Circulating miRNAs and Preeclampsia: From Implantation to Epigenetics †
Abstract
1. Introduction
2. Materials and Methods
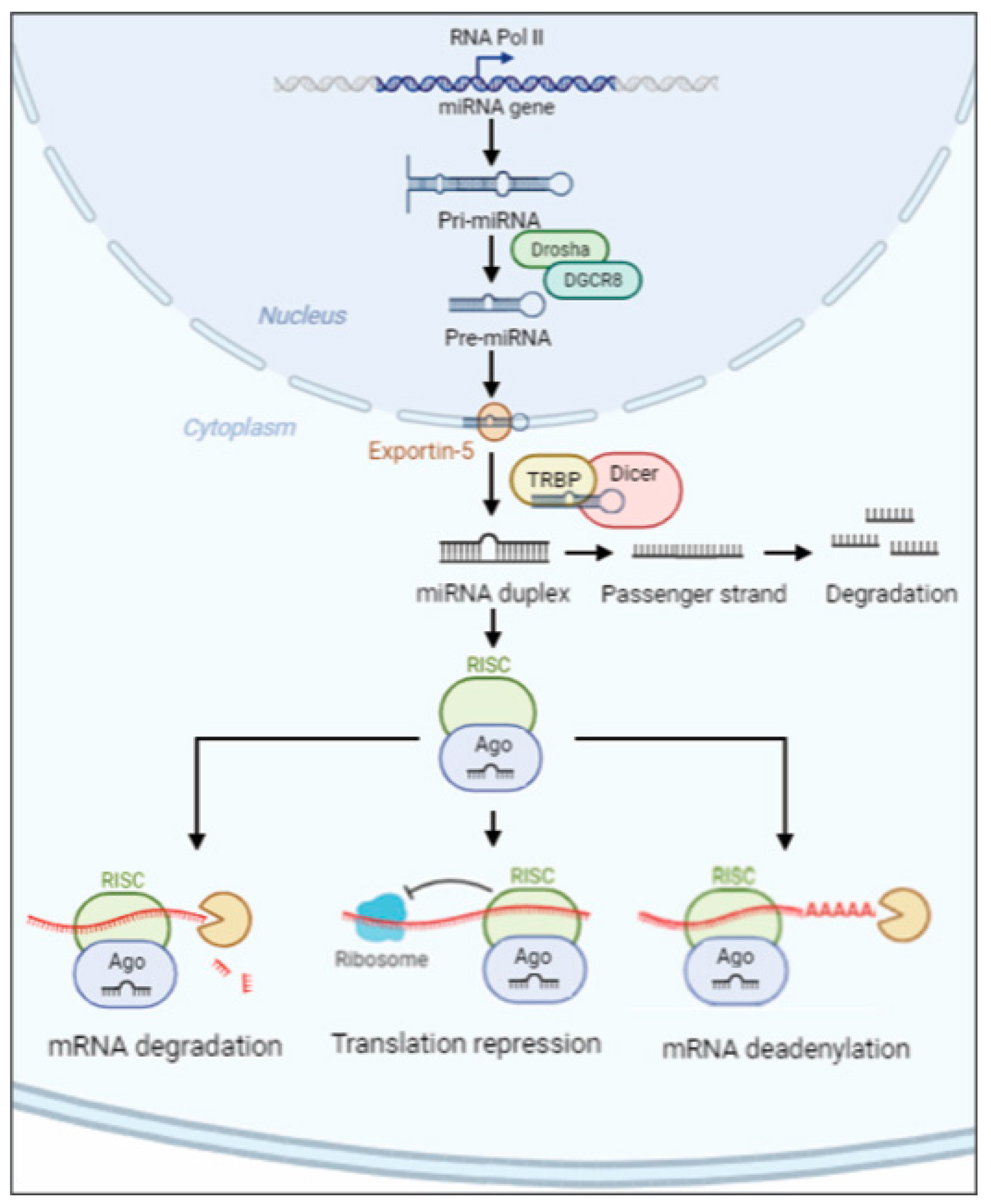
3. miRNA Biogenesis, Mechanisms of Export and Annotation Criteria
4. miRNAs, Implantation and Preeclampsia
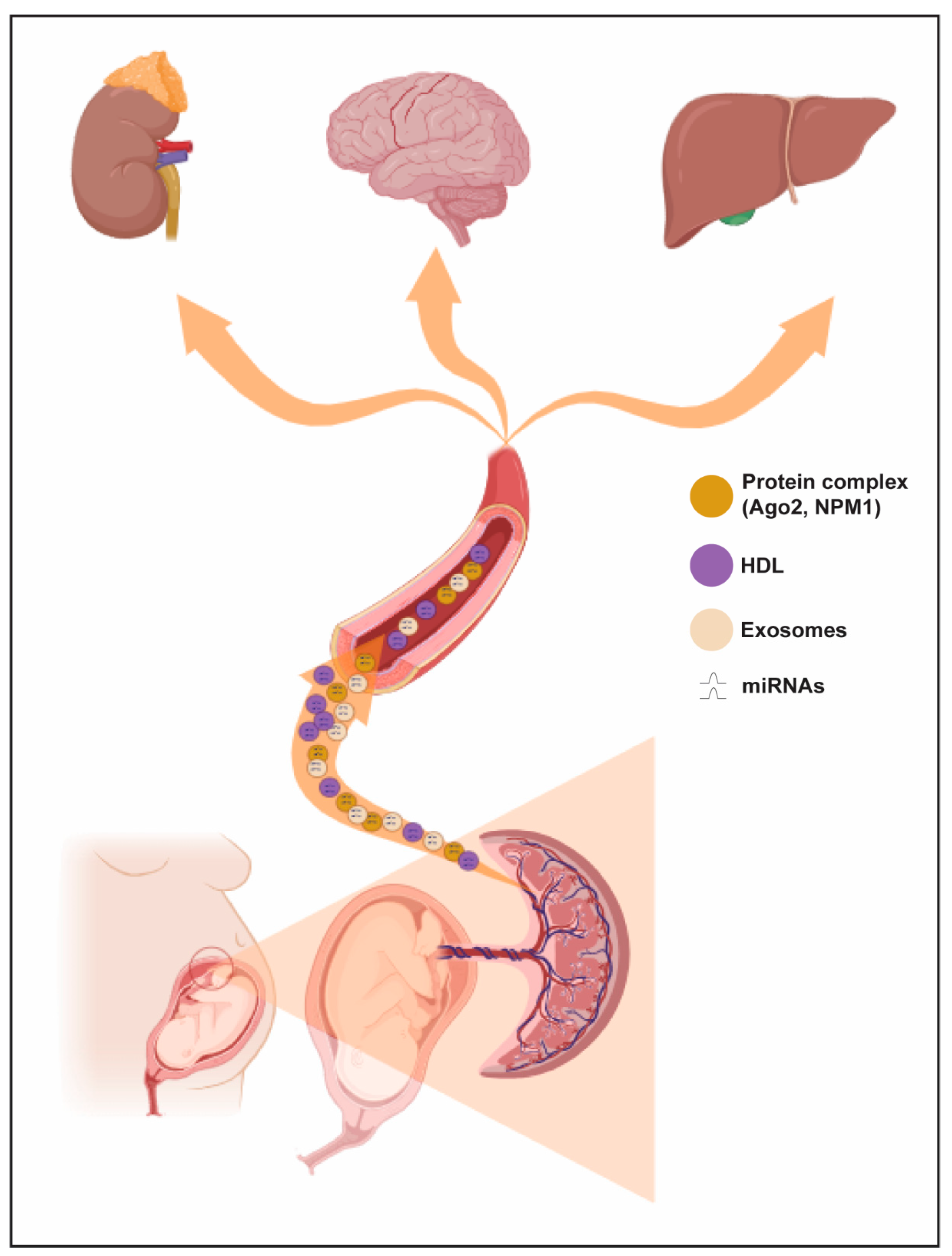
5. miRNAs and Clinical Preeclampsia
5.1. miRNAs and Diagnosed Preeclampsia
5.2. miRNAs and Onset of Preeclampsia
5.3. miRNAs and Prediction of Preeclampsia
6. miRNAs, Preeclampsia and Epigenetics
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tossetta, G.; Avellini, C.; Licini, C.; Giannubilo, S.R.; Castellucci, M.; Marzioni, D. High temperature requirement A1 and fibronectin: Two possible players in placental tissue remodelling. Eur. J. Histochem. 2016, 60, 2724. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huppertz, B. The anatomy of the normal placenta. J. Clin. Pathol. 2008, 61, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Tossetta, G.; Fantone, S.; Busilacchi, E.M.; Di Simone, N.; Giannubilo, S.R.; Scambia, G.; Giordano, A.; Marzioni, D. Modulation of matrix metalloproteases by ciliary neurotrophic factor in human placental development. Cell Tissue Res. 2022, 390, 113–129. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Giannubilo, S.R.; Marinelli Busilacchi, E.; Ciavattini, A.; Castellucci, M.; Di Simone, N.; Mattioli-Belmonte, M.; Marzioni, D. Pre-eclampsia onset and SPARC: A possible involvement in placenta development. J. Cell Physiol. 2019, 234, 6091–6098. [Google Scholar] [CrossRef] [PubMed]
- Gesuita, R.; Licini, C.; Picchiassi, E.; Tarquini, F.; Coata, G.; Fantone, S.; Tossetta, G.; Ciavattini, A.; Castellucci, M.; Di Renzo, G.C.; et al. Association between first trimester plasma htra1 level and subsequent preeclampsia: A possible early marker? Pregnancy Hypertens. 2019, 18, 58–62. [Google Scholar] [CrossRef]
- Cardaropoli, S.; Paulesu, L.; Romagnoli, R.; Ietta, F.; Marzioni, D.; Castellucci, M.; Rolfo, A.; Vasario, E.; Piccoli, E.; Todros, T. Macrophage migration inhibitory factor in fetoplacental tissues from preeclamptic pregnancies with or without fetal growth restriction. Clin. Dev. Immunol. 2012, 2012, 639342. [Google Scholar] [CrossRef]
- Marzioni, D.; Todros, T.; Cardaropoli, S.; Rolfo, A.; Lorenzi, T.; Ciarmela, P.; Romagnoli, R.; Paulesu, L.; Castellucci, M. Activating protein-1 family of transcription factors in the human placenta complicated by preeclampsia with and without fetal growth restriction. Placenta 2010, 31, 919–927. [Google Scholar] [CrossRef]
