Three-Dimensional Structural Stability and Local Electrostatic Potential at Point Mutations in Spike Protein of SARS-CoV-2 Coronavirus
Abstract
1. Introduction
2. Results and Interpretation
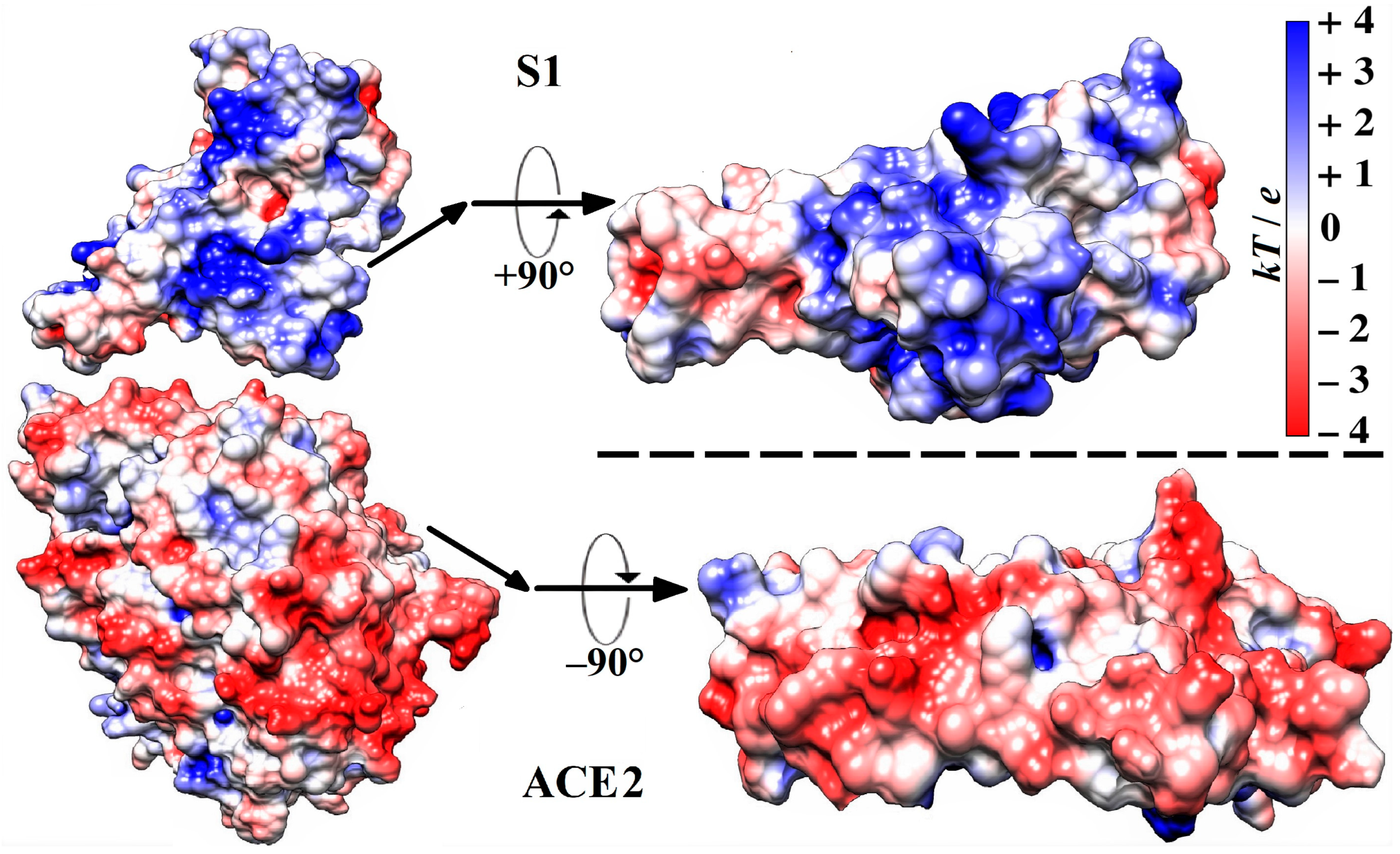
2.1. ACE2 Receptor
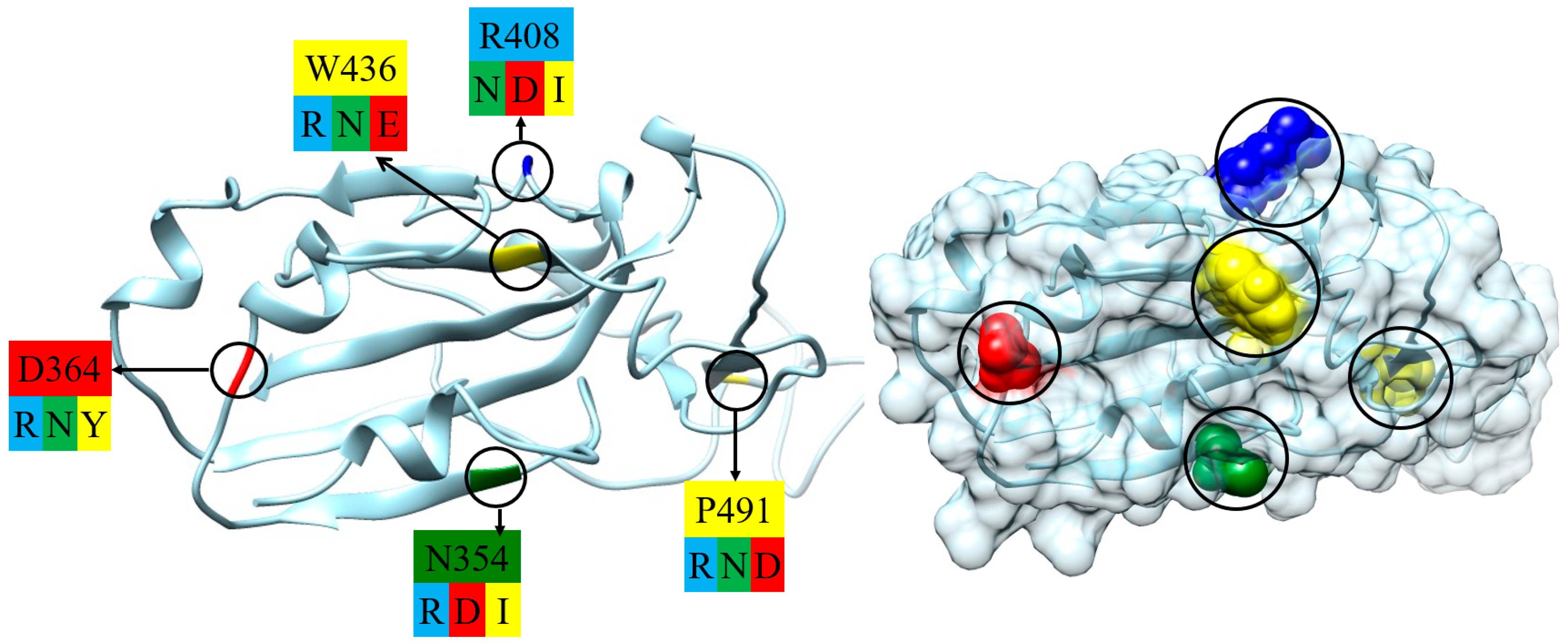
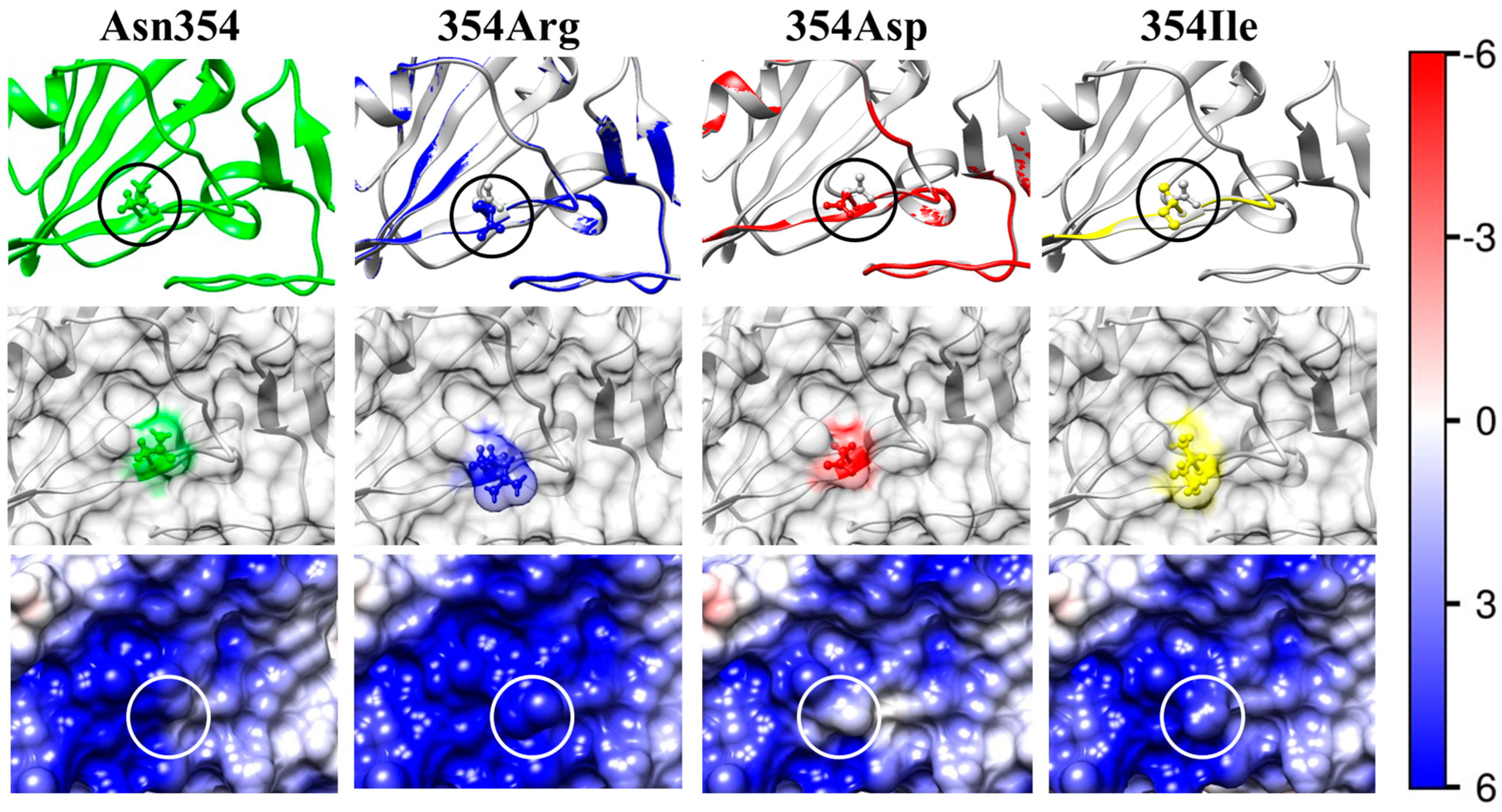
2.2. Point Mutants of S-Protein
2.3. Isoelectric Point
2.4. Local Electrostatic Potential
2.5. Free Energy of Folding
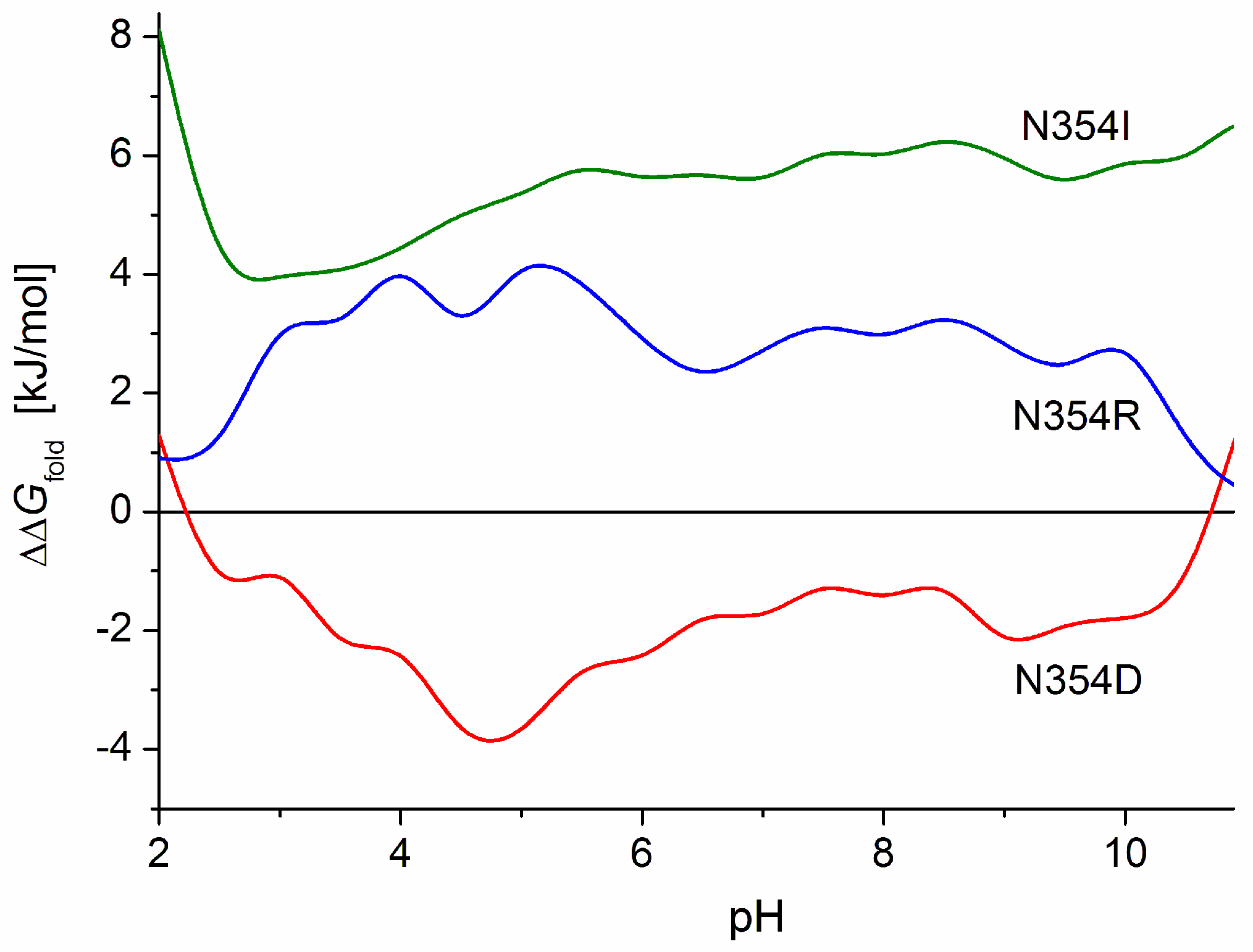
2.5.1. Folding Free Energy of S1-Point Mutants
2.5.2. Isoelectric Point as an Indicator for 3D Structural Stability
2.5.3. Effect of the Local Electrostatic Potential
2.5.4. Surface Hydrophilic Effect
2.5.5. Deep Electrostatic–Hydrophilic Effect
2.5.6. Deep Hydrophilic Effect
2.6. Peculiar Properties of S1-Mutants
3. Discussion
3.1. Gibbs Free Energy upon Folding
3.2. Computing of the Folding Free Energy
3.3. Protein–Receptor Attraction
4. Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Avdonin, P.P.; Rybakova, E.Y.; Trufanov, S.K.; Avdonin, P.V. SARS-CoV-2 receptors and their involvement in cell infection. Biochem. (Mosc.) Suppl. Ser. A Membr. Cell Biol. 2023, 17, 1–11. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.Y.; et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef]
- Hatmal, M.M.; Alshaer, W.; Al-Hatamleh, M.A.I.; Hatmal, M.; Smadi, O.; Taha, M.O.; Oweida, A.J.; Boer, J.C.; Mohamud, R.; Plebanski, M. Comprehensive structural and molecular comparison of Spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV and their interactions with ACE2. Cells 2020, 9, 2638. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef]
- Barton, M.I.; MacGowan, S.A.; Kutuzov, M.A.; Dushek, O.; Barton, G.J.; Van Der Merwe, P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. Elife 2021, 10, e70658. [Google Scholar] [CrossRef] [PubMed]
- Pascarella, S.; Ciccozzi, M.; Zella, D.; Bianchi, M.; Benedetti, F.; Benvenuto, D.; Broccolo, F.; Cauda, R.; Caruso, A.; Angeletti, S.; et al. SARS-CoV-2 B. 1.617 Indian variants: Are electrostatic potential changes responsible for a higher transmission rate? J. Med. Virol. 2021, 93, 6551–6556. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Barre, A.; Klonjkowski, B.; Benoist, H.; Rougé, P. How do point mutations enhancing the basic character of the RBDs of SARS-CoV-2 variants affect their transmissibility and infectivity capacities? Viruses 2022, 14, 783. [Google Scholar] [CrossRef]
