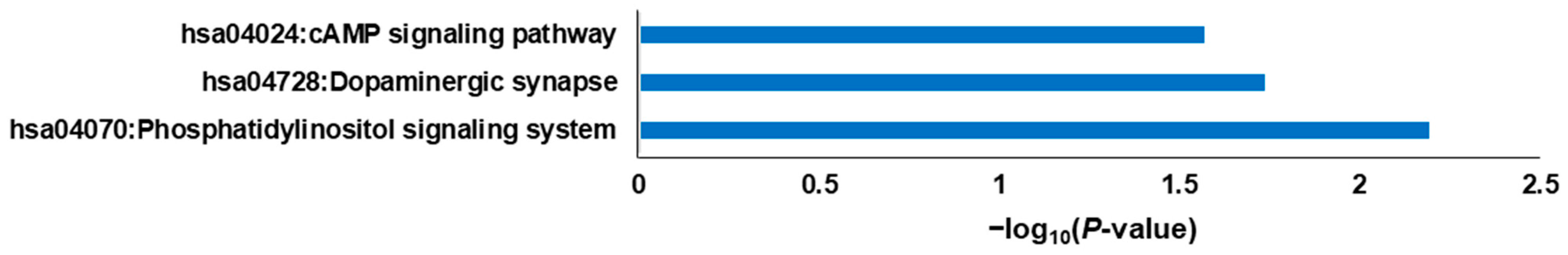
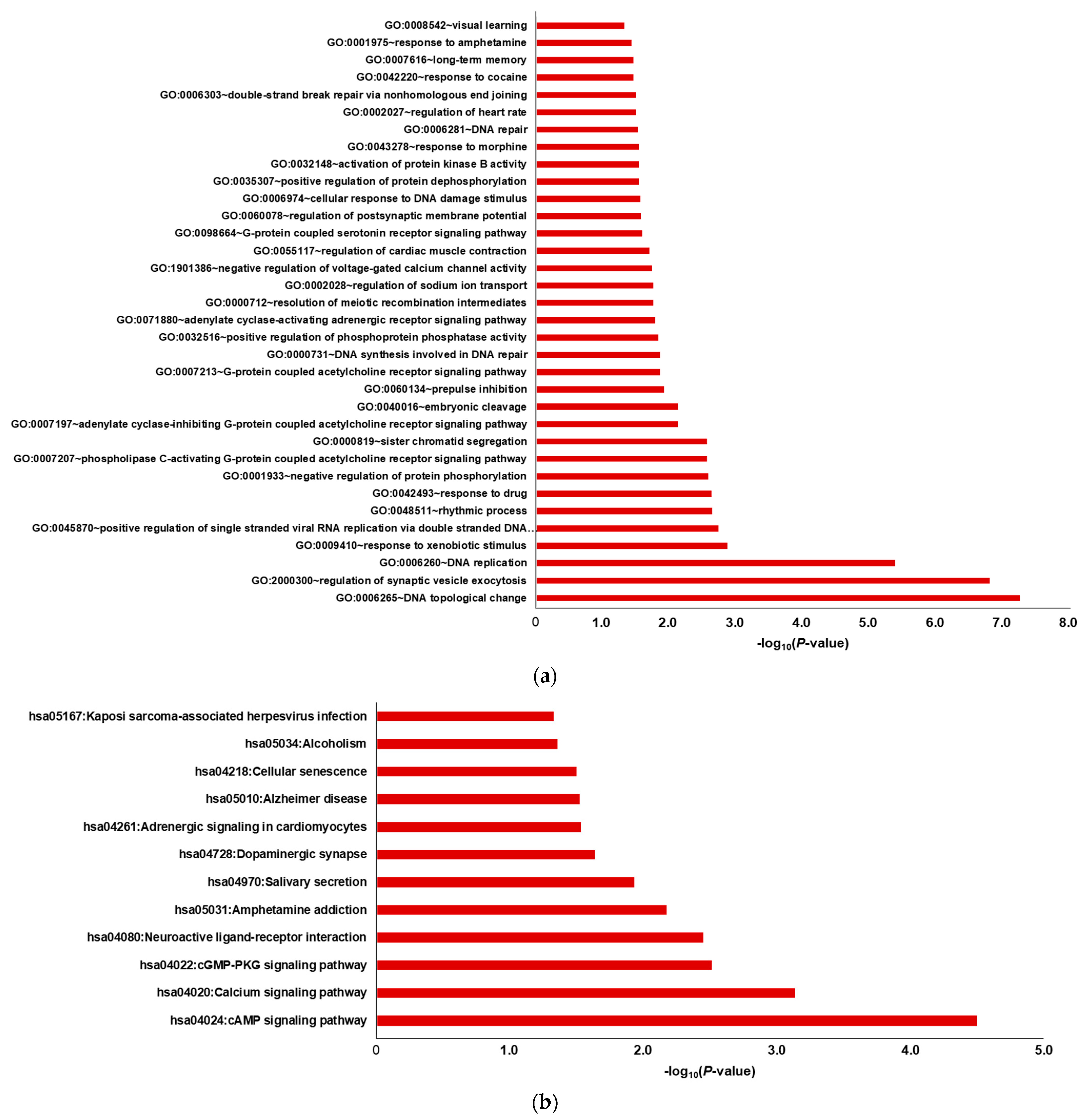
2.2. DEG Analysis through GO Mapping and KEGG Pathway Enrichment
2.3. LINCS L1000 Data-Based Expression Pattern Analysis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Cell Culture
4.3. RNA Sample Preparation
4.4. Microarray Data Analysis
4.5. Identification of the Potential Target Proteins Using the LINCS L1000 Database
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bai, J.; Zhang, Y.; Tang, C.; Hou, Y.; Ai, X.; Chen, X.; Zhang, Y.; Wang, X.; Meng, X. Gallic acid: Pharmacological activities and molecular mechanisms involved in inflammation-related diseases. Biomed. Pharmacother. 2021, 133, 110985. [Google Scholar] [CrossRef]
- Moradi, A.; Abolfathi, M.; Javadian, M.; Heidarian, E.; Roshanmehr, H.; Khaledi, M.; Nouri, A. Gallic acid exerts nephroprotective, anti-oxidative stress, and anti-inflammatory effects against diclofenac-induced renal injury in male rats. Arch. Med. Sci. 2021, 52, 380–388. [Google Scholar] [CrossRef]
- Nabavi, S.F.; Habtemariam, S.; Di Lorenzo, A.; Sureda, A.; Khanjani, S.; Nabavi, S.M.; Daglia, M. Post-stroke depression modulation and in vivo antioxidant activity of gallic acid and its synthetic derivatives in a murine model system. Nutrients 2016, 8, 248. [Google Scholar] [CrossRef]
- Variya, B.C.; Bakrania, A.K.; Patel, S.S. Antidiabetic potential of gallic acid from Emblica officinalis: Improved glucose transporters and insulin sensitivity through PPAR-γ and Akt signaling. Phytomedicine 2020, 73, 152906. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Zarrabi, A.; Mirzaei, S.; Hashemi, F.; Samarghandian, S.; Zabolian, A.; Hushmandi, K.; Ang, H.L.; Sethi, G.; Kumar, A.P.; et al. Gallic acid for cancer therapy: Molecular mechanisms and boosting efficacy by nanoscopical delivery. Food Chem. Toxicol. 2021, 157, 112576. [Google Scholar] [CrossRef]
- Kang, N.; Lee, J.H.; Lee, W.; Ko, J.Y.; Kim, E.A.; Kim, J.S.; Heu, M.S.; Kim, G.H.; Jeon, Y.J. Gallic acid isolated from Spirogyra sp. improves cardiovascular disease through a vasorelaxant and antihypertensive effect. Environ. Toxicol. Pharmacol. 