- Todros, T.; Marzioni, D.; Lorenzi, T.; Piccoli, E.; Capparuccia, L.; Perugini, V.; Cardaropoli, S.; Romagnoli, R.; Gesuita, R.; Rolfo, A.; et al. Evidence for a role of TGF-beta1 in the expression and regulation of alpha-SMA in fetal growth restricted placentae. Placenta 2007, 28, 1123–1132. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Gesuita, R.; Di Renzo, G.C.; Meyyazhagan, A.; Tersigni, C.; Scambia, G.; Di Simone, N.; Marzioni, D. HtrA1 in Gestational Diabetes Mellitus: A Possible Biomarker? Diagnostics 2022, 12, 2705. [Google Scholar] [CrossRef]
- Giannubilo, S.R.; Licini, C.; Picchiassi, E.; Tarquini, F.; Coata, G.; Fantone, S.; Tossetta, G.; Ciavattini, A.; Castellucci, M.; Giardina, I.; et al. First trimester HtrA1 maternal plasma level and spontaneous preterm birth. J. Matern. Fetal Neonatal Med. 2022, 35, 780–784. [Google Scholar] [CrossRef]
- Capparuccia, L.; Marzioni, D.; Giordano, A.; Fazioli, F.; De Nictolis, M.; Busso, N.; Todros, T.; Castellucci, M. PPARgamma expression in normal human placenta, hydatidiform mole and choriocarcinoma. Mol. Hum. Reprod. 2002, 8, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Crescimanno, C.; Marzioni, D.; Paradinas, F.J.; Schrurs, B.; Muhlhauser, J.; Todros, T.; Newlands, E.; David, G.; Castellucci, M. Expression pattern alterations of syndecans and glypican-1 in normal and pathological trophoblast. J. Pathol. 1999, 189, 600–608. [Google Scholar] [CrossRef]
- Dimitriadis, E.; Rolnik, D.L.; Zhou, W.; Estrada-Gutierrez, G.; Koga, K.; Francisco, R.P.V.; Whitehead, C.; Hyett, J.; da Silva Costa, F.; Nicolaides, K.; et al. Pre-eclampsia. Nat. Rev. Dis. Primers 2023, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Odigboegwu, O.; Pan, L.J.; Chatterjee, P. Use of Antihypertensive Drugs During Preeclampsia. Front. Cardiovasc. Med. 2018, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- Mol, B.W.J.; Roberts, C.T.; Thangaratinam, S.; Magee, L.A.; de Groot, C.J.M.; Hofmeyr, G.J. Pre-eclampsia. Lancet 2016, 387, 999–1011. [Google Scholar] [CrossRef]
- Thadhani, R.; Ecker, J.L.; Kettyle, E.; Sandler, L.; Frigoletto, F.D. Pulse pressure and risk of preeclampsia: A prospective study. Obstet. Gynecol. 2001, 97, 515–520. [Google Scholar] [CrossRef]
