Methylation Patterns of the FKBP5 Gene in Association with Childhood Maltreatment and Depressive Disorders
Abstract
1. Introduction
2. Results
2.1. Sample Characteristics
2.2. FKBP5 Methylation
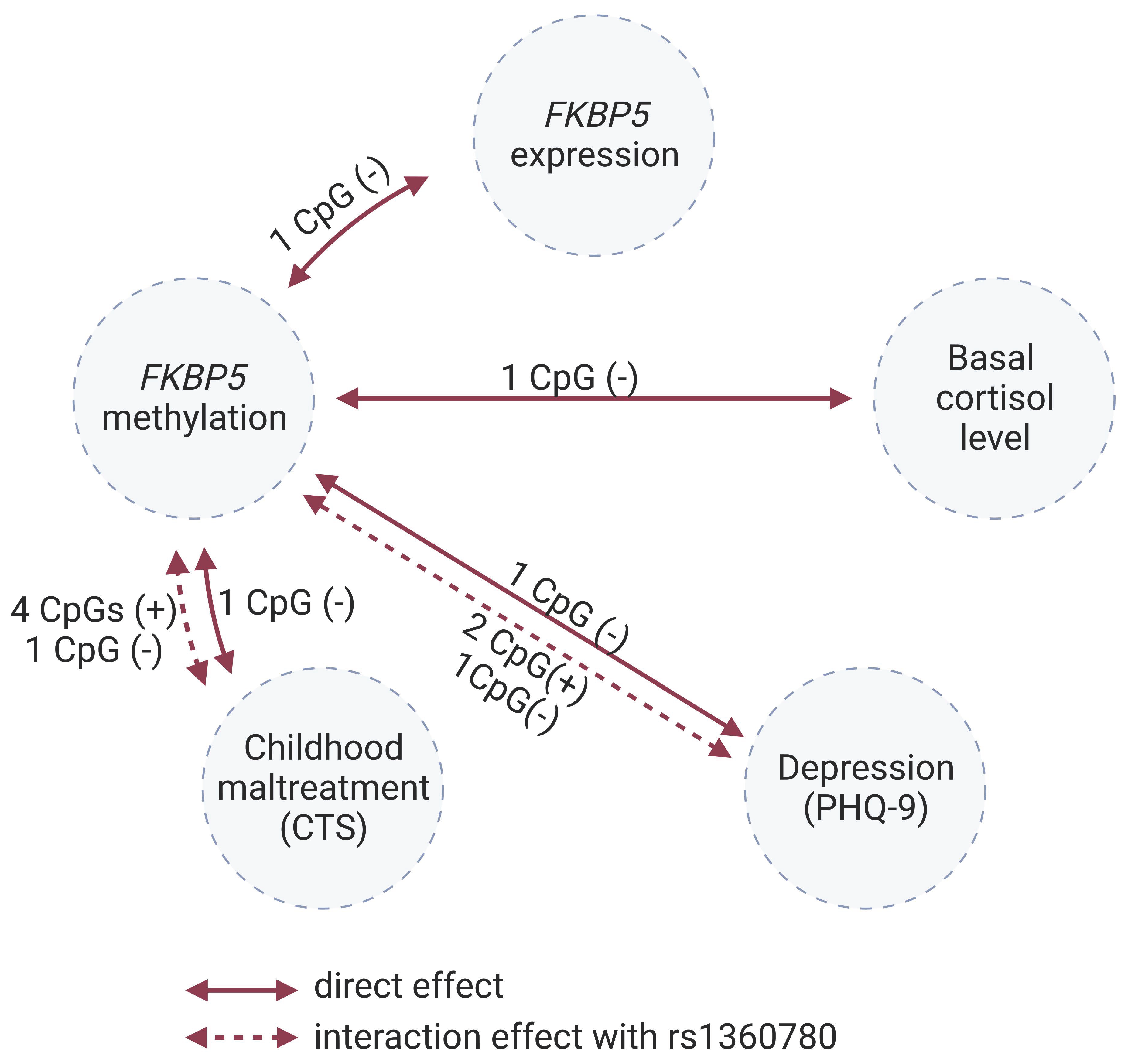
2.3. Direct Effects
2.4. Interaction Effects
3. Discussion
3.1. FKBP5 Methylation and FKBP5 Expression
3.2. FKBP5 Methylation and Cortisol
3.3. FKBP5 Methylation and Psychopathology
3.4. Interaction Models
3.5. Strengths and Limitations
3.6. Conclusion and Future Prospect
4. Materials and Methods
4.1. Study Population
4.2. Interview and Physical Examination
4.3. Assessment of Blood Measurements
4.4. FKBP5 Methylation
4.5. FKBP5 mRNA Levels
4.6. Psychometric Assessment
4.7. Genotyping
4.8. Main Statistical Analyses
4.9. Sensitivity Analysis for Genotype Interaction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CM | childhood maltreatment |
| GR | glucocorticoid receptor |
| HPA | hypothalamic pituitary adrenal |
| SNP | single-nucleotide polymorphism |
| SHIP | Study of Health in Pomerania |
| CTQ | Childhood Trauma Questionnaire (short form) |
| PHQ-9 | Patient Health Questionnaire-depression module |
| MDD | major depressive disorder |
References
- Carr, C.P.; Martins, C.M.S.; Stingel, A.M.; Lemgruber, V.B.; Juruena, M.F. The Role of Early Life Stress in Adult Psychiatric Disorders: A Systematic Review According to Childhood Trauma Subtypes. J. Nerv. Ment. Dis. 2013, 201, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Chmielewska, N.; Szyndler, J.; Maciejak, P.; Płaźnik, A. Epigenetic Mechanisms of Stress and Depression. Psychiatr. Pol. 2019, 53, 1413–1428. [Google Scholar] [CrossRef]
- Heim, C.; Newport, D.J.; Mletzko, T.; Miller, A.H.; Nemeroff, C.B. The Link between Childhood Trauma and Depression: Insights from HPA Axis Studies in Humans. Psychoneuroendocrinology 2008, 33, 693–710. [Google Scholar] [CrossRef]
- Hill, J. Childhood Trauma and Depression. Curr. Opin. Psychiatry 2003, 16, 3–6. [Google Scholar] [CrossRef]
- Mandelli, L.; Petrelli, C.; Serretti, A. The Role of Specific Early Trauma in Adult Depression: A Meta-Analysis of Published Literature. Childhood Trauma and Adult Depression. Eur. Psychiatry 2015, 30, 665–680. [Google Scholar] [CrossRef] [PubMed]
