Production of Bioactive Porcine Lactoferrin through a Novel Glucose-Inducible Expression System in Pichia pastoris: Unveiling Antimicrobial and Anticancer Functionalities
Abstract
1. Introduction
2. Results
2.1. Small- and Large-Scale Expression of rpLF in P. pastoris GS115
2.2. Purification of rpLF
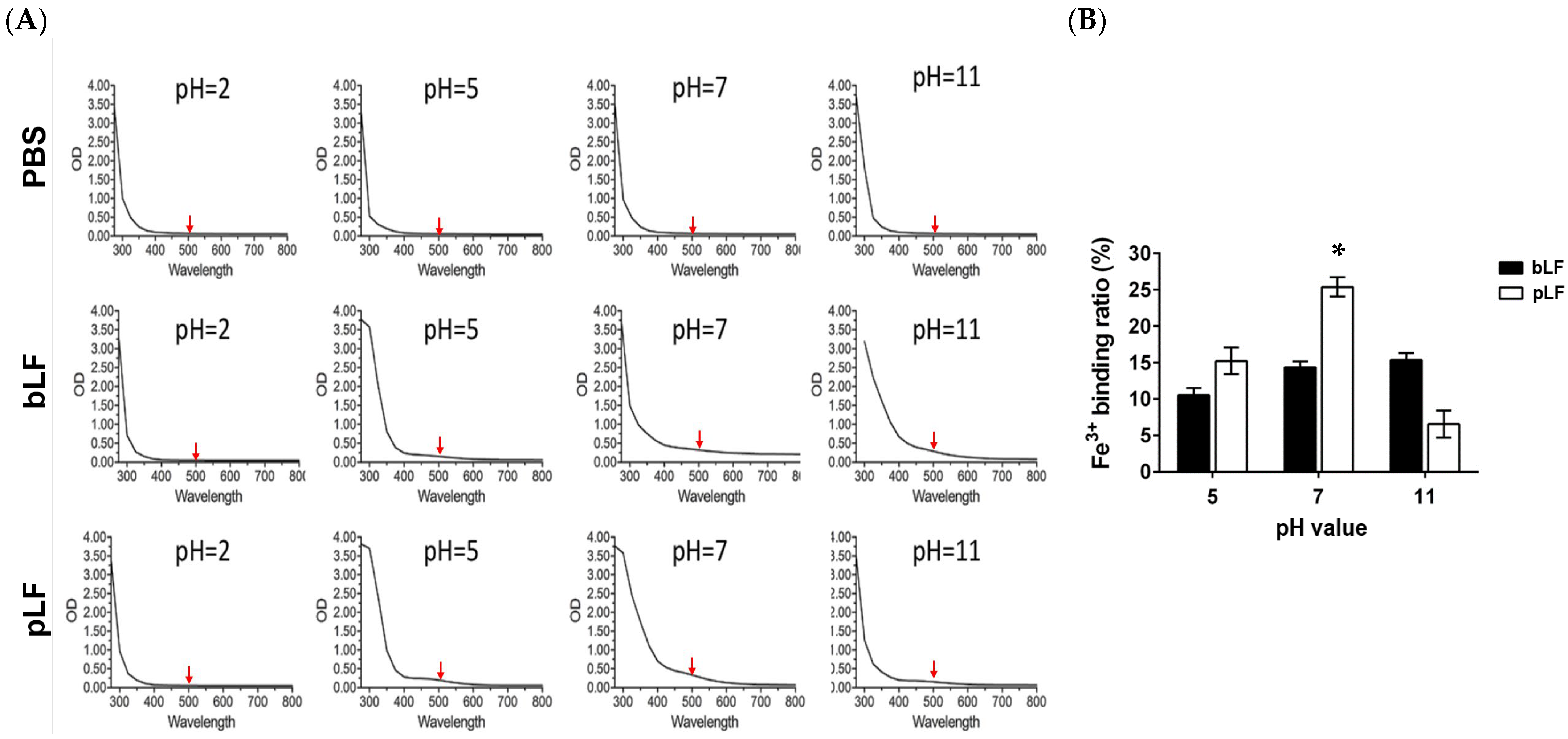
2.3. Iron (Fe3+)-Binding Activity of rpLF
2.4. Simulated Gastrointestinal Digestion of rpLF
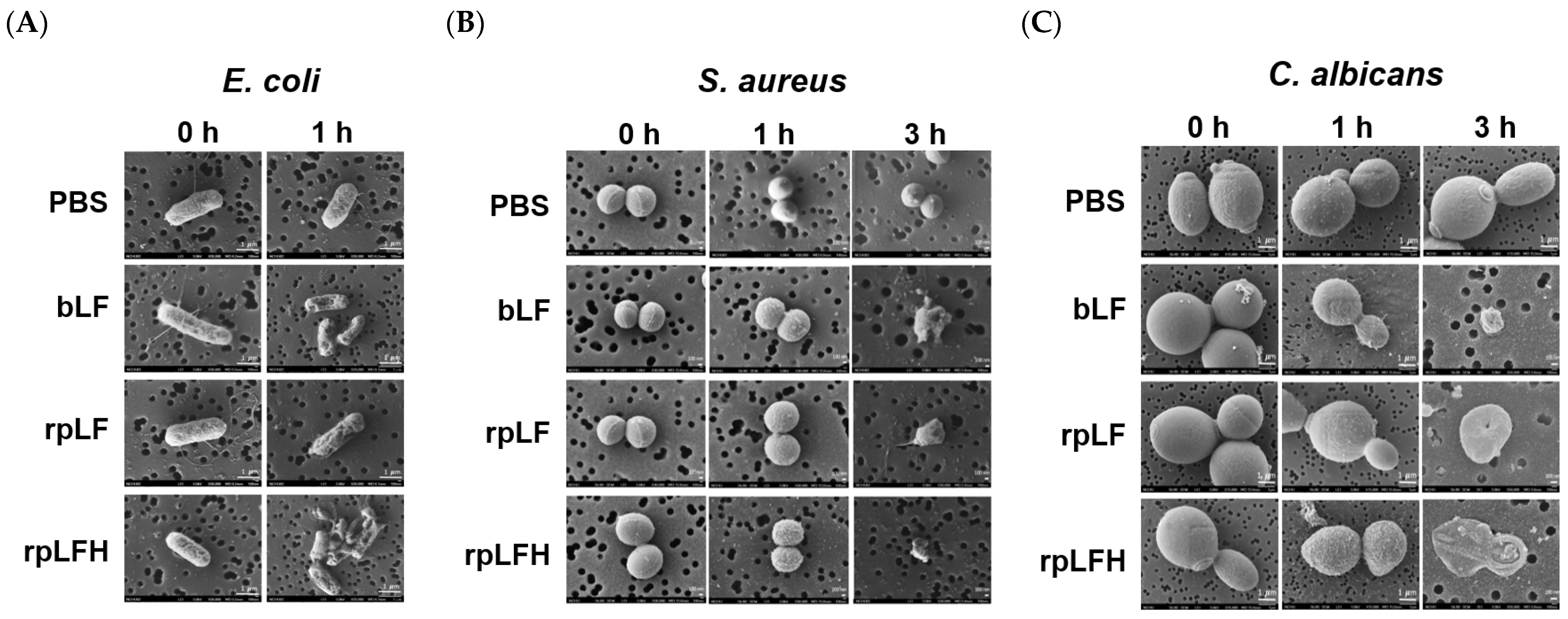
2.5. Antimicrobial Activities of rpLF and rpLFH
2.6. Anticancer Activities of rpLF and rpLFH
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction, Transformation, and High-Copy-Number Clone Selection
4.2. Small- and Large-Scale Expression of rpLF
4.3. rpLF Purification
4.4. Reverse Transcriptase–Polymerase Chain Reaction (RT-PCR)
4.5. rpLF Hydrolysate Preparation
4.6. Iron (Fe3+)-Binding Assay
4.7. Microorganisms and Cell Lines
4.8. Antimicrobial Assay and Microscopic Observation
4.9. Cell Viability and Apoptotic Assays
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bai, L.; Qiao, M.; Zheng, R.; Deng, C.; Mei, S.; Chen, W. Phylogenomic analysis of transferrin family from animals and plants. Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 17, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.B.; Iigo, M.; Yamauchi, K.; Suzui, M.; Tsuda, H. Lactoferrin: An alternative view of its role in human biological fluids. Biochem. Cell Biol. 2012, 90, 279–306. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Franco, D.A.; Vazquez-Moreno, L.; Ramos-Clamont Montfort, G. Antimicrobial mechanisms and potential clinical application of lactoferrin. Rev. Latinoam. Microbiol. 2005, 4, 102–111. [Google Scholar]
- Ward, P.P.; Paz, E.; Conneely, O.M. Multifunctional roles of lactoferrin: A critical overview. Cell Mol. Life Sci. 2005, 62, 2540–2548. [Google Scholar] [CrossRef] [PubMed]
- Berlutti, F.; Pantanella, F.; Natalizi, T.; Frioni, A.; Paesano, R.; Polimeni, A.; Valenti, P. Antiviral properties of lactoferrin—A natural immunity molecule. Molecules 2011, 16, 6992–7018. [Google Scholar] [CrossRef] [PubMed]
