Identification of Candidate Genes for Red-Eyed (Albinism) Domestic Guppies Using Genomic and Transcriptomic Analyses
Abstract
1. Introduction
2. Results
2.1. Overview of the Whole-Genome Sequencing Data
2.2. Genome-Wide Association Studies
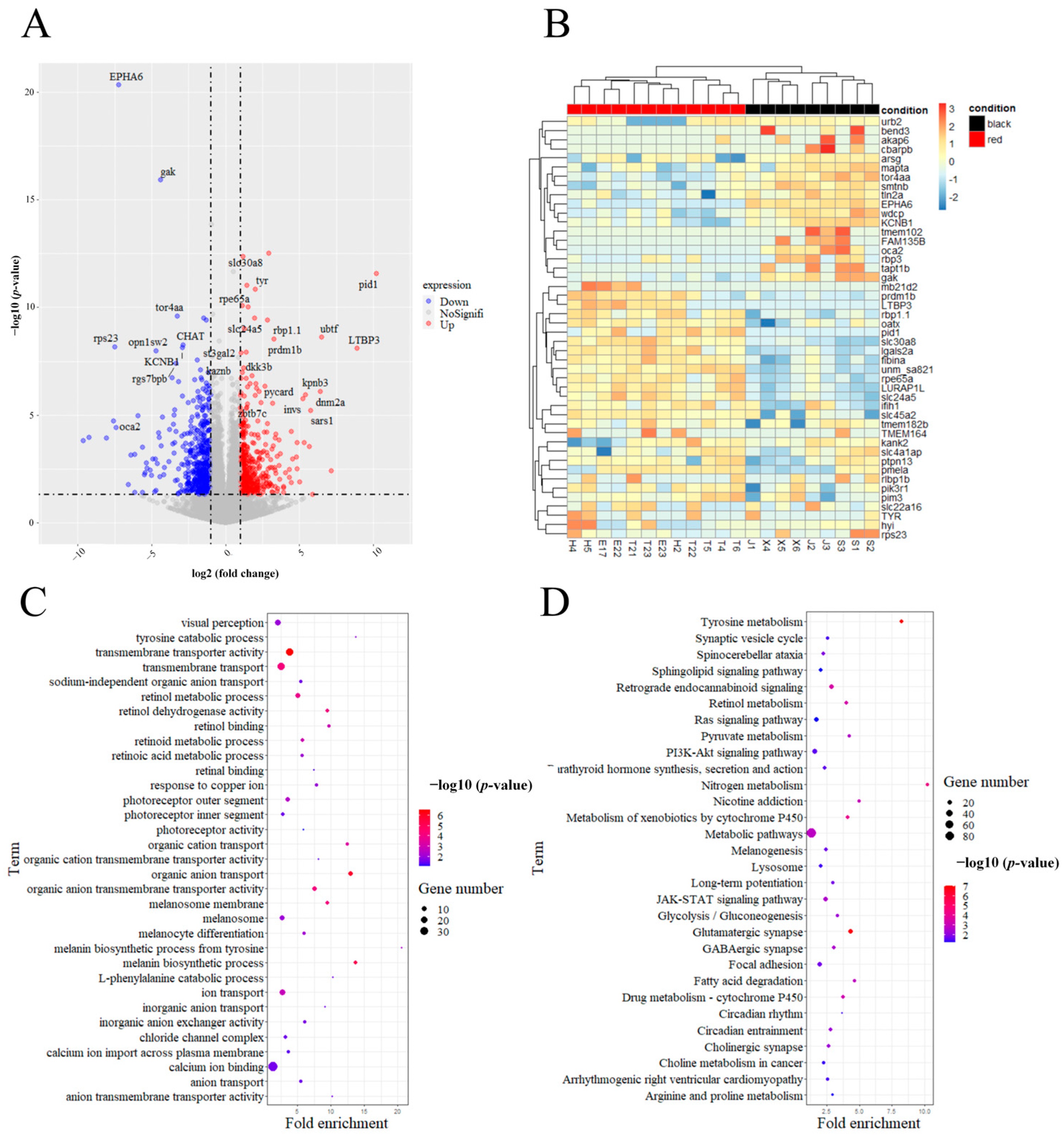
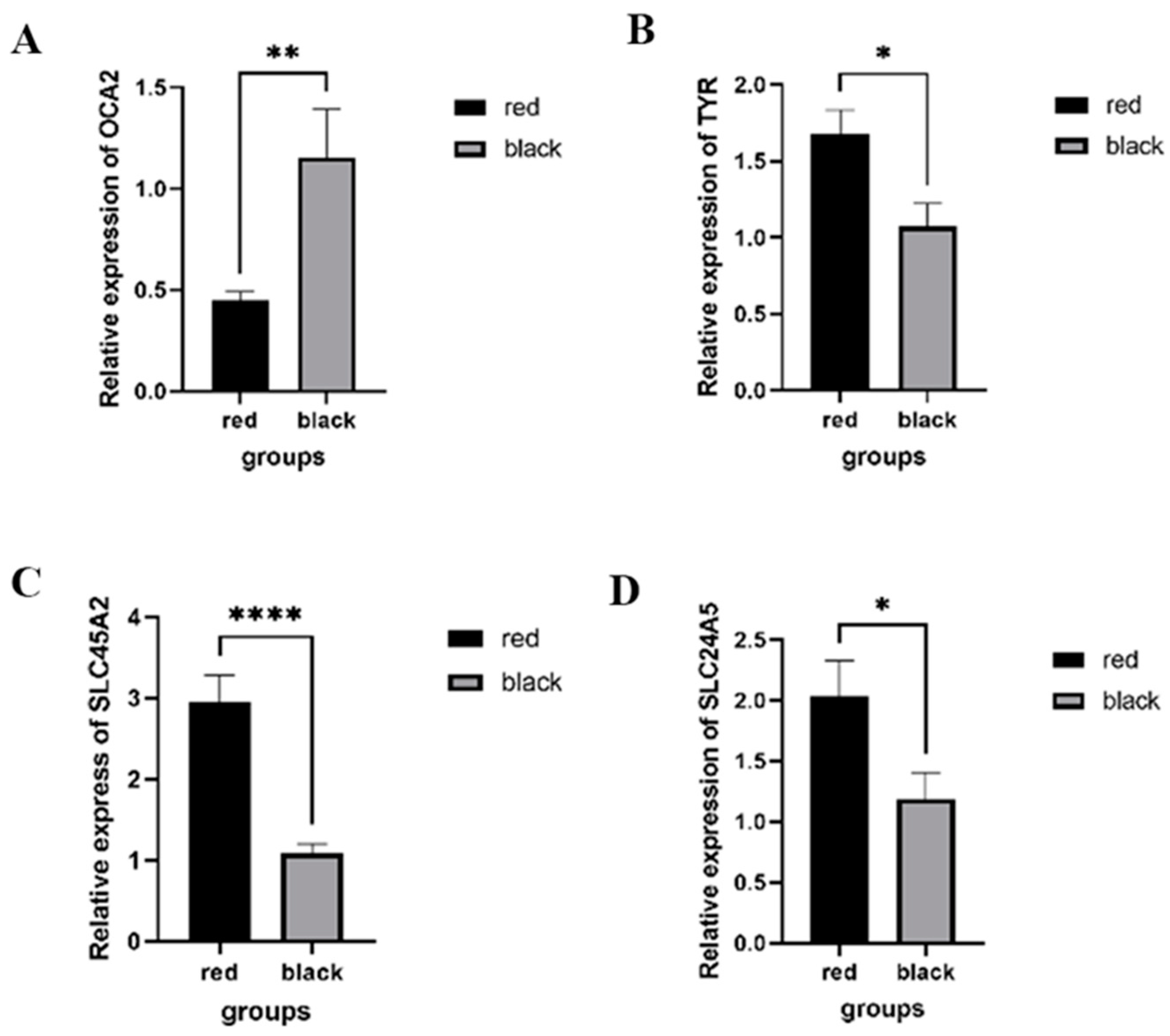
2.3. RNA Sequencing Analysis and Quantitative Real-Time PCR
3. Discussion
4. Materials and Methods
4.1. Sampling and Sequencing
4.2. Genomic Variant Calling
4.3. Genome-Wide Association Studies
4.4. RNA Sequencing Analysis
4.5. Quantitative Real-Time PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamamoto, T.-O. The Medaka, Oryzias latipes, and the Guppy, Lebistes reticularis. In Handbook of Genetics: Volume 4 Vertebrates of Genetic Interest; King, R.C., Ed.; Springer: Boston, MA, USA, 1975; pp. 133–149. [Google Scholar]
- Breden, F. Guppies. Curr. Biol. 2006, 16, R865–R866. [Google Scholar] [CrossRef][Green Version]
- Nakamura, K.; Ozaki, A.; Akutsu, T.; Iwai, K.; Sakamoto, T.; Yoshizaki, G.; Okamoto, N. Genetic mapping of the dominant albino locus in rainbow trout (Oncorhynchus mykiss). Mol. Genet. Genom. 2001, 265, 687–693. [Google Scholar] [CrossRef]
- Fernando, A.A.; Phang, V.P.E. Culture of the guppy, Poecilia reticulata, in Singapore. Aquaculture 1985, 51, 49–63. [Google Scholar] [CrossRef]
- Reznick, D.N.; Travis, J. Experimental Studies of Evolution and Eco-Evo Dynamics in Guppies (Poecilia reticulata). Annu. Rev. Ecol. Evol. Syst. 2019, 50, 335–354. [Google Scholar] [CrossRef]
- Reznick, D.N.; Shaw, F.H.; Rodd, F.H.; Shaw, R.G. Evaluation of the Rate of Evolution in Natural Populations of Guppies (Poecilia reticulata). Science 1997, 275, 1934–1937. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, M.; Taniguchi, N. Genetics of the guppy as a model for experiment in aquaculture. Genetica 2001, 111, 279–289. [Google Scholar] [CrossRef]
- Farr, J.A. The Inheritance of Quantitative Fitness Traits in Guppies, Poecilia reticulata (Pisces: Poeciliidae). Evolution 1983, 37, 1193–1209. [Google Scholar] [CrossRef]
- Whiting, J.R.; Paris, J.R.; Parsons, P.J.; Matthews, S.; Reynoso, Y.; Hughes, K.A.; Reznick, D.; Fraser, B.A. On the genetic architecture of rapidly adapting and convergent life history traits in guppies. Heredity 2022, 128, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Endler, J.A. Natural Selection on Color Patterns in Poecilia reticulata. Evolution 1980, 34, 76–91. [Google Scholar] [CrossRef]
- Reznick, D.N.; Bryga, H. Life-History Evolution in Guppies (Poecilia reticulata): 1. Phenotypic and genetic changes in an introduction experiment. Evolution 1987, 41, 1370–1385. [Google Scholar]
- Endler, J.A.; Houde, A.E. Geographic Variation in Female Preferences for Male Traits in Poecilia reticulata. Evolution 1995, 49, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Kottler, V.A.; Künstner, A.; Koch, I.; Flötenmeyer, M.; Langenecker, T.; Hoffmann, M.; Sharma, E.; Weigel, D.; Dreyer, C. Adenylate cyclase 5 is required for melanophore and male pattern development in the guppy (Poecilia reticulata). Pigment Cell Melanoma Res. 2015, 28, 545–558. [Google Scholar] [CrossRef]