- Xie, Y.; Karki, C.B.; Du, D.; Li, H.; Wang, J.; Sobitan, A.; Teng, S.; Tang, Q.; Li, L. Spike proteins of SARS-CoV and SARS-CoV-2 utilize different mechanisms to bind with human ACE2. Front. Mol. Biosci. 2020, 7, 392. [Google Scholar] [CrossRef]
- Ali, F.; Elserafy, M.; Alkordi, M.H.; Amin, M. ACE2 coding variants in different populations and their potential impact on SARS-CoV-2 binding affinity. Biochem. Biophys. Rep. 2020, 24, 100798. [Google Scholar] [CrossRef]
- Jawad, B.; Adhikari, P.; Podgornik, R.; Ching, W.Y. Key interacting residues between RBD of SARS-CoV-2 and ACE2 receptor: Combination of molecular dynamics simulation and density functional calculation. J. Chem. Inf. Model. 2021, 61, 4425–4441. [Google Scholar] [CrossRef]
- Jawad, B.; Adhikari, P.; Podgornik, R.; Ching, W.Y. Binding interactions between receptor-binding domain of spike protein and human angiotensin converting enzyme-2 in omicron variant. J. Phys. Chem. Lett. 2022, 13, 3915–3921. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Guo, W.; Lopez-Hernadez, A.; Teng, S.; Li, L. The pH effects on SARS-CoV and SARS-CoV-2 spike proteins in the process of binding to hACE2. Pathogens 2022, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Javidpour, L.; Božič, A.; Naji, A.; Podgornik, R. Electrostatic interactions between the SARS-CoV-2 virus and a charged electret fibre. Soft Matter 2021, 17, 4296–4303. [Google Scholar] [CrossRef] [PubMed]
- Sang, P.; Chen, Y.Q.; Liu, M.T.; Wang, Y.T.; Yue, T.; Li, Y.; Yin, Y.R.; Yang, L.Q. Electrostatic interactions are the primary determinant of the binding affinity of SARS-CoV-2 spike RBD to ACE2: A computational case study of omicron variants. Int. J. Mol. Sci. 2022, 23, 14796. [Google Scholar] [CrossRef]
- Biskupek, I.; Gieldon, A. Two-Stage Recognition Mechanism of the SARS-CoV-2 Receptor-Binding Domain to Angiotensin-Converting Enzyme-2 (ACE2). Int. J. Mol. Sci. 2024, 25, 679. [Google Scholar] [CrossRef]