- Donovan, S.M. The role of lactoferrin in gastrointestinal and immune development and function: A preclinical perspective. J. Pediatr. 2016, 173, S16–S28. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Chavez, S.A.; Arevalo-Gallegos, S.; Rascon-Cruz, Q. Lactoferrin: Structure, function and applications. Int. J. Antimicrob. Agents 2009, 33, 301.e1–301.e8. [Google Scholar] [CrossRef] [PubMed]
- van der Velden, W.J.; Blijlevens, N.M.; Donnelly, J.P. The potential role of lactoferrin and derivatives in the management of infectious and inflammatory complications of hematology patients receiving a hematopoietic stem cell transplantation. Transplant. Infect. Dis. 2008, 10, 80–89. [Google Scholar] [CrossRef]
- Conesa, C.; Calvo, M.; Sanchez, L. Recombinant human lactoferrin: A valuable protein for pharmaceutical products and functional foods. Biotechnol. Adv. 2010, 28, 831–838. [Google Scholar] [CrossRef]
- Arias, M.; McDonald, L.J.; Haney, E.F.; Nazmi, K.; Bolscher, J.G.; Vogel, H.J. Bovine and human lactoferricin peptides: Chimeras and new cyclic analogs. Biometals 2014, 27, 935–948. [Google Scholar] [CrossRef]
- Reyes-Cortes, R.; Acosta-Smith, E.; Mondragon-Flores, R.; Nazmi, K.; Bolscher, J.G.; Canizalez-Roman, A.; Leon-Sicairos, N. Antibacterial and cell penetrating effects of LFcin17-30, LFampin265-284, and LF chimera on enteroaggregative Escherichia coli. Biochem. Cell Biol. 2017, 95, 76–81. [Google Scholar] [CrossRef]
- Aguilar-Diaz, H.; Canizalez-Roman, A.; Nepomuceno-Mejia, T.; Gallardo-Vera, F.; Hornelas-Orozco, Y.; Nazmi, K.; Bolscher, J.G.; Carrero, J.C.; Leon-Sicairos, C.; Leon-Sicairos, N. Parasiticidal effect of synthetic bovine lactoferrin peptides on the enteric parasite Giardia intestinalis. Biochem. Cell Biol. 2017, 95, 82–90. [Google Scholar] [CrossRef]
- Alexander, D.B.; Iigo, M.; Abdelgied, M.; Ozeki, K.; Tanida, S.; Joh, T.; Takahashi, S.; Tsuda, H. Bovine lactoferrin and Crohn’s disease: A case study. Biochem. Cell Biol. 2017, 95, 133–141. [Google Scholar] [CrossRef]