- Federico, J.R.; Krishnamurthy, K. Albinism. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2023. [Google Scholar]
- Imesch, P.D.; Wallow, I.H.; Albert, D.M. The color of the human eye: A review of morphologic correlates and of some conditions that affect iridial pigmentation. Surv. Ophthalmol. 1997, 41 (Suppl. S2), S117–S123. [Google Scholar] [CrossRef] [PubMed]
- Richardson, R.; Tracey-White, D.; Webster, A.; Moosajee, M. The zebrafish eye-a paradigm for investigating human ocular genetics. Eye 2017, 31, 68–86. [Google Scholar] [CrossRef]
- Kruijt, B.; Franssen, L.; Prick, L.J.; van Vliet, J.M.; van den Berg, T.J. Ocular straylight in albinism. Optom. Vis. Sci. 2011, 88, E585–E592. [Google Scholar] [CrossRef]
- Grønskov, K.; Ek, J.; Brondum-Nielsen, K. Oculocutaneous albinism. Orphanet J. Rare Dis. 2007, 2, 43. [Google Scholar] [CrossRef]
- Bakker, R.; Wagstaff, E.L.; Kruijt, C.C.; Emri, E.; van Karnebeek, C.D.M.; Hoffmann, M.B.; Brooks, B.P.; Boon, C.J.F.; Montoliu, L.; van Genderen, M.M.; et al. The retinal pigmentation pathway in human albinism: Not so black and white. Prog. Retin. Eye Res. 2022, 91, 101091. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, H.; Bessho, Y.; Koga, A.; Hori, H. Expression of the tyrosinase-encoding gene in a colorless melanophore mutant of the medaka fish, Oryzias latipes. Gene 1994, 150, 319–324. [Google Scholar] [PubMed]
- Inagaki, H.; Koga, A.; Bessho, Y.; Hori, H. The tyrosinase gene from medakafish: Transgenic expression rescues albino mutation. Pigment Cell Res. 1998, 11, 283–290. [Google Scholar] [CrossRef]
- Koga, A.; Inagaki, H.; Bessho, Y.; Hori, H. Insertion of a novel transposable element in the tyrosinase gene is responsible for an albino mutation in the medaka fish, Oryzias latipes. Mol. Gen. Genet. 1995, 249, 400–405. [Google Scholar] [CrossRef]
- Koga, A.; Wakamatsu, Y.; Kurosawa, J.; Hori, H. Oculocutaneous albinism in the i6 mutant of the medaka fish is associated with a deletion in the tyrosinase gene. Pigment Cell Res. 1999, 12, 252–258. [Google Scholar] [CrossRef]
- Hyodo-Taguchi, Y.; Winkler, C.; Kurihara, Y.; Schartl, A.; Schartl, M. Phenotypic rescue of the albino mutation in the medakafish (Oryzias latipes) by a mouse tyrosinase transgene. Mech. Dev. 1997, 68, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Iida, A.; Inagaki, H.; Suzuki, M.; Wakamatsu, Y.; Hori, H.; Koga, A. The tyrosinase gene of the i(b) albino mutant of the medaka fish carries a transposable element insertion in the promoter region. Pigment Cell Res. 2004, 17, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Link, B.A.; Collery, R.F. Zebrafish Models of Retinal Disease. Annu. Rev. Vis. Sci. 2015, 1, 125–153. [Google Scholar] [CrossRef] [PubMed]