- Fantone, S.; Mazzucchelli, R.; Giannubilo, S.R.; Ciavattini, A.; Marzioni, D.; Tossetta, G. AT-rich interactive domain 1A protein expression in normal and pathological pregnancies complicated by preeclampsia. Histochem. Cell Biol. 2020, 154, 339–346. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Giannubilo, S.R.; Ciavattini, A.; Senzacqua, M.; Frontini, A.; Marzioni, D. HTRA1 in Placental Cell Models: A Possible Role in Preeclampsia. Curr. Issues Mol. Biol. 2023, 45, 3815–3828. [Google Scholar] [CrossRef]
- Fantone, S.; Giannubilo, S.R.; Marzioni, D.; Tossetta, G. HTRA family proteins in pregnancy outcome. Tissue Cell 2021, 72, 101549. [Google Scholar] [CrossRef]
- Ali, S.M.; Khalil, R.A. Genetic, immune and vasoactive factors in the vascular dysfunction associated with hypertension in pregnancy. Expert. Opin. Ther. Targets 2015, 19, 1495–1515. [Google Scholar] [CrossRef]
- Cecati, M.; Emanuelli, M.; Giannubilo, S.R.; Quarona, V.; Senetta, R.; Malavasi, F.; Tranquilli, A.L.; Saccucci, F. Contribution of adenosine-producing ectoenzymes to the mechanisms underlying the mitigation of maternal-fetal conflicts. J. Biol. Regul. Homeost. Agents 2013, 27, 519–529. [Google Scholar] [PubMed]
- Ferreira, L.B.; Williams, K.A.; Best, G.; Haydinger, C.D.; Smith, J.R. Inflammatory cytokines as mediators of retinal endothelial barrier dysfunction in non-infectious uveitis. Clin. Transl. Immunol. 2023, 12, e1479. [Google Scholar] [CrossRef] [PubMed]
- Mateuszuk, L.; Campagna, R.; Kutryb-Zajac, B.; Kus, K.; Slominska, E.M.; Smolenski, R.T.; Chlopicki, S. Reversal of endothelial dysfunction by nicotinamide mononucleotide via extracellular conversion to nicotinamide riboside. Biochem. Pharmacol. 2020, 178, 114019. [Google Scholar] [CrossRef] [PubMed]
- Tossetta, G.; Piani, F.; Borghi, C.; Marzioni, D. Role of CD93 in Health and Disease. Cells 2023, 12, 1778. [Google Scholar] [CrossRef] [PubMed]
- Campagna, R.; Vignini, A. NAD(+) Homeostasis and NAD(+)-Consuming Enzymes: Implications for Vascular Health. Antioxidants 2023, 12, 376. [Google Scholar] [CrossRef]
- Campagna, R.; Mateuszuk, L.; Wojnar-Lason, K.; Kaczara, P.; Tworzydlo, A.; Kij, A.; Bujok, R.; Mlynarski, J.; Wang, Y.; Sartini, D.; et al. Nicotinamide N-methyltransferase in endothelium protects against oxidant stress-induced endothelial injury. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 119082. [Google Scholar] [CrossRef]
- Szczesny-Malysiak, E.; Stojak, M.; Campagna, R.; Grosicki, M.; Jamrozik, M.; Kaczara, P.; Chlopicki, S. Bardoxolone Methyl Displays Detrimental Effects on Endothelial Bioenergetics, Suppresses Endothelial ET-1 Release, and Increases Endothelial Permeability in Human Microvascular Endothelium. Oxid. Med. Cell Longev. 2020, 2020, 4678252. [Google Scholar] [CrossRef]