- Ward, P.P.; Lo, J.Y.; Duke, M.; May, G.S.; Headon, D.R.; Conneely, O.M. Production of biologically active recombinant human lactoferrin in Aspergillus oryzae. Biotechnology (NY) 1992, 10, 784–789. [Google Scholar] [CrossRef]
- Sitaram, M.P.; Moloney, B.; McAbee, D.D. Prokaryotic expression of bovine lactoferrin deletion mutants that bind to the Ca2+-dependent lactoferrin receptor on isolated rat hepatocytes. Protein Expr. Purif. 1998, 14, 229–236. [Google Scholar] [CrossRef]
- Chang, J.H.; Chen, C.C.; Chao, M.C.; Chuang, Y.C.; Chang, M.C. Molecular cloning and characterization of Taiwan macaque lactoferrin. J. Med. Primatol. 2009, 38, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, Y.Z.; Wu, X.F. High level expression of functionally active human lactoferrin in silkworm larvae. J. Biotechnol. 2005, 118, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Lonnerdal, B. Expression of human milk proteins in plants. J. Am. Coll. Nutr. 2002, 21 (Suppl. S3), 218S–221S. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Richardson, T. Expression and characterization of human lactoferrin in yeast Saccharomyces cerevisiae. J. Agric. Food Chem. 1993, 41, 1800–1807. [Google Scholar] [CrossRef]
- Li, J.; Zhu, W.; Luo, M.; Ren, H.; Tang, L.; Liao, H.; Wang, Y. Molecular cloning, expression and purification of lactoferrin from Tibetan sheep mammary gland using a yeast expression system. Protein Expr. Purif. 2015, 109, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Waterham, H.R.; Digan, M.E.; Koutz, P.J.; Lair, S.V.; Cregg, J.M. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 1997, 186, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Hong, J.; Lee, H.; Park, M.; Lee, E.; Kim, C.; Choi, E.; Jung, J.; Lee, H. Translation elongation factor 1-alpha gene from Pichia pastoris: Molecular cloning, sequence, and use of its promoter. Appl. Microbiol. Biotechnol. 