- Wiersma, J.E.; Hovens, J.G.; van Oppen, P.; Giltay, E.J.; van Schaik, D.J.; Penninx, B.W. The Importance of Childhood Trauma and Childhood Life Events for Chronicity of Depression in Adults. J. Clin. Psychiatry 2009, 70, 8733. [Google Scholar] [CrossRef]
- Ingram, R.E.; Luxton, D.D. Vulnerability-Stress Models. Development of Psychopathology: A Vulnerability-Stress Perspective; Hankin, B.L., Abela, J.R.Z., Eds.; Sage Publications, Inc.: Thousand Oaks, CA, USA, 2005; pp. 32–46. [Google Scholar]
- Hammen, C. Stress and Depression. Annu. Rev. Clin. Psychol. 2005, 1, 293–319. [Google Scholar] [CrossRef]
- Höhne, N.; Poidinger, M.; Merz, F.; Pfister, H.; Bruckl, T.; Zimmermann, P.; Uhr, M.; Holsboer, F.; Ising, M. FKBP5 Genotype-Dependent DNA Methylation and mRNA Regulation After Psychosocial Stress in Remitted Depression and Healthy Controls. Int. J. Neuropsychopharmacol. 2015, 18, pyu087. [Google Scholar] [CrossRef]
- Klengel, T.; Mehta, D.; Anacker, C.; Rex-Haffner, M.; Pruessner, J.C.; Pariante, C.M.; Pace, T.W.W.; Mercer, K.B.; Mayberg, H.S.; Bradley, B.; et al. Allele-Specific FKBP5 DNA Demethylation Mediates Gene–Childhood Trauma Interactions. Nat. Neurosci. 2013, 16, 33–41. [Google Scholar] [CrossRef]
- Klinger-König, J.; Hertel, J.; Van der Auwera, S.; Frenzel, S.; Pfeiffer, L.; Waldenberger, M.; Golchert, J.; Teumer, A.; Nauck, M.; Homuth, G.; et al. Methylation of the FKBP5 Gene in Association with FKBP5 Genotypes, Childhood Maltreatment and Depression. Neuropsychopharmacology 2019, 44, 930–938. [Google Scholar] [CrossRef]
- Yehuda, R.; Daskalakis, N.P.; Bierer, L.M.; Bader, H.N.; Klengel, T.; Holsboer, F.; Binder, E.B. Holocaust Exposure Induced Intergenerational Effects on FKBP5 Methylation. Biol. Psychiatry 2016, 80, 372–380. [Google Scholar] [CrossRef]
- Binder, E.B. The Role of FKBP5, a Co-Chaperone of the Glucocorticoid Receptor in the Pathogenesis and Therapy of Affective and Anxiety Disorders. Psychoneuroendocrinology 2009, 34, 186–195. [Google Scholar] [CrossRef]
- Storer, C.L.; Dickey, C.A.; Galigniana, M.D.; Rein, T.; Cox, M.B. FKBP51 and FKBP52 in Signaling and Disease. Trends Endocrinol. Metab. 2011, 22, 481–490. [Google Scholar] [CrossRef]
- Matosin, N.; Halldorsdottir, T.; Binder, E.B. Understanding the Molecular Mechanisms Underpinning Gene by Environment Interactions in Psychiatric Disorders: The FKBP5 Model. Biol. Psychiatry 2018, 83, 821–830. [Google Scholar] [CrossRef]