- Pascarella, S.; Ciccozzi, M.; Bianchi, M.; Benvenuto, D.; Cauda, R.; Cassone, A. The value of electrostatic potentials of the spike receptor binding and N-terminal domains in addressing transmissibility and infectivity of SARS-CoV-2 variants of concern. J. Infect. 2022, 84, 62–63. [Google Scholar] [CrossRef]
- Nie, C.; Sahoo, A.K.; Netz, R.R.; Herrmann, A.; Ballauff, M.; Haag, R. Charge matters: Mutations in omicron variant favor binding to cells. ChemBioChem 2022, 23, e202100681. [Google Scholar] [CrossRef]
- Ishikawa, T.; Ozono, H.; Akisawa, K.; Hatada, R.; Okuwaki, K.; Mochizuki, Y. Interaction analysis on the SARS-CoV-2 Spike protein receptor binding domain using visualization of the interfacial electrostatic complementarity. J. Phys. Chem. Lett. 2021, 12, 11267–11272. [Google Scholar] [CrossRef]
- Zhao, X.; Xiong, D.; Luo, S.; Duan, L. Origin of the tight binding mode to ACE2 triggered by multi-point mutations in the omicron variant: A dynamic insight. Phys. Chem. Chem. Phys. 2022, 24, 8724–8737. [Google Scholar] [CrossRef]
- Goher, S.S.; Ali, F.; Amin, M. The Delta variant mutations in the receptor binding domain of SARS-CoV-2 show enhanced electrostatic interactions with the ACE2. Med. Drug Discov. 2022, 13, 100114. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, S.; Bhattacharyya, R.; Sengupta, J. Dynamics and electrostatics define an allosteric druggable site within the receptor-binding domain of SARS-CoV-2 spike protein. FEBS Lett. 2021, 595, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.L.; Padhi, A.K.; Mandal, N. Scanning the RBD-ACE2 molecular interactions in Omicron variant. Biochem. Biophys. Res. Commun. 2022, 592, 18–23. [Google Scholar] [CrossRef]
- Fazekas, Z.; Menyhárd, D.K.; Perczel, A. Omicron binding mode: Contact analysis and dynamics of the omicron receptor-binding domain in complex with ACE2. J. Chem. Inf. Model. 2022, 62, 3844–3853. [Google Scholar] [CrossRef]
- Lupala, C.S.; Ye, Y.; Chen, H.; Su, X.D.; Liu, H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun. 2022, 590, 34–41. [Google Scholar] [CrossRef] [PubMed]
- Giron, C.C.; Laaksonen, A.; Barroso da Silva, F.L. Differences between Omicron SARS-CoV-2 RBD and other variants in their ability to interact with cell receptors and monoclonal antibodies. J. Biomol. Struct. Dyn. 2023, 41, 5707–5727. [Google Scholar] [CrossRef]