2007, 74, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Prielhofer, R.; Maurer, M.; Klein, J.; Wenger, J.; Kiziak, C.; Gasser, B.; Mattanovich, D. Induction without methanol: Novel regulated promoters enable high-level expression in Pichia pastoris. Microb. Cell Fact. 2013, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Tschopp, J.F.; Brust, P.F.; Cregg, J.M.; Stillman, C.A.; Gingeras, T.R. Expression of the lacZ gene from two methanol-regulated promoters in Pichia pastoris. Nucleic Acids Res. 1987, 15, 3859–3876. [Google Scholar] [CrossRef]
- Kobayashi, K.; Kuwae, S.; Ohya, T.; Ohda, T.; Ohyama, M.; Tomomitsu, K. Addition of oleic acid increases expression of recombinant human serum albumin by the AOX2 promoter in Pichia pastoris. J. Biosci. Bioeng. 2000, 89, 479–484. [Google Scholar] [CrossRef]
- Resina, D.; Serrano, A.; Valero, F.; Ferrer, P. Expression of a Rhizopus oryzae lipase in Pichia pastoris under control of the nitrogen source-regulated formaldehyde dehydrogenase promoter. J. Biotechnol. 2004, 109, 103–113. [Google Scholar] [CrossRef]
- Garrigós-Martínez, J.; Vuoristo, K.; Nieto-Taype, M.A.; Tähtiharju, J.; Uusitalo, J.; Tukiainen, P.; Schmid, C.; Tolstorukov, I.; Madden, K.; Penttilä, M.; et al. Bioprocess performance analysis of novel methanol-independent promoters for recombinant protein production with Pichia pastoris. Microb. Cell Fact. 2021, 20, 74. [Google Scholar] [CrossRef]
- Chen, H.L.; Lai, Y.W.; Yen, C.C.; Lin, Y.Y.; Lu, C.Y.; Yang, S.H.; Tsai, T.C.; Lin, Y.J.; Lin, C.W.; Chen, C.M. Production of recombinant porcine lactoferrin exhibiting antibacterial activity in methylotrophic yeast, Pichia pastoris. J. Mol. Microbiol. Biotechnol. 2004, 8, 141–149. [Google Scholar] [CrossRef]
- Buckholz, R.G.; Gleeson, M.A. Yeast systems for the commercial production of heterologous proteins. Biotechnology 1991, 9, 1067–1072. [Google Scholar] [CrossRef]
- Wang, S.H.; Yang, T.S.; Lin, S.M.; Tsai, M.S.; Wu, S.C.; Mao, S.J. Expression, characterization, and purification of recombinant porcine lactoferrin in Pichia pastoris. Protein Expr. Purif. 2002, 25, 41–49. [Google Scholar] [CrossRef]