- Neuffer, S.J.; Cooper, C.D. Zebrafish Syndromic Albinism Models as Tools for Understanding and Treating Pigment Cell Disease in Humans. Cancers 2022, 14, 1752. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, T.; O’Connor, M.N.; Seabra, M.C.; Cutler, D.F.; Futter, C.E. Regulation of melanosome number, shape and movement in the zebrafish retinal pigment epithelium by OA1 and PMEL. J. Cell Sci. 2015, 128, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Page-McCaw, P.S.; Chung, S.C.; Muto, A.; Roeser, T.; Staub, W.; Finger-Baier, K.C.; Korenbrot, J.I.; Baier, H. Retinal network adaptation to bright light requires tyrosinase. Nat. Neurosci. 2004, 7, 1329–1336. [Google Scholar] [CrossRef] [PubMed]
- Haffter, P.; Odenthal, J.; Mullins, M.C.; Lin, S.; Farrell, M.J.; Vogelsang, E.; Haas, F.; Brand, M.; van Eeden, F.J.; Furutani-Seiki, M.; et al. Mutations affecting pigmentation and shape of the adult zebrafish. Dev. Genes Evol. 1996, 206, 260–276. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Liedtke, D.; Volff, J.N.; Schartl, M. Pigmentary function and evolution of tyrp1 gene duplicates in fish. Pigment Cell Melanoma Res. 2009, 22, 839–850. [Google Scholar] [CrossRef]
- Beirl, A.J.; Linbo, T.H.; Cobb, M.J.; Cooper, C.D. oca2 Regulation of chromatophore differentiation and number is cell type specific in zebrafish. Pigment Cell Melanoma Res. 2014, 27, 178–189. [Google Scholar] [CrossRef]
- Tsetskhladze, Z.R.; Canfield, V.A.; Ang, K.C.; Wentzel, S.M.; Reid, K.P.; Berg, A.S.; Johnson, S.L.; Kawakami, K.; Cheng, K.C. Functional assessment of human coding mutations affecting skin pigmentation using zebrafish. PLoS ONE 2012, 7, e47398. [Google Scholar] [CrossRef]
- Lamason, R.L.; Mohideen, M.A.; Mest, J.R.; Wong, A.C.; Norton, H.L.; Aros, M.C.; Jurynec, M.J.; Mao, X.; Humphreville, V.R.; Humbert, J.E.; et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 2005, 310, 1782–1786. [Google Scholar] [CrossRef] [PubMed]
- Grønskov, K.; Dooley, C.M.; Østergaard, E.; Kelsh, R.N.; Hansen, L.; Levesque, M.P.; Vilhelmsen, K.; Møllgård, K.; Stemple, D.L.; Rosenberg, T. Mutations in c10orf11, a melanocyte-differentiation gene, cause autosomal-recessive albinism. Am. J. Hum. Genet. 2013, 92, 415–421. [Google Scholar] [CrossRef]
- Daly, C.M.; Willer, J.; Gregg, R.; Gross, J.M. snow white, a zebrafish model of Hermansky-Pudlak Syndrome type 5. Genetics 2013, 195, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Van Gele, M.; Dynoodt, P.; Lambert, J. Griscelli syndrome: A model system to study vesicular trafficking. Pigment Cell Melanoma Res. 2009, 22, 268–282. [Google Scholar] [CrossRef] [PubMed]
- Sheets, L.; Ransom, D.G.; Mellgren, E.M.; Johnson, S.L.; Schnapp, B.J. Zebrafish melanophilin facilitates melanosome dispersion by regulating dynein. Curr. Biol. 2007, 17, 1721–1734. [Google Scholar] [CrossRef]
- Maldonado, E.; Hernandez, F.; Lozano, C.; Castro, M.E.; Navarro, R.E. The zebrafish mutant vps18 as a model for vesicle-traffic related hypopigmentation diseases. Pigment Cell Res. 2006, 19, 315–326. [Google Scholar] [CrossRef]
- Bahadori, R.; Rinner, O.; Schonthaler, H.B.; Biehlmaier, O.; Makhankov, Y.V.; Rao, P.; Jagadeeswaran, P.; Neuhauss, S.C. The Zebrafish fade out mutant: A novel genetic model for Hermansky-Pudlak syndrome. Investig. Ophthalmol. Vis. Sci. 2006, 47, 4523–4531. [Google Scholar] [CrossRef]