- Gan, H.H.; Zinno, J.; Piano, F.; Gunsalus, K.C. Omicron Spike protein has a positive electrostatic surface that promotes ACE2 recognition and antibody escape. Front. Virol. 2022, 23, 14796. [Google Scholar] [CrossRef]
- Shah, M.; Ahmad, B.; Choi, S.; Woo, H.G. Mutations in the SARS-CoV-2 spike RBD are responsible for stronger ACE2 binding and poor anti-SARS-CoV mAbs cross-neutralization. Comput. Struct. Biotechnol. J. 2020, 18, 3402–3414. [Google Scholar] [CrossRef]
- Dung, D.N.; Phan, A.D.; Nguyen, T.T.; Lam, V.D. Effects of surface charge and environmental factors on the electrostatic interaction of fiber with virus-like particle: A case of coronavirus. AIP Adv. 2021, 11, 10508. [Google Scholar] [CrossRef] [PubMed]
- Barroso da Silva, F.L.; Giron, C.C.; Laaksonen, A. Electrostatic features for the receptor binding domain of SARS-COV-2 wildtype and its variants. Compass to the severity of the future variants with the charge-rule. J. Phys. Chem. B 2022, 126, 6835–6852. [Google Scholar] [CrossRef]
- Hristova, S.H.; Zhivkov, A.M. Omicron coronavirus: pH-dependent electric potential and energy of association of spike protein to ACE2 receptor. Viruses 2023, 15, 1752. [Google Scholar] [CrossRef]
- Pandurangan, A.P.; Ochoa-Montaño, B.; Ascher, D.B.; Blundell, T.L. SDM: A server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017, 45, W229–W235. [Google Scholar] [CrossRef] [PubMed]
- Worth, C.L.; Preissner, R.; Blundell, T.L. SDM—A server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011, 39 (Suppl. S2), W215–W222. [Google Scholar] [CrossRef]
- Kantardjiev, A.A.; Atanasov, B.P. PHEMTO: Protein pH-dependent electric moment tools. Nucleic Acids Res. 2009, 37, W422–W427. [Google Scholar] [CrossRef][Green Version]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Li, H.; Robertson, A.D.; Jensen, J.H. Very fast empirical prediction and rationalization of protein pKa values. Proteins Struct. Funct. Bioinform. 2005, 61, 704–721. [Google Scholar] [CrossRef]