- Choi, B.K.; Actor, J.K.; Rios, S.; d’Anjou, M.; Stadheim, T.A.; Warburton, S.; Giaccone, E.; Cukan, M.; Li, H.; Kull, A.; et al. Recombinant human lactoferrin expressed in glycoengineered Pichia pastoris: Effect of terminal N-acetylneuraminic acid on in vitro secondary humoral immune response. Glycoconj. J. 2008, 25, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.H.; Yin, L.J.; Chiang, I.H.; Jiang, S.T. Expression and purification of goat lactoferrin from Pichia pastoris expression system. J. Food Sci. 2007, 72, M67–M71. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.Y.; Zhang, Y.Z. Molecular cloning and expression of yak (Bos grunniens) lactoferrin cDNA in Pichia pastoris. Biotechnol. Lett. 2006, 28, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, M.; Saravanan, K.; Uma, K.; Sharma, S.; Singh, T.P.; Srinivasan, A. Expression, purification, and characterization of equine lactoferrin in Pichia pastoris. Protein Expr. Purif. 2002, 26, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.H.; Chen, W.M.; Huang, G.T.; Chen, Y.W.; Jiang, S.T. Expression of recombinant antibacterial lactoferricin-related peptides from Pichia pastoris expression system. J. Agri. Food Chem. 2009, 57, 9509–9515. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Figueroa, B.; Valdiviezo-Godina, N.; Siqueiros-Cendon, T.; Sinagawa-Garcia, S.; Arevalo-Gallegos, S.; Rascon-Cruz, Q. High-level expression of recombinant bovine lactoferrin in Pichia pastoris with antimicrobial activity. Int. J. Mol. Sci. 2016, 17, 902. [Google Scholar] [CrossRef] [PubMed]
- Sienkiewicz, M.; Jaskiewicz, A.; Tarasiuk, A.; Fichna, J. Lactoferrin: An overview of its main functions, immunomodulatory and antimicrobial role, and clinical significance. Crit. Rev. Food Sci. Nutr. 2022, 62, 6016–6033. [Google Scholar] [CrossRef] [PubMed]
- Tomita, M.; Bellamy, W.; Takase, M.; Yamauchi, K.; Wakabayashi, H.; Kawase, K. Potent antibacterial peptides generated by pepsin digestion of bovine lactoferrin. J. Dairy Sci. 1991, 74, 4137–4142. [Google Scholar] [CrossRef]