- Schonthaler, H.B.; Fleisch, V.C.; Biehlmaier, O.; Makhankov, Y.; Rinner, O.; Bahadori, R.; Geisler, R.; Schwarz, H.; Neuhauss, S.C.; Dahm, R. The zebrafish mutant lbk/vam6 resembles human multisystemic disorders caused by aberrant trafficking of endosomal vesicles. Development 2008, 135, 387–399. [Google Scholar] [CrossRef]
- Thomas, J.L.; Vihtelic, T.S.; denDekker, A.D.; Willer, G.; Luo, X.; Murphy, T.R.; Gregg, R.G.; Hyde, D.R.; Thummel, R. The loss of vacuolar protein sorting 11 (vps11) causes retinal pathogenesis in a vertebrate model of syndromic albinism. Investig. Ophthalmol. Vis. Sci. 2011, 52, 3119–3128. [Google Scholar] [CrossRef]
- Kon, T.; Omori, Y.; Fukuta, K.; Wada, H.; Watanabe, M.; Chen, Z.; Iwasaki, M.; Mishina, T.; Matsuzaki, S.S.; Yoshihara, D.; et al. The Genetic Basis of Morphological Diversity in Domesticated Goldfish. Curr. Biol. 2020, 30, 2260–2274.e2266. [Google Scholar] [CrossRef]
- Yu, P.; Wang, Y.; Li, Z.; Jin, H.; Li, L.L.; Han, X.; Wang, Z.W.; Yang, X.L.; Li, X.Y.; Zhang, X.J.; et al. Causal gene identification and desirable trait recreation in goldfish. Sci. China Life Sci. 2022, 65, 2341–2353. [Google Scholar] [CrossRef] [PubMed]
- Haskins, C.P.; Haskins, E.F. Albinism, a semi-lethal autosomal mutation in Lebistes reticulatus. Heredity 1948, 2, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Wiriyasermkul, P.; Moriyama, S.; Nagamori, S. Membrane transport proteins in melanosomes: Regulation of ions for pigmentation. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183318. [Google Scholar] [CrossRef]
- Xu, B.; Chen, X.; Li, H. Mutational Analysis of TYR, OCA2, SLC45A2, and TYRP1 Genes Identifies Novel and Reported Mutations in Chinese Families with Oculocutaneous Albinism. Altern. Ther. Health Med. 2023, 29, 278–283. [Google Scholar]
- Schnetkamp, P.P. The SLC24 gene family of Na⁺/Ca²⁺-K⁺ exchangers: From sight and smell to memory consolidation and skin pigmentation. Mol. Aspects Med. 2013, 34, 455–464. [Google Scholar] [CrossRef]
- Dessinioti, C.; Antoniou, C.; Katsambas, A.; Stratigos, A.J. Melanocortin 1 receptor variants: Functional role and pigmentary associations. Photochem. Photobiol. 2011, 87, 978–987. [Google Scholar] [CrossRef] [PubMed]
- Oetting, W.S.; King, R.A. Molecular basis of albinism: Mutations and polymorphisms of pigmentation genes associated with albinism. Hum. Mutat. 1999, 13, 99–115. [Google Scholar] [CrossRef]
- Sun, L.; Hu, L.; Zhang, P.; Li, H.; Sun, J.; Wang, H.; Xie, X.; Hu, J. Silencing of PMEL attenuates melanization via activating lysosomes and degradation of tyrosinase by lysosomes. Biochem. Biophys. Res. Commun. 2018, 503, 2536–2542. [Google Scholar] [CrossRef]
- Lai, X.; Wichers, H.J.; Soler-Lopez, M.; Dijkstra, B.W. Structure and Function of Human Tyrosinase and Tyrosinase-Related Proteins. Chemistry 2018, 24, 47–55. [Google Scholar] [CrossRef]
- Hearing, V.J.; Tsukamoto, K. Enzymatic control of pigmentation in mammals. FASEB J. 1991, 5, 2902–2909. [Google Scholar] [CrossRef] [PubMed]
- Ancans, J.; Hoogduijn, M.J.; Thody, A.J. Melanosomal pH, pink locus protein and their roles in melanogenesis. J. Investig. Dermatol. 2001, 117, 158–159. [Google Scholar] [CrossRef] [PubMed]