- Walsh, I.; Minervini, G.; Corazza, A.; Esposito, G.; Tosatto, S.C.E.; Fogolari, F. Bluues server: Electrostatic properties of wild-type and mutated protein structures. Bioinformatics 2012, 28, 2189–2190. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Vargyas, M.; Vasko-Szedlar, J.; Roux, B.; Im, W. PBEQ-Solver for online visualization of electrostatic potential of Biomolecules. Nucleic Acids Res. 2008, 36, W270–W275. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Fauchere, I.I.; Pliska, N. Hydrophobic parameters II of amino acid side-chains from the partitioning of N-acetyl-amino acid amides. Eur. J. Med. Chem-Chim. 1983, 18, 369–375. [Google Scholar]
- Shulz, G.R.; Schirmer, R.H. Principles of Protein Structure; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 1979. [Google Scholar]
- England, R.J.A.; Homer, J.J.; Knight, L.C.; Ell, S.R. Nasal pH measurement: A reliable and repeatable parameter. Clin. Otoringol. 1999, 24, 67–68. [Google Scholar] [CrossRef]
- Hristova, S.; Zhivkov, A.; Atanasov, B. Electrostatics of horse heart cytochrome c and montmorillonite monolamellar plate. Biotechnol. Biotechnol. Equip. 2009, 23 (Suppl. S1), 568–571. [Google Scholar] [CrossRef]
- Ge, M.; Pan, X.M. The contribution of proline residues to protein stability is associated with isomerization equilibrium in both unfolded and folded states. Extremophiles 2009, 13, 481–489. [Google Scholar] [CrossRef]
- Phan, T. Genetic diversity and evolution of SARS-CoV-2. Infect. Genet. Evol. 2020, 81, 104260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; He, X.; Zhai, J.; Ji, B.; Man, V.H.; Wang, J. In silico binding profile characterization of SARS-CoV-2 spike protein and its mutants bound to human ACE2 receptor. Brief. Bioinform. 2021, 22, bbab188. [Google Scholar] [CrossRef]
- Ahamad, S.; Kanipakam, H.; Gupta, D. Insights into the structural and dynamical changes of spike glycoprotein mutations associated with SARS-CoV-2 host receptor binding. J. Biomol. Struct. Dyn. 2020, 40, 263–275. [Google Scholar] [CrossRef]
- Ou, J.; Zhou, Z.; Zhang, J.; Lan, W.; Zhao, S.; Wu, J.; Seto, D.; Zhang, G.; Zhang, Q. RBD mutations from circulating SARS-CoV-2 strains enhance the structural stability and human ACE2 affinity of the spike protein. J. Virol. 2021, 95, 16–40. [Google Scholar]