- Binder, E.B.; Salyakina, D.; Lichtner, P.; Wochnik, G.M.; Ising, M.; Pütz, B.; Papiol, S.; Seaman, S.; Lucae, S.; Kohli, M.A.; et al. Polymorphisms in FKBP5 Are Associated with Increased Recurrence of Depressive Episodes and Rapid Response to Antidepressant Treatment. Nat. Genet. 2004, 36, 1319–1325. [Google Scholar] [CrossRef]
- Appel, K.; Schwahn, C.; Mahler, J.; Schulz, A.; Spitzer, C.; Fenske, K.; Stender, J.; Barnow, S.; John, U.; Teumer, A.; et al. Moderation of Adult Depression by a Polymorphism in the FKBP5 Gene and Childhood Physical Abuse in the General Population. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 2011, 36, 1982–1991. [Google Scholar] [CrossRef]
- Grabe, H.J.; Wittfeld, K.; Van der Auwera, S.; Janowitz, D.; Hegenscheid, K.; Habes, M.; Homuth, G.; Barnow, S.; John, U.; Nauck, M.; et al. Effect of the Interaction between Childhood Abuse and Rs1360780 of the FKBP5 Gene on Gray Matter Volume in a General Population Sample. Hum. Brain Mapp. 2016, 37, 1602–1613. [Google Scholar] [CrossRef]
- Hertel, J.; König, J.; Homuth, G.; Van der Auwera, S.; Wittfeld, K.; Pietzner, M.; Kacprowski, T.; Pfeiffer, L.; Kretschmer, A.; Waldenberger, M.; et al. Evidence for Stress-like Alterations in the HPA-Axis in Women Taking Oral Contraceptives. Sci. Rep. 2017, 7, 14111. [Google Scholar] [CrossRef]
- Matthews, S.G. Early Programming of the Hypothalamo–Pituitary–Adrenal Axis. Trends Endocrinol. Metab. 2002, 13, 373–380. [Google Scholar] [CrossRef]
- Bustamante, A.C.; Aiello, A.E.; Guffanti, G.; Galea, S.; Wildman, D.E.; Uddin, M. FKBP5 DNA Methylation Does Not Mediate the Association between Childhood Maltreatment and Depression Symptom Severity in the Detroit Neighborhood Health Study. J. Psychiatr. Res. 2018, 96, 39–48. [Google Scholar] [CrossRef]
- Wiechmann, T.; Röh, S.; Sauer, S.; Czamara, D.; Arloth, J.; Ködel, M.; Beintner, M.; Knop, L.; Menke, A.; Binder, E.B.; et al. Identification of Dynamic Glucocorticoid-Induced Methylation Changes at the FKBP5 Locus. Clin. Epigenetics 2019, 11, 83. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA Methylation and Human Disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef]