- Ancans, J.; Tobin, D.J.; Hoogduijn, M.J.; Smit, N.P.; Wakamatsu, K.; Thody, A.J. Melanosomal pH controls rate of melanogenesis, eumelanin/phaeomelanin ratio and melanosome maturation in melanocytes and melanoma cells. Exp. Cell Res. 2001, 268, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Suzuki, N.; Takebayashi, S.; Commo, S.; Wakamatsu, K. Neutral pH and copper ions promote eumelanogenesis after the dopachrome stage. Pigment Cell Melanoma Res. 2013, 26, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Rzepka, Z.; Buszman, E.; Beberok, A.; Wrześniok, D. From tyrosine to melanin: Signaling pathways and factors regulating melanogenesis. Postepy Hig. Med. Dosw. 2016, 70, 695–708. [Google Scholar] [CrossRef] [PubMed]
- Riley, P.A. Mechanistic aspects of the control of tyrosinase activity. Pigment Cell Res. 1993, 6, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Pavan, W.J.; Sturm, R.A. The Genetics of Human Skin and Hair Pigmentation. Annu. Rev. Genom. Hum. Genet. 2019, 20, 41–72. [Google Scholar] [CrossRef]
- Solano, F. On the Metal Cofactor in the Tyrosinase Family. Int. J. Mol. Sci. 2018, 19, 633. [Google Scholar] [CrossRef]
- Pfefferkorn, C.M.; McGlinchey, R.P.; Lee, J.C. Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc. Natl. Acad. Sci. USA 2010, 107, 21447–21452. [Google Scholar] [CrossRef]
- Le, L.; Sirés-Campos, J.; Raposo, G.; Delevoye, C.; Marks, M.S. Melanosome Biogenesis in the Pigmentation of Mammalian Skin. Integr. Comp. Biol. 2021, 61, 1517–1545. [Google Scholar] [CrossRef]
- Bellono, N.W.; Oancea, E.V. Ion transport in pigmentation. Arch. Biochem. Biophys. 2014, 563, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Bellono, N.W.; Escobar, I.E.; Lefkovith, A.J.; Marks, M.S.; Oancea, E. An intracellular anion channel critical for pigmentation. eLife 2014, 3, e04543. [Google Scholar] [CrossRef]
- Chao, Y.K.; Schludi, V.; Chen, C.C.; Butz, E.; Nguyen, O.N.P.; Müller, M.; Krüger, J.; Kammerbauer, C.; Ben-Johny, M.; Vollmar, A.M.; et al. TPC2 polymorphisms associated with a hair pigmentation phenotype in humans result in gain of channel function by independent mechanisms. Proc. Natl. Acad. Sci. USA 2017, 114, E8595–E8602. [Google Scholar] [CrossRef]
- Bartölke, R.; Heinisch, J.J.; Wieczorek, H.; Vitavska, O. Proton-associated sucrose transport of mammalian solute carrier family 45: An analysis in Saccharomyces cerevisiae. Biochem. J. 2014, 464, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Dooley, C.M.; Schwarz, H.; Mueller, K.P.; Mongera, A.; Konantz, M.; Neuhauss, S.C.; Nüsslein-Volhard, C.; Geisler, R. Slc45a2 and V-ATPase are regulators of melanosomal pH homeostasis in zebrafish, providing a mechanism for human pigment evolution and disease. Pigment Cell Melanoma Res. 2013, 26, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Rinchik, E.M.; Bultman, S.J.; Horsthemke, B.; Lee, S.T.; Strunk, K.M.; Spritz, R.A.; Avidano, K.M.; Jong, M.T.; Nicholls, R.D. A gene for the mouse pink-eyed dilution locus and for human type II oculocutaneous albinism. Nature 1993, 361, 72–76. [Google Scholar] [CrossRef]