- Kim, J.S.; Jang, J.H.; Kim, J.M.; Chung, Y.S.; Yoo, C.K.; Han, M.G. Genome-Wide Identification and Characterization of Point Mutations in the SARS-CoV-2 Genome. Osong Public Health Res. Perspect. 2020, 11, 101–111. [Google Scholar] [CrossRef]
- Antosiewicz, J.; McCammon, J.A.; Gilson, M.K. Prediction of pH-dependent properties of proteins. J. Mol. Biol. 1994, 283, 415–436. [Google Scholar] [CrossRef]
- Schutz, C.N.; Warshel, A. What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins 2001, 44, 400–417. [Google Scholar] [CrossRef] [PubMed]
- Varhač, R.; Antalík, M.; Bánó, M. Effect of temperature and guanidine hydrochloride on ferrocytochrome c at neutral pH. J. Biol. Inorg. Chem. 2004, 9, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Baba, T.; Kidokoro, S.I. A molten globule-like intermediate state detected in the thermal transition of cytochrome c under low salt concentration. Biophys. Chem. 2007, 127, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Kumar, R.; Kumar, S.; Chhabra, R.; Agarwal, M.C. Analysis of the pH-dependent stability and millisecond folding kinetics of horse cytochrome c. Arch. Biochem. Biophys. 2015, 585, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Seki, Y.; Katoh, E.; Kidokoro, S.I. Thermodynamic and structural properties of the acid molten globule state of horse cytochrome c. Biochemistry 2011, 50, 3116–3126. [Google Scholar] [CrossRef]


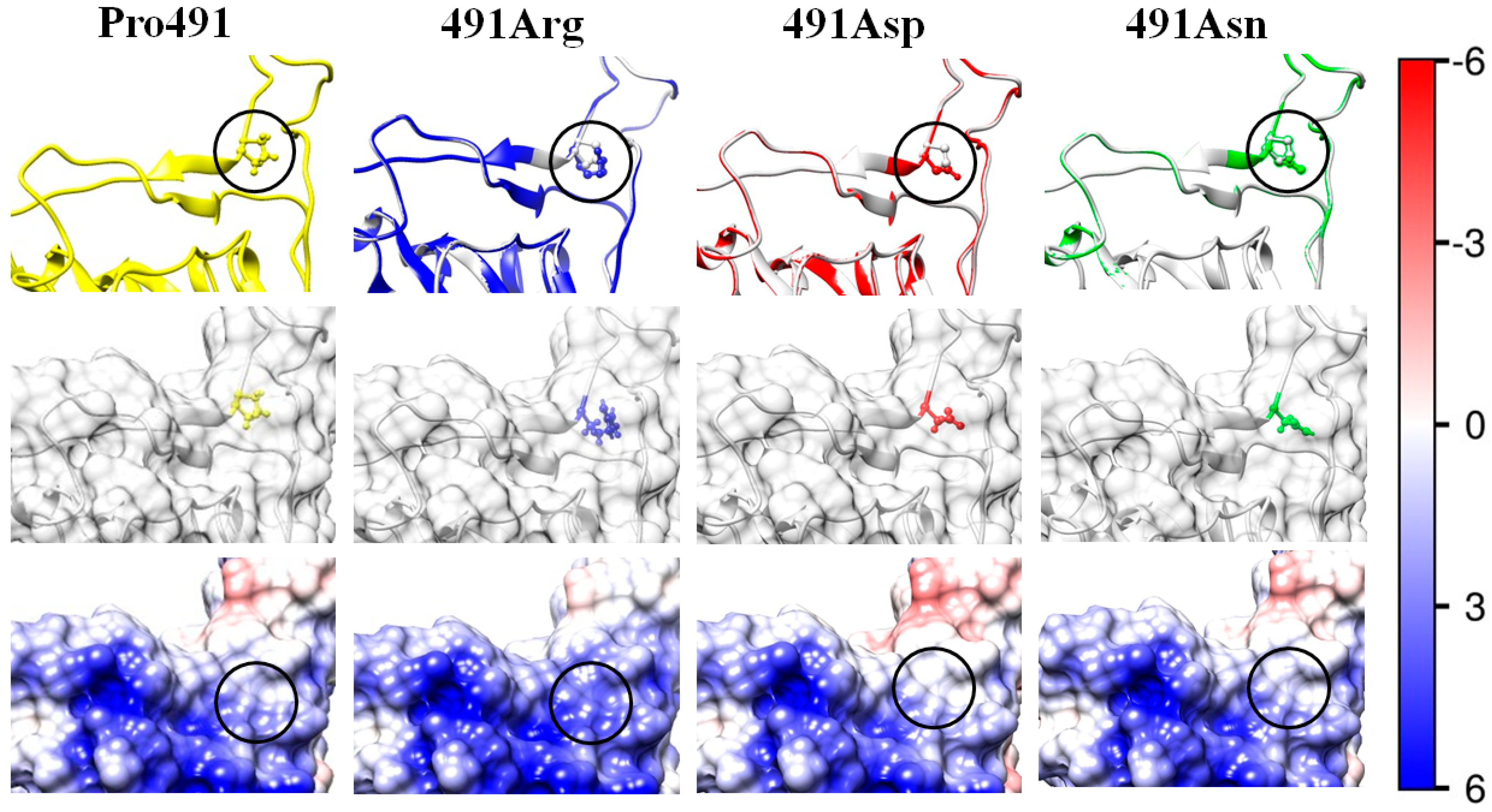
| Mutant | Replaced Residue | Substituting Residue | ΔΔGtrans | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| № | Code | Name | Mres | Vres | ΔGtrans | Name | Mres | Vres | ΔGtrans | |
| 1 | N354R | asparagine | 114 | 0.15 | −3.3 | arginine | 157 | 0.21 | −6.3 | −3.0 |
| 2 | N354D | asparagine | 114 | 0.15 | −3.3 | aspartate | 114 | 0.15 | −4.6 | −1.3 |
| 3 | N354I | asparagine | 114 | 0.15 | −3.3 | isoleucine | 113 | 0.21 | +10.0 | +13.3 |
| 4 | D364R | aspartate | 114 | 0.15 | −4.6 | arginine | 157 | 0.21 | −6.3 | −1.7 |
| 5 | D364N | aspartate | 114 | 0.15 | −4.6 | asparagine | 114 | 0.15 | −3.3 | +1.3 |
| 6 | D364Y | aspartate | 114 | 0.15 | −4.6 | tyrosine | 163 | 0.15 | +5.4 | +10.0 |
| 7 | R408D | arginine | 157 | 0.21 | −6.3 | aspartate | 114 | 0.15 | −4.6 | +1.7 |