- Lee, R.S.; Tamashiro, K.L.K.; Yang, X.; Purcell, R.H.; Harvey, A.; Willour, V.L.; Huo, Y.; Rongione, M.; Wand, G.S.; Potash, J.B. Chronic Corticosterone Exposure Increases Expression and Decreases Deoxyribonucleic Acid Methylation of Fkbp5 in Mice. Endocrinology 2010, 151, 4332–4343. [Google Scholar] [CrossRef]
- Yang, X.; Ewald, E.R.; Huo, Y.; Tamashiro, K.L.; Salvatori, R.; Sawa, A.; Wand, G.S.; Lee, R.S. Glucocorticoid-Induced Loss of DNA Methylation in Non-Neuronal Cells and Potential Involvement of DNMT1 in Epigenetic Regulation of Fkbp5. Biochem. Biophys. Res. Commun. 2012, 420, 570–575. [Google Scholar] [CrossRef]
- Tarullo, A.R.; Gunnar, M.R. Child Maltreatment and the Developing HPA Axis. Horm. Behav. 2006, 50, 632–639. [Google Scholar] [CrossRef]
- Spiga, F.; Walker, J.J.; Terry, J.R.; Lightman, S.L. HPA Axis-Rhythms. In Comprehensive Physiology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2014; pp. 1273–1298. ISBN 978-0-470-65071-4. [Google Scholar]
- Tyrka, A.R.; Ridout, K.K.; Parade, S.H.; Paquette, A.; Marsit, C.J.; Seifer, R. Childhood Maltreatment and Methylation of FKBP5. Dev. Psychopathol. 2015, 27, 1637–1645. [Google Scholar] [CrossRef]
- Mihaljevic, M.; Franic, D.; Soldatovic, I.; Lukic, I.; Petrovic, S.A.; Mirjanic, T.; Stankovic, B.; Zukic, B.; Zeljic, K.; Gasic, V.; et al. The FKBP5 Genotype and Childhood Trauma Effects on FKBP5 DNA Methylation in Patients with Psychosis, Their Unaffected Siblings, and Healthy Controls. Psychoneuroendocrinology 2021, 128, 105205. [Google Scholar] [CrossRef]
- Kang, J.I.; Kim, T.Y.; Choi, J.H.; So, H.S.; Kim, S.J. Allele-Specific DNA Methylation Level of FKBP5 Is Associated with Post-Traumatic Stress Disorder. Psychoneuroendocrinology 2019, 103, 1–7. [Google Scholar] [CrossRef]
- Saito, T.; Shinozaki, G.; Koga, M.; Tanichi, M.; Takeshita, S.; Nakagawa, R.; Nagamine, M.; Cho, H.R.; Morimoto, Y.; Kobayashi, Y.; et al. Effect of Interaction between a Specific Subtype of Child Abuse and the FKBP5 Rs1360780 SNP on DNA Methylation among Patients with Bipolar Disorder. J. Affect. Disord. 2020, 272, 417–422. [Google Scholar] [CrossRef]
- Murphy, F.; Nasa, A.; Cullinane, D.; Raajakesary, K.; Gazzaz, A.; Sooknarine, V.; Haines, M.; Roman, E.; Kelly, L.; O’Neill, A.; et al. Childhood Trauma, the HPA Axis and Psychiatric Illnesses: A Targeted Literature Synthesis. Front. Psychiatry 2022, 13. [Google Scholar] [CrossRef]