- van der Kraan, M.I.A.; Groenink, J.; Nazmi, K.; Veerman, E.C.I.; Bolscher, J.G.M.; Nieuw Amerongen, A.V. Lactoferrampin: A novel antimicrobial peptide in the N1-domain of bovine lactoferrin. Peptides 2004, 25, 177–183. [Google Scholar] [CrossRef]
- Chen, H.L.; Yen, C.C.; Lu, C.Y.; Yu, C.H.; Chen, C.M. Synthetic porcine lactoferricin with a 20-residue peptide exhibits antimicrobial activity against Escherichia coli, Staphylococcus aureus, and Candida albicans. J. Agri. Food Chem. 2006, 54, 3277–3282. [Google Scholar] [CrossRef]
- Ushida, Y.; Sekine, K.; Kuhara, T.; Takasuka, N.; Iigo, M.; Maeda, M.; Tsuda, H. Possible chemopreventive effects of bovine lactoferrin on esophagus and lung carcinogenesis in the rat. Japan. J. Cancer Res. 1999, 90, 262–267. [Google Scholar] [CrossRef]
- Hegazy, R.R.; Mansour, D.F.; Salama, A.A.; Abdel-Rahman, R.F.; Hassan, A.M. Regulation of PKB/Akt-pathway in the chemopreventive effect of lactoferrin against diethylnitrosamine-induced hepatocarcinogenesis in rats. Pharmacol. Rep. 2019, 71, 879–891. [Google Scholar] [CrossRef]
- Sugihara, Y.; Zuo, X.; Takata, T.; Jin, S.; Miyauti, M.; Isikado, A.; Imanaka, H.; Tatsuka, M.; Qi, G.; Shimamoto, F. Inhibition of DMH-DSS-induced colorectal cancer by liposomal bovine lactoferrin in rats. Oncol. Lett. 2017, 14, 5688–5694. [Google Scholar] [CrossRef]
- Tung, Y.T.; Chen, H.L.; Yen, C.C.; Lee, P.Y.; Tsai, H.C.; Lin, M.F.; Chen, C.M. Bovine lactoferrin inhibits lung cancer growth through suppression of both inflammation and expression of vascular endothelial growth factor. J. Dairy Sci. 2013, 96, 2095–2106. [Google Scholar] [CrossRef]
- Iglesias-Figueroa, B.F.; Siqueiros-Cendón, T.S.; Gutierrez, D.A.; Aguilera, R.J.; Espinoza-Sánchez, E.A.; Arévalo-Gallegos, S.; Varela-Ramirez, A.; Rascón-Cruz, Q. Recombinant human lactoferrin induces apoptosis, disruption of F-actin structure and cell cycle arrest with selective cytotoxicity on human triple negative breast cancer cells. Apoptosis 2019, 24, 562–577. [Google Scholar] [CrossRef]