- Chi, A.; Valencia, J.C.; Hu, Z.Z.; Watabe, H.; Yamaguchi, H.; Mangini, N.J.; Huang, H.; Canfield, V.A.; Cheng, K.C.; Yang, F.; et al. Proteomic and bioinformatic characterization of the biogenesis and function of melanosomes. J. Proteome Res. 2006, 5, 3135–3144. [Google Scholar] [CrossRef]
- Ambrosio, A.L.; Boyle, J.A.; Aradi, A.E.; Christian, K.A.; Di Pietro, S.M. TPC2 controls pigmentation by regulating melanosome pH and size. Proc. Natl. Acad. Sci. USA 2016, 113, 5622–5627. [Google Scholar] [CrossRef]
- Brilliant, M.H. The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigment Cell Res. 2001, 14, 86–93. [Google Scholar] [CrossRef]
- Lyon, M.F.; King, T.R.; Gondo, Y.; Gardner, J.M.; Nakatsu, Y.; Eicher, E.M.; Brilliant, M.H. Genetic and molecular analysis of recessive alleles at the pink-eyed dilution (p) locus of the mouse. Proc. Natl. Acad. Sci. USA 1992, 89, 6968–6972. [Google Scholar] [CrossRef]
- Loftus, S.K.; Lundh, L.; Watkins-Chow, D.E.; Baxter, L.L.; Pairo-Castineira, E.; Nisc Comparative Sequencing, P.; Jackson, I.J.; Oetting, W.S.; Pavan, W.J.; Adams, D.R. A custom capture sequence approach for oculocutaneous albinism identifies structural variant alleles at the OCA2 locus. Hum. Mutat. 2021, 42, 1239–1253. [Google Scholar] [CrossRef]
- Sturm, R.A.; Frudakis, T.N. Eye colour: Portals into pigmentation genes and ancestry. Trends Genet. 2004, 20, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Sturm, R.A.; Larsson, M. Genetics of human iris colour and patterns. Pigment Cell Melanoma Res. 2009, 22, 544–562. [Google Scholar] [CrossRef] [PubMed]
- Eiberg, H.; Troelsen, J.; Nielsen, M.; Mikkelsen, A.; Mengel-From, J.; Kjaer, K.W.; Hansen, L. Blue eye color in humans may be caused by a perfectly associated founder mutation in a regulatory element located within the HERC2 gene inhibiting OCA2 expression. Hum. Genet. 2008, 123, 177–187. [Google Scholar] [CrossRef]
- Sturm, R.A.; Duffy, D.L.; Zhao, Z.Z.; Leite, F.P.; Stark, M.S.; Hayward, N.K.; Martin, N.G.; Montgomery, G.W. A single SNP in an evolutionary conserved region within intron 86 of the HERC2 gene determines human blue-brown eye color. Am. J. Hum. Genet. 2008, 82, 424–431. [Google Scholar] [CrossRef] [PubMed]
- White, D.; Rabago-Smith, M. Genotype-phenotype associations and human eye color. J. Hum. Genet. 2011, 56, 5–7. [Google Scholar] [CrossRef]
- Visser, M.; Kayser, M.; Palstra, R.J. HERC2 rs12913832 modulates human pigmentation by attenuating chromatin-loop formation between a long-range enhancer and the OCA2 promoter. Genome Res. 2012, 22, 446–455. [Google Scholar] [CrossRef]
- Sun, D.; Qi, X.; Wen, H.; Li, C.; Li, J.; Chen, J.; Tao, Z.; Zhu, M.; Zhang, X.; Li, Y. The genetic basis and potential molecular mechanism of yellow-albino northern snakehead (Channa argus). Open Biol. 2023, 13, 220235. [Google Scholar] [CrossRef]
- Fu, C.; Chen, J.; Lu, J.; Pei, S.; Hu, S.; Jiang, L.; Ding, Y.; Huang, L.; Xiang, H.; Huang, J.; et al. Downregulation of TUG1 promotes melanogenesis and UVB-induced melanogenesis. Exp. Dermatol. 2019, 28, 730–733. [Google Scholar] [CrossRef]