- Fogelman, N.; Canli, T. Early Life Stress and Cortisol: A Meta-Analysis. Horm. Behav. 2018, 98, 63–76. [Google Scholar] [CrossRef]
- Braun, P.R.; Han, S.; Hing, B.; Nagahama, Y.; Gaul, L.N.; Heinzman, J.T.; Grossbach, A.J.; Close, L.; Dlouhy, B.J.; Howard, M.A.; et al. Genome-Wide DNA Methylation Comparison between Live Human Brain and Peripheral Tissues within Individuals. Transl. Psychiatry 2019, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Hardt, J.; Rutter, M. Validity of Adult Retrospective Reports of Adverse Childhood Experiences: Review of the Evidence. J. Child Psychol. Psychiatry 2004, 45, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Schiele, M.A.; Gottschalk, M.G.; Domschke, K. The Applied Implications of Epigenetics in Anxiety, Affective and Stress-Related Disorders - A Review and Synthesis on Psychosocial Stress, Psychotherapy and Prevention. Clin. Psychol. Rev. 2020, 77, 101830. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, C.; Richter, J.; Mahr, M.; Gajewska, A.; Schiele, M.A.; Gehrmann, A.; Schmidt, B.; Lesch, K.-P.; Lang, T.; Helbig-Lang, S.; et al. MAOA Gene Hypomethylation in Panic Disorder—Reversibility of an Epigenetic Risk Pattern by Psychotherapy. Transl. Psychiatry 2016, 6, e773. [Google Scholar] [CrossRef]
- Szyf, M. Prospects for the Development of Epigenetic Drugs for CNS Conditions. Nat. Rev. Drug Discov. 2015, 14, 461–474. [Google Scholar] [CrossRef]
- Kumsta, R. The Role of Epigenetics for Understanding Mental Health Difficulties and Its Implications for Psychotherapy Research. Psychol. Psychother. Theory Res. Pract. 2019, 92, 190–207. [Google Scholar] [CrossRef]
- Yehuda, R.; Daskalakis, N.; Desarnaud, F.; Makotkine, I.; Lehrner, A.; Koch, E.; Flory, J.; Buxbaum, J.; Meaney, M.; Bierer, L. Epigenetic Biomarkers as Predictors and Correlates of Symptom Improvement Following Psychotherapy in Combat Veterans with PTSD. Front. Psychiatry 2013, 4. [Google Scholar] [CrossRef]
- Völzke, H.; Schössow, J.; Schmidt, C.O.; Jürgens, C.; Richter, A.; Werner, A.; Werner, N.; Radke, D.; Teumer, A.; Ittermann, T.; et al. Cohort Profile Update: The Study of Health in Pomerania (SHIP). Int. J. Epidemiol. 2022, 51, e372–e383. [Google Scholar] [CrossRef]