- Piani, F.; Tossetta, G.; Cara-Fuentes, G.; Agnoletti, D.; Marzioni, D.; Borghi, C. Diagnostic and Prognostic Role of CD93 in Cardiovascular Disease: A Systematic Review. Biomolecules 2023, 13, 910. [Google Scholar] [CrossRef]
- Ristovska, E.C.; Genadieva-Dimitrova, M.; Todorovska, B.; Milivojevic, V.; Rankovic, I.; Samardziski, I.; Bojadzioska, M. The Role of Endothelial Dysfunction in the Pathogenesis of Pregnancy-Related Pathological Conditions: A Review. Pril (Makedon. Akad. Nauk. Umet. Odd. Med. Nauki) 2023, 44, 113–137. [Google Scholar] [CrossRef]
- Shen, J.; San, W.; Zheng, Y.; Zhang, S.; Cao, D.; Chen, Y.; Meng, G. Different types of cell death in diabetic endothelial dysfunction. Biomed. Pharmacother. 2023, 168, 115802. [Google Scholar] [CrossRef]
- Tossetta, G.; Fantone, S.; Delli Muti, N.; Balercia, G.; Ciavattini, A.; Giannubilo, S.R.; Marzioni, D. Preeclampsia and severe acute respiratory syndrome coronavirus 2 infection: A systematic review. J. Hypertens. 2022, 40, 1629–1638. [Google Scholar] [CrossRef] [PubMed]
- Fantone, S.; Tossetta, G.; Di Simone, N.; Tersigni, C.; Scambia, G.; Marcheggiani, F.; Giannubilo, S.R.; Marzioni, D. CD93 a potential player in cytotrophoblast and endothelial cell migration. Cell Tissue Res. 2022, 387, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Cevher Akdulum, M.F.; Demirdag, E.; Arik, S.I.; Safarova, S.; Erdem, M.; Bozkurt, N.; Erdem, A. Is the First-Trimester Systemic Immune-Inflammation Index Associated With Preeclampsia? Cureus 2023, 15, e44063. [Google Scholar] [CrossRef] [PubMed]
- Tossetta, G.; Fantone, S.; Piani, F.; Crescimanno, C.; Ciavattini, A.; Giannubilo, S.R.; Marzioni, D. Modulation of NRF2/KEAP1 Signaling in Preeclampsia. Cells 2023, 12, 1545. [Google Scholar] [CrossRef] [PubMed]
- Mazzanti, L.; Cecati, M.; Vignini, A.; D’Eusanio, S.; Emanuelli, M.; Giannubilo, S.R.; Saccucci, F.; Tranquilli, A.L. Placental expression of endothelial and inducible nitric oxide synthase and nitric oxide levels in patients with HELLP syndrome. Am. J. Obstet. Gynecol. 2011, 205, 236.e1-7. [Google Scholar] [CrossRef] [PubMed]
- Fantone, S.; Ermini, L.; Piani, F.; Di Simone, N.; Barbaro, G.; Giannubilo, S.R.; Gesuita, R.; Tossetta, G.; Marzioni, D. Downregulation of argininosuccinate synthase 1 (ASS1) is associated with hypoxia in placental development. Hum. Cell 2023, 36, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Cecati, M.; Giannubilo, S.R.; Saccucci, F.; Sartini, D.; Ciavattini, A.; Emanuelli, M.; Tranquilli, A.L. Potential Role of Placental Klotho in the Pathogenesis of Preeclampsia. Cell Biochem. Biophys. 2016, 74, 49–57. [Google Scholar] [CrossRef]