- Chea, C.; Miyauchi, M.; Inubushi, T.; Febriyanti Ayuningtyas, N.; Subarnbhesaj, A.; Nguyen, P.T.; Shrestha, M.; Haing, S.; Ohta, K.; Takata, T. Molecular mechanism of inhibitory effects of bovine lactoferrin on the growth of oral squamous cell carcinoma. PLoS ONE 2018, 13, e0191683. [Google Scholar] [CrossRef]
- Xu, X.X.; Jiang, H.R.; Li, H.B.; Zhang, T.N.; Zhou, Q.; Liu, N. Apoptosis of stomach cancer cell SGC-7901 and regulation of Akt signaling way induced by bovine lactoferrin. J. Dairy Sci. 2010, 93, 2344–2350. [Google Scholar] [CrossRef] [PubMed]
- Arredondo-Beltran, I.G.; Ramirez-Sanchez, D.A.; Zazueta-Garcia, J.R.; Canizalez-Roman, A.; Angulo-Zamudio, U.A.; Velazquez-Roman, J.A.; Bolscher, J.G.M.; Nazmi, K.; Leon-Sicairos, N. Antitumor activity of bovine lactoferrin and its derived peptides against HepG2 liver cancer cells and Jurkat leukemia cells. Biometals 2023, 36, 639–655. [Google Scholar] [CrossRef] [PubMed]
- Filippi, S.; Paccosi, E.; Balzerano, A.; Ferretti, M.; Poli, G.; Taborri, J.; Brancorsini, S.; Proietti-De-Santis, L. CSA antisense targeting enhances anticancer drug sensitivity in breast cancer cells, including the triple-negative subtype. Cancers 2022, 14, 1687. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-M.; Cheng, W.T.K.; Chang, Y.-C.; Chang, T.-J.; Chen, H.-L. Growth enhancement of fowls by dietary administration of recombinant yeast cultures containing enriched growth hormone. Life Sci. 2000, 67, 2103–2115. [Google Scholar] [CrossRef] [PubMed]
- Kube, D.M.; Srivastava, A. Quantitative DNA slot blot analysis: Inhibition of DNA binding to membranes by magnesium ions. Nucleic Acids Res. 1997, 25, 3375–3376. [Google Scholar] [CrossRef] [PubMed]
- Chang, G.R.; Tu, M.Y.; Chen, Y.H.; Chang, K.Y.; Chen, C.F.; Lai, J.C.; Tung, Y.T.; Chen, H.L.; Fan, H.C.; Chen, C.M. KFP-1, a novel calcium-binding peptide isolated from kefir, promotes calcium influx through TRPV6 channels. Mol. Nutr. Food Res. 2021, 65, e2100182. [Google Scholar] [CrossRef] [PubMed]

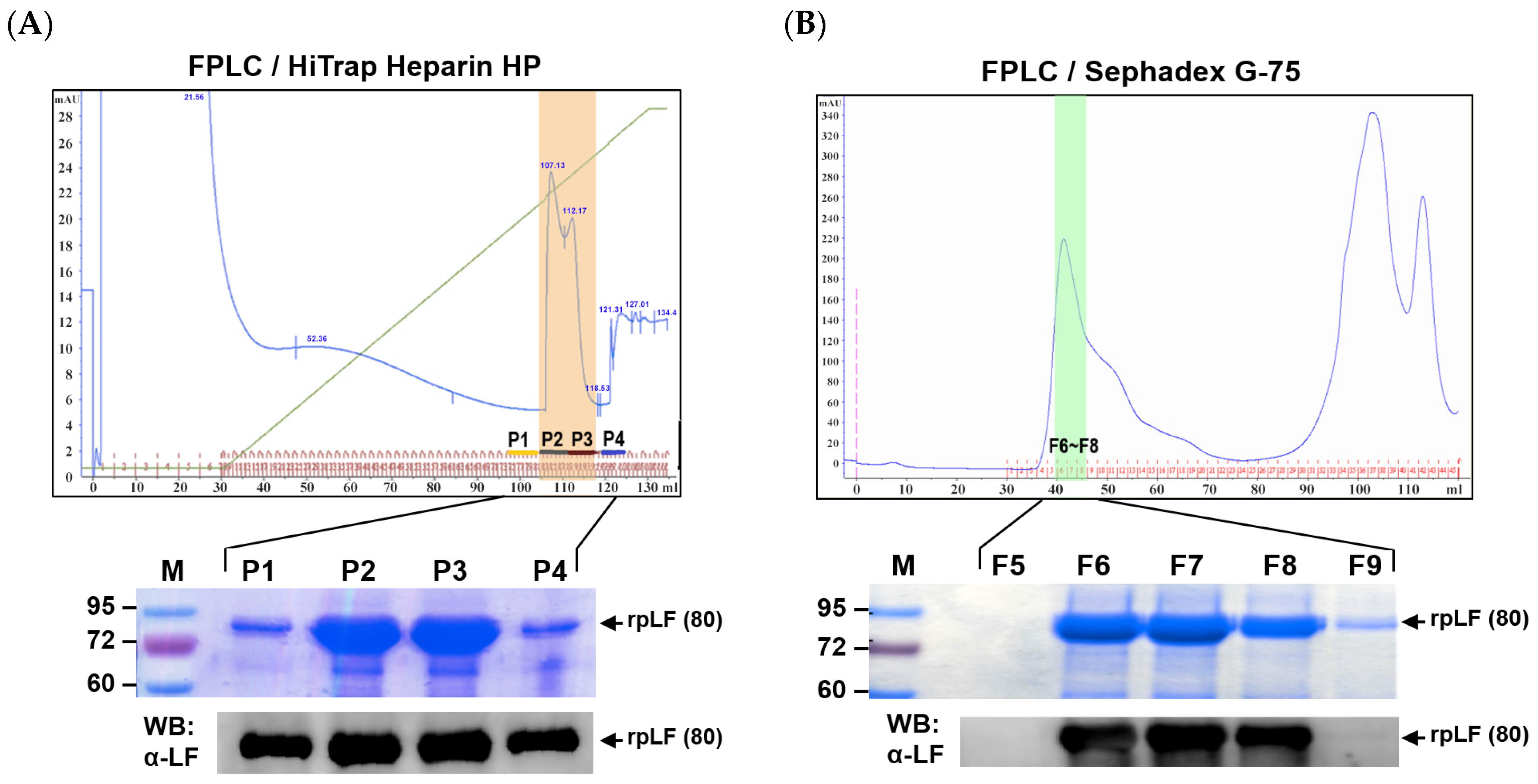


| Step | Start Volume (mL) | End Volume (mL) | Total Protein (mg/mL) | rpLF (mg/mL) | Purity (%) | Recovery (%) |