- Völzke, H.; Alte, D.; Schmidt, C.O.; Radke, D.; Lorbeer, R.; Friedrich, N.; Aumann, N.; Lau, K.; Piontek, M.; Born, G.; et al. Cohort Profile: The Study of Health in Pomerania. Int. J. Epidemiol. 2011, 40, 294–307. [Google Scholar] [CrossRef]
- Klinger-König, J.; Frenzel, S.; Hannemann, A.; Wittfeld, K.; Bülow, R.; Friedrich, N.; Nauck, M.; Völzke, H.; Grabe, H.J. Sex Differences in the Association between Basal Serum Cortisol Concentrations and Cortical Thickness. Neurobiol. Stress 2021, 15, 100416. [Google Scholar] [CrossRef]
- Masuch, A.; Budde, K.; Kastenmüller, G.; Artati, A.; Adamski, J.; Völzke, H.; Nauck, M.; Pietzner, M. Metabolic Signature Associated with Parameters of the Complete Blood Count in Apparently Healthy Individuals. J. Cell. Mol. Med. 2019, 23, 5144–5153. [Google Scholar] [CrossRef] [PubMed]
- Roeh, S.; Wiechmann, T.; Sauer, S.; Ködel, M.; Binder, E.B.; Provençal, N. HAM-TBS: High-Accuracy Methylation Measurements via Targeted Bisulfite Sequencing. Epigenetics Chromatin 2018, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zhang, X.; Huang, C.-C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-Value and M-Value Methods for Quantifying Methylation Levels by Microarray Analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef]
- Oscanoa, J.; Sivapalan, L.; Gadaleta, E.; Dayem Ullah, A.Z.; Lemoine, N.R.; Chelala, C. SNPnexus: A Web Server for Functional Annotation of Human Genome Sequence Variation (2020 Update). Nucleic Acids Res. 2020, 48, W185–W192. [Google Scholar] [CrossRef]
- Liu, Y.; Siegmund, K.D.; Laird, P.W.; Berman, B.P. Bis-SNP: Combined DNA Methylation and SNP Calling for Bisulfite-Seq Data. Genome Biol. 2012, 13, R61. [Google Scholar] [CrossRef]
- Schurmann, C.; Heim, K.; Schillert, A.; Blankenberg, S.; Carstensen, M.; Dörr, M.; Endlich, K.; Felix, S.B.; Gieger, C.; Grallert, H.; et al. Analyzing Illumina Gene Expression Microarray Data from Different Tissues: Methodological Aspects of Data Analysis in the Metaxpress Consortium. PLoS ONE 2012, 7, e50938. [Google Scholar] [CrossRef]
- Kroenke, K.; Spitzer, R.L.; Williams, J.B. The PHQ-9: Validity of a Brief Depression Severity Measure. J. Gen. Intern. Med. 2001, 16, 606–613. [Google Scholar] [CrossRef]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders, 4th ed.; American Psychiatric Association: Washington, DC, USA, 1994; Volume 4, ISBN 978-0-89042-061-4. [Google Scholar]