- Maynard, S.E.; Karumanchi, S.A. Angiogenic factors and preeclampsia. Semin. Nephrol. 2011, 31, 33–46. [Google Scholar] [CrossRef]
- Van Niekerk, G.; Christowitz, C.; Engelbrecht, A.M. Insulin-mediated immune dysfunction in the development of preeclampsia. J. Mol. Med. 2021, 99, 889–897. [Google Scholar] [CrossRef]
- Leavey, K.; Benton, S.J.; Grynspan, D.; Kingdom, J.C.; Bainbridge, S.A.; Cox, B.J. Unsupervised Placental Gene Expression Profiling Identifies Clinically Relevant Subclasses of Human Preeclampsia. Hypertension 2016, 68, 137–147. [Google Scholar] [CrossRef]
- Benton, S.J.; Leavey, K.; Grynspan, D.; Cox, B.J.; Bainbridge, S.A. The clinical heterogeneity of preeclampsia is related to both placental gene expression and placental histopathology. Am. J. Obstet. Gynecol. 2018, 219, 604.e1–604.e25. [Google Scholar] [CrossRef]
- Rana, S.; Lemoine, E.; Granger, J.P.; Karumanchi, S.A. Preeclampsia: Pathophysiology, Challenges, and Perspectives. Circ. Res. 2019, 124, 1094–1112. [Google Scholar] [CrossRef] [PubMed]
- Licini, C.; Avellini, C.; Picchiassi, E.; Mensa, E.; Fantone, S.; Ramini, D.; Tersigni, C.; Tossetta, G.; Castellucci, C.; Tarquini, F.; et al. Pre-eclampsia predictive ability of maternal miR-125b: A clinical and experimental study. Transl. Res. 2021, 228, 13–27. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Pei, J.; Zhang, X.; Wang, C.; Tang, Y.; Liu, H.; Yu, Y.; Luo, S.; Gu, W. APOA1 Is a Novel Marker for Preeclampsia. Int. J. Mol. Sci. 2023, 24, 16363. [Google Scholar] [CrossRef] [PubMed]
- Piani, F.; Agnoletti, D.; Baracchi, A.; Scarduelli, S.; Verde, C.; Tossetta, G.; Montaguti, E.; Simonazzi, G.; Degli Esposti, D.; Borghi, C.; et al. Serum uric acid to creatinine ratio and risk of preeclampsia and adverse pregnancy outcomes. J. Hypertens. 2023, 41, 1333–1338. [Google Scholar] [CrossRef]
- Soobryan, N.; Reddy, K.; Ibrahim, U.H.; Moodley, J.; Kumar, A.; Mackraj, I. Identification of gene signature markers in gestational hypertension and early-onset pre-eclampsia. Placenta 2023, 145, 1–8. [Google Scholar] [CrossRef]
- Piani, F.; Tossetta, G.; Fantone, S.; Agostinis, C.; Di Simone, N.; Mandala, M.; Bulla, R.; Marzioni, D.; Borghi, C. First Trimester CD93 as a Novel Marker of Preeclampsia and Its Complications: A Pilot Study. High. Blood Press. Cardiovasc. Prev. 2023, 30, 591–594. [Google Scholar] [CrossRef]