|---|---|---|---|---|---|---|
| Hollow fiber cartridge 0.45 micron | 2900 | 2400 | 8.3 | 4.3 | 52.4 | 100 |
| Biomax 100 kD | 2400 | 1350 | 10.8 | 7.3 | 67.9 | 95.5 |
| Biomax 30 kD | 1350 | 240 | 27.3 | 23.1 | 84.9 | 53.7 |
| Protein | E. coli | S. aureus | C. albicans | |||
|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | |
| Empty | >960 | >960 | >960 | >960 | >960 | >960 |
| bLF | 480 | >960 | 720 | >960 | 480 | >960 |
| rpLF | 720 | >960 | 960 | >960 | 720 | >960 |
| rpLFH | 120 | 240 | 180 | 360 | 240 | 480 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yen, C.-C.; Wu, P.-Y.; Ou-Yang, H.; Chen, H.-L.; Chong, K.-Y.; Chang, R.-L.; Chen, C.-M. Production of Bioactive Porcine Lactoferrin through a Novel Glucose-Inducible Expression System in Pichia pastoris: Unveiling Antimicrobial and Anticancer Functionalities. Int. J. Mol. Sci. 2024, 25, 1818. https://doi.org/10.3390/ijms25031818
Yen C-C, Wu P-Y, Ou-Yang H, Chen H-L, Chong K-Y, Chang R-L, Chen C-M. Production of Bioactive Porcine Lactoferrin through a Novel Glucose-Inducible Expression System in Pichia pastoris: Unveiling Antimicrobial and Anticancer Functionalities. International Journal of Molecular Sciences. 2024; 25(3):1818. https://doi.org/10.3390/ijms25031818
Chicago/Turabian StyleYen, Chih-Ching, Pei-Ying Wu, Huan Ou-Yang, Hsiao-Ling Chen, Kowit-Yu Chong, Ro-Lin Chang, and Chuan-Mu Chen. 2024. "Production of Bioactive Porcine Lactoferrin through a Novel Glucose-Inducible Expression System in Pichia pastoris: Unveiling Antimicrobial and Anticancer Functionalities" International Journal of Molecular Sciences 25, no. 3: 1818. https://doi.org/10.3390/ijms25031818
APA StyleYen, C.-C., Wu, P.-Y., Ou-Yang, H., Chen, H.-L., Chong, K.-Y., Chang, R.-L., & Chen, C.-M. (2024). Production of Bioactive Porcine Lactoferrin through a Novel Glucose-Inducible Expression System in Pichia pastoris: Unveiling Antimicrobial and Anticancer Functionalities. International Journal of Molecular Sciences, 25(3), 1818. https://doi.org/10.3390/ijms25031818