2015, 39, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Daglia, M.; Di Lorenzo, A.; Nabavi, S.F.; Talas, Z.S.; Nabavi, S.M. Polyphenols: Well beyond the antioxidant capacity: Gallic acid and related compounds as neuroprotective agents: You are what you eat! Curr. Pharm. Biotechnol. 2014, 15, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhu, X.; Zhang, K.; Zhu, L.; Zhou, F. Investigation of gallic acid induced anticancer effect in human breast carcinoma MCF-7 cells. J. Biochem. Mol. Toxicol. 2014, 28, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.N.; Hua, T.N.; Kim, M.K.; Vo, V.T.; Choi, J.W.; Kim, H.W.; Rho, J.K.; Kim, K.W.; Jeong, Y. Gallic acid inhibition of Src-Stat3 signaling overcomes acquired resistance to EGF receptor tyrosine kinase inhibitors in advanced non-small cell lung cancer. Oncotarget 2016, 7, 54702–54713. [Google Scholar] [CrossRef] [PubMed]
- Lima, K.G.; Krause, G.C.; Schuster, A.D.; Catarina, A.V.; de Souza Basso, B.; de Mesquita, F.C.; Pedrazza, L.; Marczak, E.S.; Martha, B.A.; Nunes, F.B.; et al. Gallic acid reduces cell growth by induction of apoptosis and reduction of IL-8 in HepG2 cells. Biomed. Pharmacother. 2016, 84, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Badhani, B.; Sharma, N.; Kakkar, R. Gallic acid: A versatile antioxidant with promising therapeutic and industrial applications. RSC Adv. 2015, 5, 27540–27557. [Google Scholar] [CrossRef]
- Sun, G.; Zhang, S.; Xie, Y.; Zhang, Z.; Zhao, W. Gallic acid as a selective anticancer agent that induces apoptosis in SMMC-7721 human hepatocellular carcinoma cells. Oncol. Lett. 2016, 11, 150–158. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. WIREs RNA 2017, 8, e1364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.H.; Jhaveri, D.J.; Marshall, V.M.; Bauer, D.C.; Edson, J.; Narayanan, R.K. A comparative study of techniques for differential expression analysis on RNA-Seq data. PLoS ONE 2014, 9, e103207. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.; Ozburn, A.; Ponomarev, I. Genome-wide expression profiles drive discovery of novel compounds that reduce binge drinking in mice. Neuropsychopharmacology 2018, 43, 1257–1266. [Google Scholar] [CrossRef]
- Coelho, E.D.; Arrais, J.P.; Oliveira, J.L. Computational discovery of putative leads for drug repositioning through drug-target interaction prediction. PLoS Comput. Biol. 2016, 12, e1005219. [Google Scholar] [CrossRef]