- Qin, S.; Sun, N.; Xu, L.; Xu, Y.; Tang, Q.; Tan, L.; Chen, A.; Zhang, L.; Liu, S. The Value of Circulating microRNAs for Diagnosis and Prediction of Preeclampsia: A Meta-analysis and Systematic Review. Reprod. Sci. 2022, 29, 3078–3090. [Google Scholar] [CrossRef]
- Ping, Z.; Feng, Y.; Lu, Y.; Ai, L.; Jiang, H. Integrated analysis of microRNA and mRNA expression profiles in Preeclampsia. BMC Med. Genom. 2023, 16, 309. [Google Scholar] [CrossRef]
- Kimura, T. Non-coding Natural Antisense RNA: Mechanisms of Action in the Regulation of Target Gene Expression and Its Clinical Implications. Yakugaku Zasshi 2020, 140, 687–700. [Google Scholar] [CrossRef]
- Zeng, Q.; Wan, H.; Zhao, S.; Xu, H.; Tang, T.; Oware, K.A.; Qu, S. Role of PIWI-interacting RNAs on cell survival: Proliferation, apoptosis, and cycle. IUBMB Life 2020, 72, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yu, Y.; Xu, B.; Zhang, M.; Li, Q.; Miao, L. Pivotal prognostic and diagnostic role of the long non-coding RNA colon cancer-associated transcript 1 expression in human cancer (Review). Mol. Med. Rep. 2019, 19, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; An, Y.; Liang, Y.; Xie, X.W. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 1930–1936. [Google Scholar] [PubMed]
- Shields, E.J.; Petracovici, A.F.; Bonasio, R. lncRedibly versatile: Biochemical and biological functions of long noncoding RNAs. Biochem. J. 2019, 476, 1083–1104. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Gahan, P.B. MicroRNA Shuttle from Cell-To-Cell by Exosomes and Its Impact in Cancer. Noncoding RNA 2019, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Avellini, C.; Licini, C.; Lazzarini, R.; Gesuita, R.; Guerra, E.; Tossetta, G.; Castellucci, C.; Giannubilo, S.R.; Procopio, A.; Alberti, S.; et al. The trophoblast cell surface antigen 2 and miR-125b axis in urothelial bladder cancer. Oncotarget 2017, 8, 58642–58653. [Google Scholar] [CrossRef]
- Filipow, S.; Laczmanski, L. Blood Circulating miRNAs as Cancer Biomarkers for Diagnosis and Surgical Treatment Response. Front. Genet. 2019, 10, 169. [Google Scholar] [CrossRef]
- Cui, L.; Zhang, X.; Ye, G.; Zheng, T.; Song, H.; Deng, H.; Xiao, B.; Xia, T.; Yu, X.; Le, Y.; et al. Gastric juice MicroRNAs as potential biomarkers for the screening of gastric cancer. Cancer 2013, 119, 1618–1626. [Google Scholar] [CrossRef]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef] [PubMed]