- Bernstein, D.P.; Stein, J.A.; Newcomb, M.D.; Walker, E.; Pogge, D.; Ahluvalia, T.; Stokes, J.; Handelsman, L.; Medrano, M.; Desmond, D.; et al. Development and Validation of a Brief Screening Version of the Childhood Trauma Questionnaire. Child Abuse Negl. 2003, 27, 169–190. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013.
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A Flexible Statistical Power Analysis Program for the Social, Behavioral, and Biomedical Sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Völzke, H.; Schmidt, C.O.; Hoffmann, W. Research: Increasing Value, Reducing Waste. Lancet Lond. Engl. 2014, 383, 1124. [Google Scholar] [CrossRef]

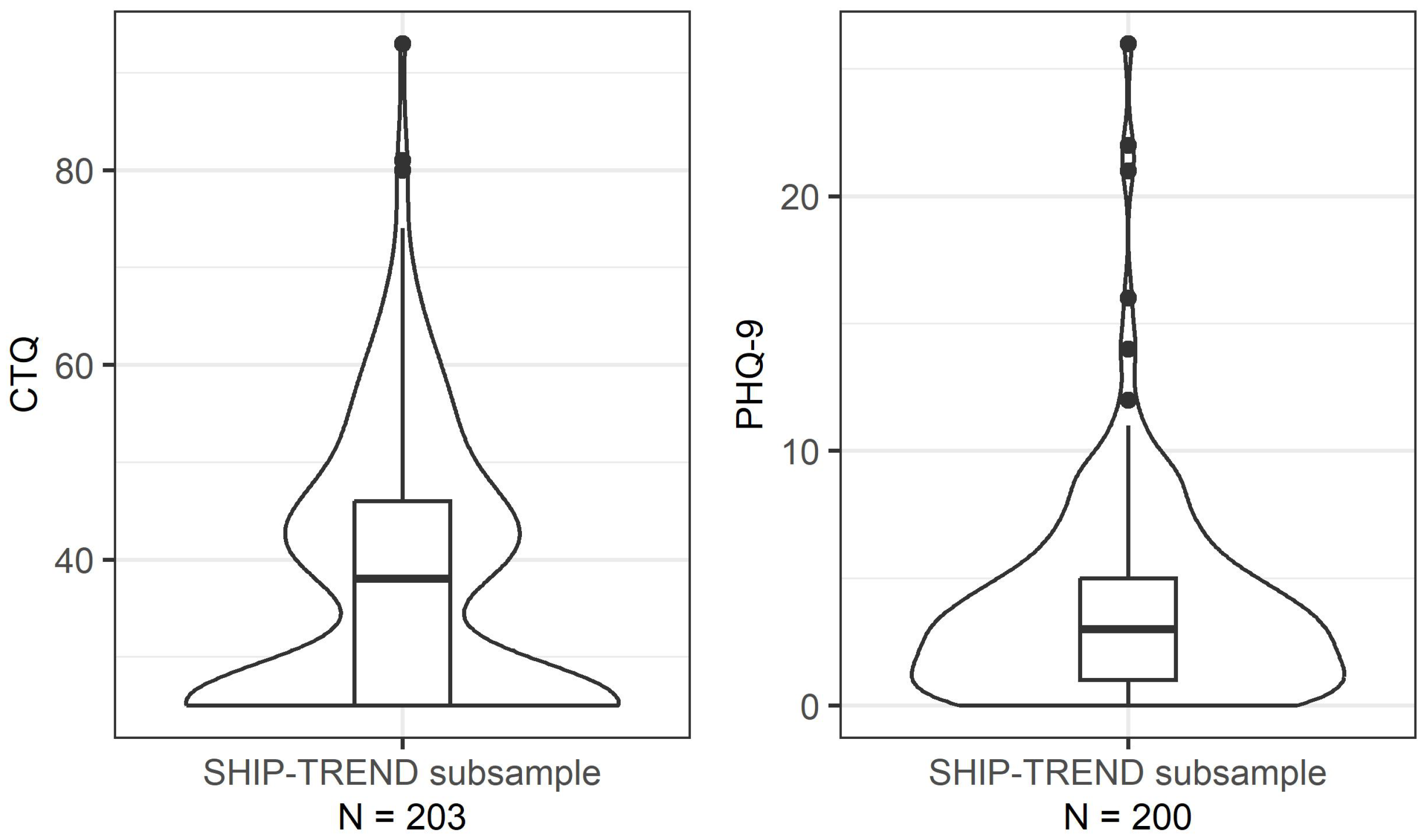



| Parameter | SHIP-TREND | Herein Analysed Subsample |
|---|---|---|
| N | 4420 | 203 |
| Sex | (0 × NA) | (0 × NA) |
| Female | 2275 (51%) | 97 (48%) |
| Male | 2145 (49%) | 106 (52%) |
| Age [years] | 51.96 ± 15.46 (0 × NA) | 49.00 ± 12.66 |
| Body mass index [kg/m2] | 28.12 ± 5.23 (7 × NA) | 27.15 ± 4.53 |
| Smoking | (22 × NA) | (1 × NA) |
| Never | 1605 (36%) | 79 (39%) |
| Ex | 1610 (37%) | 70 (34%) |
| Current | 1183 (27%) | 53 (26%) |
| Haematocrit | 0.42 ± 0.033 (15 × NA) | 0.42 ± 0.03 |
| Thrombocytes [gpt/l] | 228.10 ± 54.57 (15 × NA) | 5.62 ± 1.27 |
| Leukocytes [gpt/l] | 6.18 ± 2.55 (15 × NA) | 225 ± 49.6 |
| Neutrophils [%] | 58.34 ± 8.62 (31 × NA) | 57.26 ± 7.44 |
| Eosinophils [%] | 2.70 ± 1.83 (201 × NA) | 2.53 ± 1.56 |
| Basophils [%] | 0.49 ± 0.31 (203 × NA) | 12.23 ± 0.30 |
| Lymphocytes [%] | 29.28 ± 7.57 (31 × NA) | 30.60 ± 6.57 |
| Monocytes [%] | 8.78 ± 2.51 (31 × NA) | 9.01 ± 2.11 |
| mRNA | 9.03 ± 0.46 (3429 × NA) | 8.98 ± 0.42 |
| Cortisol [nmol/mL] | 334.4 ± 131.1 (80 × NA) | 333.0 ± 135.0 |
| PHQ-9 | 3.91 ± 3.59 (249 × NA) | 3.87 ± 3.93 (3 × NA) |
| CTQ | 33.34 ± 9.68 (324 × NA) | 37.26 ± 13.99 (0 × NA) |
| rs1360780 [CC/CT + TT] | (300 × NA) | (0 × NA) |
| CC | 2068 (50%) | 103 (51%) |
| CT + TT | 2052 (50%) | 100 (49%) |
| Model | Position | Effect Size | S.E. | t-Value | d.f. | p-Value | Location |
|---|---|---|---|---|---|---|---|
| Gene expression | chr6:35693949 | −0.133 | 0.065 | −2.054 | 174 | 0.041 | 3′-end |
| Cortisol | chr6:35558438 | −0.436 | 0.204 | −2.137 | 178 | 0.034 | intron 7 |