- Wang, K.; Sun, J.; Zhou, S.; Wan, C.; Qin, S.; Li, C.; He, L.; Yang, L. Prediction of drug-target interactions for drug repositioning only based on genomic expression similarity. PLoS Comput. Biol. 2013, 9, e1003315. [Google Scholar] [CrossRef]
- Luo, Y.; Zhao, X.; Zhou, J.; Yang, J.; Zhang, Y.; Kuang, W.; Peng, J.; Chen, L.; Zeng, J. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 2017, 8, 573. [Google Scholar] [CrossRef]
- Lim, G.; Lim, C.J.; Lee, J.H.; Lee, B.H.; Ryu, J.Y.; Oh, K.S. Identification of new target proteins of a Urotensin-II receptor antagonist using transcriptome-based drug repositioning approach. Sci. Rep. 2021, 11, 17138. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Tong, W.; Roberts, R.; Liu, Z.; Thakkar, S. Deep learning on high-throughput transcriptomics to predict drug-induced liver injury. Front. Bioeng. Biotechnol. 2020, 8, 562677. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Wang, S.H.; Ji, Y.M.; Tong, S.; Li, D.; Ding, X.C.; Wu, C.Y. The roles and mechanisms of TRAT1 in the progression of non-small cell lung cancer. Curr. Med. Sci. 2022, 42, 1186–1200. [Google Scholar] [CrossRef]
- Nozawa, R.S.; Nagao, K.; Masuda, H.T.; Iwasaki, O.; Hirota, T.; Nozaki, N.; Kimura, H.; Obuse, C. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 2010, 12, 719–727. [Google Scholar] [CrossRef]
- Li, N.; Mruk, D.D.; Tang, E.I.; Lee, W.M.; Wong, C.K.; Cheng, C.Y. Formin 1 regulates microtubule and F-Actin organization to support spermatid transport during spermatogenesis in the rat testis. Endocrinology 2016, 157, 2894–2908. [Google Scholar] [CrossRef]
- Heath, J.; Cheyou, E.S.; Findlay, S.; Luo, V.M.; Carpio, E.P.; Lee, J.; Djerir, B.; Chen, X.; Morin, T.; Lebeau, B.; et al. POGZ promotes homology-directed DNA repair in an HP1-dependent manner. EMBO Rep. 2022, 23, e51041. [Google Scholar] [CrossRef]
- Labat-de-Hoz, L.; Alonso, M.A. Formins in human disease. Cells 2021, 10, 2554. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Ryu, J.Y.; Lee, J.H.; Lee, B.H.; Song, J.S.; Ahn, S.; Oh, K.S. PredMS: A random forest model for predicting metabolic stability of drug candidates in human liver microsomes. Bioinformatics 2022, 38, 364–368. [Google Scholar] [CrossRef]