- Hu, S.; Huang, J.; Pei, S.; Ouyang, Y.; Ding, Y.; Jiang, L.; Lu, J.; Kang, L.; Huang, L.; Xiang, H.; et al. Ganoderma lucidum polysaccharide inhibits UVB-induced melanogenesis by antagonizing cAMP/PKA and ROS/MAPK signaling pathways. J. Cell Physiol. 2019, 234, 7330–7340. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction: Twenty-something years on. Nat. Protoc. 2006, 1, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Sandkam, B.; Young, C.M.; Breden, F. Beauty in the eyes of the beholders: Colour vision is tuned to mate preference in the Trinidadian guppy (Poecilia reticulata). Mol. Ecol. 2015, 24, 596–609. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Chr | Position (bp) | N_Sig a | Lead Variant b | p c | Genomic Location | Corresponding Genes |
|---|---|---|---|---|---|---|
| 4 | 13,919,679 | 1 | 13,919,679 | 2.13 × 10−8 | intergenic | NLGN1-NAALADL2 |
| 4 | 15,579,097–157,04,885 | 2 | 155,79,097 | 3.34 × 10−9 | intergenic | BARHL2-LRRC8DB |
| 4 | 15,728,116 | 1 | 157,28,116 | 3.05 × 10−8 | upstream | LRRC8C |
| 4 | 15,735,128 | 1 | 15,735,128 | 3.23 × 10−8 | intergenic | LRRC8C-KYAT3 |
| 4 | 15,815,011–15,929,479 | 28 | 15,908,422 | 2.87 × 10−13 | intergenic | KYAT3-LMO4B |
| 4 | 16,159,568 | 1 | 16,159,568 | 2.36 × 10−8 | intergenic | SOX14-HDLBPA |
| 4 | 16,365,953–16,388,992 | 5 | 16,379,136 | 1.89 × 10−10 | upstream; intron; intergenic; downstream | BOKA |
| 4 | 16,384,579–16,391,411 | 3 | 16,385,964 | 1.03 × 10−10 | upstream; intron; | ATG4B |
| 4 | 16,394,845–16,403,441 | 2 | 16,403,441 | 8.71 × 10−10 | upstream; downstream | DTYMK |
| 4 | 16,410,980–16,411,725 | 2 | 16,411,725 | 1.31 × 10−8 | upstream | AGXTA |
| 4 | 16,430,204 | 1 | 16,430,204 | 2.34 × 10−8 | downstream | TTC39C |
| 4 | 16,431,698–16,455,327 | 6 | 16,431,698 | 9.91 × 10−11 | upstream; intron | KIF1AA |
| 4 | 16,525,371–16,566,144 | 3 | 16,525,371 | 1.41 × 10−9 | intergenic | KLHL30-SNED1 |
| 4 | 16,574,314–16,600,468 | 3 | 16,574,314 | 6.58 × 10−9 | intron; CDS | SNED1 |
| 4 | 16,613,965–16,632,460 | 2 | 16,632,460 | 1.25 × 10−9 | intron | RYK |
| 4 | 16,675,710 | 1 | 16,675,710 | 3.32 × 10−9 | intron | SLCO2A1 |
| 4 | 16,722,251 | 1 | 16,722,251 | 3.54 × 10−8 | intron | RAB6BA |
| 4 | 16,749,558–16,756,105 | 7 | 16,752,127 | 5.82 × 10−11 | intron; downstream | CEP63 |
| 4 | 16,814,130 | 1 | 16,814,130 | 2.90 × 10−8 | upstream | SAP130A |
| 4 | 16,920,982–16,976,594 | 2 | 16,920,982 | 3.33 × 10−8 | intergenic | BCL6AA-LPP |
| 4 | 17,001,755 | 1 | 17,001,755 | 3.76 × 10−8 | CDS | LPP |
| 4 | 17,192,862 | 1 | 17,192,862 | 6.27 × 10−9 | intron | TPRG1 |
| 4 | 17,243,325–17,250,077 | 2 | 17,250,077 | 9.00 × 10−9 | intergenic | TPRG1-TP63 |
| 4 | 17,250,825–17,282,572 | 3 | 17,282,572 | 1.05 × 10−10 | upstream; intron; downstream | TP63 |
| 4 | 17,276,094–17,292,722 | 5 | 17,289,603 | 6.20 × 10−15 | upstream; downstream | ZBTB11 |
| 4 | 17,295,315–17,299,997 | 2 | 17,299,997 | 2.71 × 10−8 | intergenic | ZBTB11-DPT |
| 4 | 17,302,046–17,310,860 | 3 | 17,302,046 | 2.30 × 10−17 | downstream; CDS; intron | DPT |