| CTQ | chr6:35693881 | −0.063 | 0.032 | −1.986 | 180 | 0.049 | 3′-end |
| PHQ-9 | chr6:35592141 | 0.043 | 0.019 | 2.303 | 177 | 0.022 | intron 3 |
| Model | Position | Interaction Effect | S.E. | t-Value | d.f. | p-Value | Location |
|---|---|---|---|---|---|---|---|
| CTQ × SNP | chr6:35490599 | −0.070 | 0.033 | −2.133 | 179 | 0.034 | 5′-end |
| chr6:35694724 | 0.082 | 0.034 | 2.387 | 181 | 0.018 | 3′-end | |
| chr6:35694748 | 0.097 | 0.034 | 2.829 | 179 | 0.005 | 3′-end | |
| chr6:35694756 | 0.103 | 0.035 | 2.955 | 179 | 0.004 | 3′-end | |
| chr6:35697759 | 0.138 | 0.064 | 2.151 | 181 | 0.033 | 3′-end | |
| PHQ-9 × SNP | chr6:35558386 | 0.122 | 0.046 | 2.62 | 178 | 0.010 | intron 7 |
| chr6:35558438 | 0.114 | 0.045 | 2.502 | 177 | 0.013 | intron 7 | |
| chr6:35694056 | −0.093 | 0.045 | −2.047 | 176 | 0.042 | 3′-end |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Großmann, N.L.; Weihs, A.; Kühn, L.; Sauer, S.; Röh, S.; Wiechmann, T.; Rex-Haffner, M.; Völzke, H.; Völker, U.; Binder, E.B.; et al. Methylation Patterns of the FKBP5 Gene in Association with Childhood Maltreatment and Depressive Disorders. Int. J. Mol. Sci. 2024, 25, 1485. https://doi.org/10.3390/ijms25031485
Großmann NL, Weihs A, Kühn L, Sauer S, Röh S, Wiechmann T, Rex-Haffner M, Völzke H, Völker U, Binder EB, et al. Methylation Patterns of the FKBP5 Gene in Association with Childhood Maltreatment and Depressive Disorders. International Journal of Molecular Sciences. 2024; 25(3):1485. https://doi.org/10.3390/ijms25031485
Chicago/Turabian StyleGroßmann, Nora L., Antoine Weihs, Luise Kühn, Susann Sauer, Simone Röh, Tobias Wiechmann, Monika Rex-Haffner, Henry Völzke, Uwe Völker, Elisabeth B. Binder, and et al. 2024. "Methylation Patterns of the FKBP5 Gene in Association with Childhood Maltreatment and Depressive Disorders" International Journal of Molecular Sciences 25, no. 3: 1485. https://doi.org/10.3390/ijms25031485
APA StyleGroßmann, N. L., Weihs, A., Kühn, L., Sauer, S., Röh, S., Wiechmann, T., Rex-Haffner, M., Völzke, H., Völker, U., Binder, E. B., Teumer, A., Homuth, G., Klinger-König, J., & Grabe, H. J. (2024). Methylation Patterns of the FKBP5 Gene in Association with Childhood Maltreatment and Depressive Disorders. International Journal of Molecular Sciences, 25(3), 1485. https://doi.org/10.3390/ijms25031485