- Zhang, S.D.; Gant, T.W. A simple and robust method for connecting small-molecule drugs using gene-expression signatures. BMC Bioinformatics 2008, 9, 258. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Fayaz, S.M.; Suvanish Kumar, V.S.; Rajanikant, K.G. Finding needles in a haystack: Application of network analysis and target enrichment studies for the identification of potential anti-diabetic phytochemicals. PLoS ONE 2014, 9, e112911. [Google Scholar] [CrossRef]
- Chang, T.-L.; Chiang, H.-Y.; Shen, J.-Y.; Lin, S.-W.; Tsai, P.-J. Phenolic compounds stage an interplay between the ubiquitin–proteasome system and ubiquitin signal autophagic degradation for the ubiquitin-based cancer chemoprevention. J. Funct. Foods 2015, 17, 857–871. [Google Scholar] [CrossRef]
- Kang, D.Y.; Sp, N.; Jo, E.S.; Rugamba, A.; Hong, D.Y.; Lee, H.G.; Yoo, J.-S.; Liu, Q.; Jang, K.-J.; Yang, Y.M. The inhibitory mechanisms of tumor PD-L1 expression by natural bioactive gallic acid in non-small-cell lung cancer (NSCLC) cells. Cancers 2020, 12, 727. [Google Scholar] [CrossRef]
- Gu, R.; Zhang, M.; Meng, H.; Xu, D.; Xie, Y. Gallic acid targets acute myeloid leukemia via Akt/mTOR-dependent mitochondrial respiration inhibition. Biomed. Pharmacother. 2018, 105, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhang, Y.L.; Zhang, L.; Zou, L.X.; Chen, C.; Liu, Y.; Xia, Y.L.; Li, H.H. Gallic acid suppresses cardiac hypertrophic remodeling and heart failure. Mol. Nutr. Food Res. 2019, 63, e1800807. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, M.T.; Farbood, Y.; Sameri, M.J.; Sarkaki, A.; Naghizadeh, B.; Rafeirad, M. Neuroprotective effects of oral gallic acid against oxidative stress induced by 6-hydroxydopamine in rats. Food Chem. 2013, 138, 1028–1033. [Google Scholar] [CrossRef]
- Urrunaga, N.H.; Jadeja, R.N.; Rachakonda, V.; Ahmad, D.; McLean, L.P.; Cheng, K.; Shah, V.; Twaddell, W.S.; Raufman, J.P.; Khurana, S. M1 muscarinic receptors modify oxidative stress response to acetaminophen-induced acute liver injury. Free Radic. Biol. Med. 2015, 78, 66–81. [Google Scholar] [CrossRef]
- Maya, S.; Prakash, T.; Madhu, K. Assessment of neuroprotective effects of gallic acid against glutamate-induced neurotoxicity in primary rat cortex neuronal culture. Neurochem. Int. 2018, 121, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Can, Ö.D.; Turan, N.; Demir Özkay, Ü.; Öztürk, Y. Antidepressant-like effect of gallic acid in mice: Dual involvement of serotonergic and catecholaminergic systems. Life Sci. 2017, 190, 110–117. [Google Scholar] [CrossRef]