| 8 | R408N | arginine | 157 | 0.21 | −6.3 | asparagine | 114 | 0.15 | −3.3 | +3.0 |
| 9 | R408I | arginine | 157 | 0.21 | −6.3 | isoleucine | 113 | 0.21 | +10.0 | +16.3 |
| 10 | W436R | tryptophan | 186 | 0.25 | +12.6 | arginine | 157 | 0.15 | −6.3 | −18.9 |
| 11 | W436E | tryptophan | 186 | 0.25 | +12.6 | glutamate | 128 | 165 | −1.3 | −13.9 |
| 12 | W436N | tryptophan | 186 | 0.25 | +12.6 | asparagine | 97 | 0.13 | −3.3 | −8.4 |
| 13 | P491R | proline | 97 | 0.13 | +4.2 | arginine | 157 | 0.21 | −6.3 | −10.5 |
| 14 | P491D | proline | 97 | 0.13 | +4.2 | aspartate | 114 | 0.15 | −4.6 | −8.8 |
| 15 | P491N | proline | 97 | 0.13 | +4.2 | asparagine | 114 | 0.15 | −3.3 | −7.5 |
| Mutant | Replaced Residue | Substituting | S1-Subunit | RBD | |||||
|---|---|---|---|---|---|---|---|---|---|
| № | Code | Name | +/− | Name | +/− | pI | ΔpI | pI | ΔpI |
| 0 | Wild | – | 0 | – | 0 | 8.70 | 0 | 9.02 | 0 |
| 1 | N354R | asparagine | 0 | arginine | P | 8.87 | +0.17 | 9.27 | +0.25 |
| 2 | N354D | asparagine | 0 | aspartate | N | 8.50 | −0.20 | 8.68 | −0.34 |
| 3 | N354I | asparagine | 0 | isoleucine | 0 | 8.70 | 0 | 9.01 | −0.01 |
| 4 | D364R | aspartate | N | arginine | P | 9.01 | +0.31 | 9.48 | +0.46 |
| 5 | D364N | aspartate | N | asparagine | 0 | 8.87 | +0.17 | 9.28 | +0.26 |
| 6 | D364Y | aspartate | N | tyrosine | 0 | 8.86 | +0.17 | 9.25 | +0.23 |
| 7 | R408D | arginine | P | aspartate | N | 8.21 | −0.49 | 8.07 | −0.95 |
| 8 | R408N | arginine | P | asparagine | 0 | 8.49 | −0.21 | 8.66 | −0.36 |
| 9 | R408I | arginine | P | isoleucine | 0 | 8.49 | −0.21 | 8.66 | −0.36 |
| 10 | W436R | tryptophan | 0 | arginine | P | 8.87 | +0.17 | 9.28 | +0.26 |
| 11 | W436E | tryptophan | 0 | glutamate | N | 8.49 | -0.21 | 8.65 | −0.37 |
| 12 | W436N | tryptophan | 0 | asparagine | 0 | 8.70 | 0 | 9.02 | 0 |
| 13 | P491R | proline | 0 | arginine | P | 8.87 | +0.17 | 9.27 | +0.25 |
| 14 | P491D | proline | 0 | aspartate | N | 8.49 | −0.21 | 8.66 | −0.36 |
| 15 | P491N | proline | 0 | asparagine | 0 | 8.7 | 0 | 9.02 | 0 |
| S1-Subunit | pH 6 | pH 7 | pH 9 | ||||
|---|---|---|---|---|---|---|---|
| № | Mutant | ΔGfold [kJ/mol] | ΔΔGfold [kJ/mol] | ΔGfold [kJ/mol] | ΔΔGfold [kJ/mol] | ΔGfold [kJ/mol] | ΔΔGfold [kJ/mol] |