| 4 | 17,317,551–17,328,988 | 8 | 17,320,328 | 1.89 × 10−16 | intergenic | DPT-ATP1B1A |
| 4 | 17,331,317–17,348,850 | 7 | 17,348,850 | 1.66 × 10−15 | upstream; intron; downstream | ATP1B1A |
| 4 | 17,356,626–17,372,435 | 8 | 17,369,146 | 2.26 × 10−17 | downstream | NME7 |
| 4 | 17,366,454–17,378,171 | 3 | 17,378,171 | 7.54 × 10−16 | downstream | ZGC:172121 |
| 4 | 17,381,538–17,383,905 | 3 | 17,381,538 | 4.39 × 10−13 | intron; CDS | BIVM |
| 4 | 17,398,644 | 1 | 17,398,644 | 1.57 × 10−11 | downstream | SI:CH211-201H21.5 |
| 4 | 17,402,571–17,433,938 | 12 | 17,433,938 | 4.80 × 10−13 | intergenic; upstream | TMEM131 |
| 4 | 17,446,682–17,446,995 | 2 | 17,446,682 | 2.20 × 10−9 | intergenic | TMEM131-CNGA3A |
| 4 | 17,458,944 | 1 | 17,458,944 | 3.36 × 10−8 | downstream | UBE3A |
| 4 | 17,461,407–17,462,566 | 2 | 17,462,566 | 3.36 × 10−8 | downstream | CNGA3A |
| 4 | 17,511,176–17,516,034 | 3 | 17,514,805 | 2.46 × 10−8 | intron | ATP10A |
| 4 | 17,548,494 | 1 | 17,548,494 | 4.67 × 10−9 | intergenic | ATP10A-GABRB3 |
| 4 | 17,558,221 | 1 | 17,558,221 | 2.94 × 10−8 | intron | GABRB3 |
| 4 | 17,594,093 | 1 | 17,594,093 | 3.29 × 10−8 | intron | GABRA5 |
| 4 | 17,626,784–17,660,211 | 2 | 17,660,211 | 2.66 × 10−9 | intron | GABRG3 |
| 4 | 17,703,618–17,709,085 | 2 | 17,703,618 | 1.26 × 10−8 | intron | OCA2 |
| 4 | 17,770,644 | 1 | 17,770,644 | 1.25 × 10−8 | CDS | HERC2 |
| 4 | 18,167,098 | 1 | 18,167,098 | 2.68 × 10−9 | intron | ENSPREG00000018047 |
| Primers | Forward Primer | Reverse Primer | References | Tm | Product Size (bp) |
|---|---|---|---|---|---|
| β-actin | gcttgtgcgggatatcatttg | gaatccggctttgcacatac | NM_001297475.1 | 60 °C | 137 |
| OCA2-2 | cagactttcgggataacgcct | gagcactcctcctccgct | XM_008407632.2 | 60 °C | 141 |
| TYR-2 | ctccatgtccaacgtccagg | catttgctcgtgggtagctg | XM_008425495.2 | 60 °C | 131 |
| SLC45A2-2 | gagaggtctgcactaccacg | gtactcggagcccaacagac | XM_008423772.2 | 60 °C | 115 |
| SLC24A5-2 | ttctcaggatgtggcaggag | tgctgattccaatgtccccc | XM_008404588.2 | 60 °C | 110 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.; Wu, S.; Li, J.; Bao, H.; Wu, C. Identification of Candidate Genes for Red-Eyed (Albinism) Domestic Guppies Using Genomic and Transcriptomic Analyses. Int. J. Mol. Sci. 2024, 25, 2175. https://doi.org/10.3390/ijms25042175
Chang Y, Wu S, Li J, Bao H, Wu C. Identification of Candidate Genes for Red-Eyed (Albinism) Domestic Guppies Using Genomic and Transcriptomic Analyses. International Journal of Molecular Sciences. 2024; 25(4):2175. https://doi.org/10.3390/ijms25042175
Chicago/Turabian StyleChang, Ying, Shenjun Wu, Junying Li, Haigang Bao, and Changxin Wu. 2024. "Identification of Candidate Genes for Red-Eyed (Albinism) Domestic Guppies Using Genomic and Transcriptomic Analyses" International Journal of Molecular Sciences 25, no. 4: 2175. https://doi.org/10.3390/ijms25042175
APA StyleChang, Y., Wu, S., Li, J., Bao, H., & Wu, C. (2024). Identification of Candidate Genes for Red-Eyed (Albinism) Domestic Guppies Using Genomic and Transcriptomic Analyses. International Journal of Molecular Sciences, 25(4), 2175. https://doi.org/10.3390/ijms25042175