- Jin, L.; Piao, Z.H.; Liu, C.P.; Sun, S.; Liu, B.; Kim, G.R.; Choi, S.Y.; Ryu, Y.; Kee, H.J.; Jeong, M.H. Gallic acid attenuates calcium calmodulin-dependent kinase II-induced apoptosis in spontaneously hypertensive rats. J. Cell. Mol. Med. 2018, 22, 1517–1526. [Google Scholar] [CrossRef] [PubMed]
- López-Lázaro, M.; Calderón-Montaño, J.M.; Burgos-Morón, E.; Austin, C.A. Green tea constituents (-)-epigallocatechin-3-gallate (EGCG) and gallic acid induce topoisomerase I- and topoisomerase II-DNA complexes in cells mediated by pyrogallol-induced hydrogen peroxide. Mutagenesis 2011, 26, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Zago, A.M.; Carvalho, F.B.; Gutierres, J.M.; Bohnert, C.; Fernandes, M.D.C.; Morandini, L.M.; Coelho, H.S.; Fogaça, A.O.; Andrade, C.M.; Mostardeiro, M.A.; et al. A phytochemical study of the Cuphea glutinosa from Southern Brazil: Na+, K+-ATPase activity inhibition and antioxidant properties. Nat. Prod. Res. 2019, 33, 3426–3431. [Google Scholar] [CrossRef]
- Sherin, L.; Sohail, A.; Shujaat, S. Time-dependent AI-Modeling of the anticancer efficacy of synthesized gallic acid analogues. Comput. Biol. Chem. 2019, 79, 137–146. [Google Scholar] [CrossRef]
- AL Zahrani, N.A.; El-Shishtawy, R.M.; Asiri, A.M. Recent developments of gallic acid derivatives and their hybrids in medicinal chemistry: A review. Eur. J. Med. Chem. 2020, 204, 112609. [Google Scholar] [CrossRef]
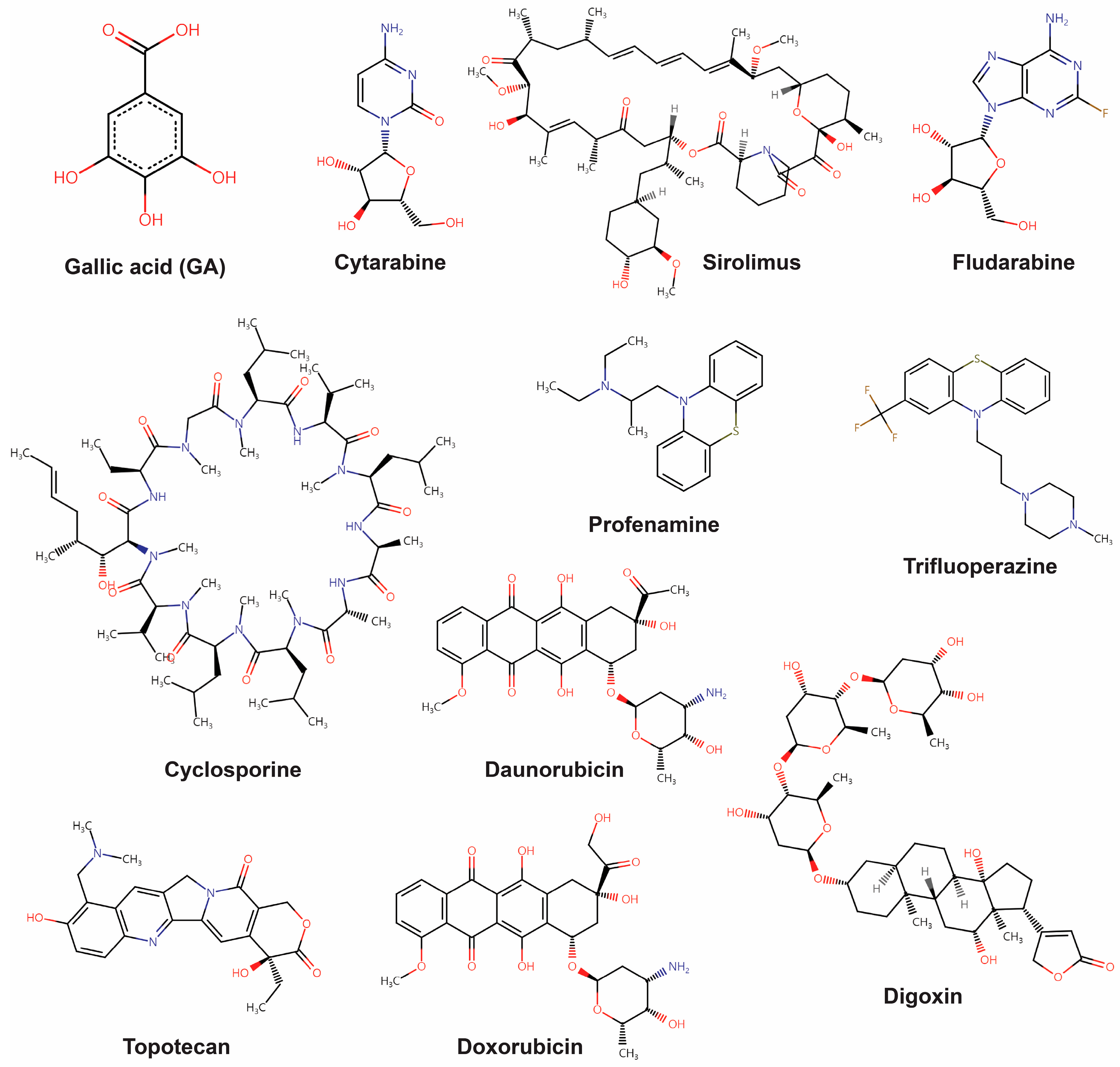
| Perturbagen ID | Drug Name | Mean of the Zhang Score | Targets | |||
|---|---|---|---|---|---|---|
| Gene Symbol | UniProt ID | Name of the Target Proteins | ||||
| 1 | BRD-K33106058_DB00987 | Cytarabine | 0.402339 | POLB | P06746 | DNA polymerase beta |
| 2 | BRD-K89626439_DB00877 | Sirolimus (rapamycin) | 0.396491 | MTOR | P42345 | Serine/threonine-protein kinase mTOR |
| 3 | BRD-K72238567_DB01073 | Fludarabine | 0.20117 | POLA1, RRM1 | P09884, P23921 | DNA polymerase alpha catalytic subunit Ribonucleoside-diphosphate reductase large subunit |