| 0 | Wild | −95.17 | 0 | −122.47 | 0 | −138.28 | 0 |
| 1 | N354R | −92.30 | 2.87 | −119.47 | 2.73 | −135.46 | 2.82 |
| 2 | N354D | −96.69 | −1.52 | −124.62 | −2.15 | −140.97 | −2.69 |
| 3 | N354I | −89.60 | 5.57 | −117.00 | 5.47 | −132.29 | 5.99 |
| 4 | D364R | −85.79 | 9.38 | −113.54 | 8.93 | −129.21 | 9.07 |
| 5 | D364N | −88.94 | 6.23 | −117.21 | 5.26 | −132.91 | 5.37 |
| 6 | D364Y | −88.67 | 6.5 | −117.39 | 5.08 | −133.39 | 4.89 |
| 7 | R408D | −85.08 | 10.09 | −113.49 | 8.98 | −129.86 | 8.42 |
| 8 | R408N | −84.26 | 10.91 | −111.97 | 10.5 | −128.97 | 9.37 |
| 9 | R408I | −84.03 | 11.14 | −113.28 | 9.19 | −129.43 | 8.85 |
| 10 | W436R | −91 | 4.17 | −120.26 | 2.21 | −136.59 | 1.69 |
| 11 | W436E | −85.24 | 9.93 | −113.37 | 9.91 | −128.98 | 9.3 |
| 12 | W436N | −90.25 | 4.92 | −118.87 | 5.75 | −133.87 | 7.1 |
| 13 | P491R | −80.88 | 14.29 | −109.64 | 12.83 | −123.3 | 14.98 |
| 14 | P491D | −83.64 | 11.53 | −111.85 | 10.62 | −128.44 | 9.84 |
| 15 | P491N | −90.83 | 4.34 | −119.01 | 3.46 | −134.33 | 3.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hristova, S.H.; Zhivkov, A.M. Three-Dimensional Structural Stability and Local Electrostatic Potential at Point Mutations in Spike Protein of SARS-CoV-2 Coronavirus. Int. J. Mol. Sci. 2024, 25, 2174. https://doi.org/10.3390/ijms25042174
Hristova SH, Zhivkov AM. Three-Dimensional Structural Stability and Local Electrostatic Potential at Point Mutations in Spike Protein of SARS-CoV-2 Coronavirus. International Journal of Molecular Sciences. 2024; 25(4):2174. https://doi.org/10.3390/ijms25042174
Chicago/Turabian StyleHristova, Svetlana H., and Alexandar M. Zhivkov. 2024. "Three-Dimensional Structural Stability and Local Electrostatic Potential at Point Mutations in Spike Protein of SARS-CoV-2 Coronavirus" International Journal of Molecular Sciences 25, no. 4: 2174. https://doi.org/10.3390/ijms25042174
APA StyleHristova, S. H., & Zhivkov, A. M. (2024). Three-Dimensional Structural Stability and Local Electrostatic Potential at Point Mutations in Spike Protein of SARS-CoV-2 Coronavirus. International Journal of Molecular Sciences, 25(4), 2174. https://doi.org/10.3390/ijms25042174