| 4 | BRD-A38030642_DB00091 | Cyclosporine | 0.192982 | PPP3R2, PPIA | Q96LZ3, P62937 | Calcineurin subunit B type 2 Peptidyl-prolyl cis-trans isomerase A |
| 5 | BRD-A16311756_DB00392 | Profenamine | 0.166082 | CHRM1, CHRM2, GRIN3A | P11229, P08172, Q8TCU5 | Muscarinic acetylcholine receptor M1 Muscarinic acetylcholine receptor M2 Glutamate receptor ionotropic, NMDA 3A |
| 6 | BRD-K89732114_DB00831 | Trifluoperazine | 0.153216 | DRD2, CALY, ADRA1A, CALM, S100A4 | P14416, Q9NYX4, P35348, P0DP23, P26447 | Dopamine D2 receptor Neuron-specific vesicular protein calcyon Alpha-1A adrenergic receptor Calmodulin Protein S100-A4 |
| 7 | BRD-K43389675_DB00694 | Daunorubicin | 0.145029 | TOP2A, TOP2B | P11388, Q02880 | DNA topoisomerase 2-alpha DNA topoisomerase 2-beta |
| 8 | BRD-K23478508_DB00390 | Digoxin | 0.138012 | ATP1A1 | P05023 | Sodium/potassium-transporting ATPase subunit alpha-1 |
| 9 | BRD-A59985574_DB01030 | Topotecan | 0.127485 | TOP1, TOP1MT | P11387, Q969P6 | DNA topoisomerase 1 DNA topoisomerase I, mitochondrial |
| 10 | BRD-K04548931_DB00997 | Doxorubicin | 0.100585 | TOP1, TOP2A, TOP2B | P11387, P11388, Q02880 | DNA topoisomerase 1 DNA topoisomerase 2-alpha DNA topoisomerase 2-beta |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Kim, B.K.; Moh, S.H.; Jang, G.; Ryu, J.Y. Investigation of the General Molecular Mechanisms of Gallic Acid via Analyses of Its Transcriptome Profile. Int. J. Mol. Sci. 2024, 25, 2303. https://doi.org/10.3390/ijms25042303
Kim J, Kim BK, Moh SH, Jang G, Ryu JY. Investigation of the General Molecular Mechanisms of Gallic Acid via Analyses of Its Transcriptome Profile. International Journal of Molecular Sciences. 2024; 25(4):2303. https://doi.org/10.3390/ijms25042303
Chicago/Turabian StyleKim, Jiyeon, Bo Kyung Kim, Sang Hyun Moh, Goo Jang, and Jae Yong Ryu. 2024. "Investigation of the General Molecular Mechanisms of Gallic Acid via Analyses of Its Transcriptome Profile" International Journal of Molecular Sciences 25, no. 4: 2303. https://doi.org/10.3390/ijms25042303
APA StyleKim, J., Kim, B. K., Moh, S. H., Jang, G., & Ryu, J. Y. (2024). Investigation of the General Molecular Mechanisms of Gallic Acid via Analyses of Its Transcriptome Profile. International Journal of Molecular Sciences, 25(4), 2303. https://doi.org/10.3390/ijms25042303