- Hanke, M.; Hoefig, K.; Merz, H.; Feller, A.C.; Kausch, I.; Jocham, D.; Warnecke, J.M.; Sczakiel, G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. 2010, 28, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- Mayya, V.K.; Duchaine, T.F. Ciphers and Executioners: How 3′-Untranslated Regions Determine the Fate of Messenger RNAs. Front. Genet. 2019, 10, 6. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef]
- Chen, Y.M.; Zheng, Y.L.; Su, X.; Wang, X.Q. Crosstalk Between MicroRNAs and Circular RNAs in Human Diseases: A Bibliographic Study. Front. Cell Dev. Biol. 2021, 9, 754880. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Quintero, B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif. 2016, 49, 281–303. [Google Scholar] [CrossRef] [PubMed]
- Simionescu, N.; Niculescu, L.S.; Sanda, G.M.; Margina, D.; Sima, A.V. Analysis of circulating microRNAs that are specifically increased in hyperlipidemic and/or hyperglycemic sera. Mol. Biol. Rep. 2014, 41, 5765–5773. [Google Scholar] [CrossRef]
- Axmann, M.; Meier, S.M.; Karner, A.; Strobl, W.; Stangl, H.; Plochberger, B. Serum and Lipoprotein Particle miRNA Profile in Uremia Patients. Genes 2018, 9, 533. [Google Scholar] [CrossRef]
- Watts, G.F.; Barrett, P.H. High-density lipoprotein metabolism in familial hypercholesterolaemia: Significance, mechanisms, therapy. Nutr. Metab. Cardiovasc. Dis. 2002, 12, 36–41. [Google Scholar]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Sabanovic, B.; Piva, F.; Cecati, M.; Giulietti, M. Promising Extracellular Vesicle-Based Vaccines against Viruses, Including SARS-CoV-2. Biology 2021, 10, 94. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Li, C.; Zhang, Y.; Zhang, D.; Otterbein, L.E.; Jin, Y. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 2019, 216, 2202–2220. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V.; Bartel, B.; Bartel, D.P.; Burge, C.B.; Carrington, J.C.; Chen, X.; Dreyfuss, G.; Eddy, S.R.; Griffiths-Jones, S.; Marshall, M.; et al. A uniform system for microRNA annotation. RNA 2003, 9, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Chirshev, E.; Oberg, K.C.; Ioffe, Y.J.; Unternaehrer, J.J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 2019, 8, 24. [Google Scholar] [CrossRef] [PubMed]
- Donker, R.B.; Mouillet, J.F.; Chu, T.; Hubel, C.A.; Stolz, D.B.; Morelli, A.E.; Sadovsky, Y. The expression profile of C19MC microRNAs in primary human trophoblast cells and exosomes. Mol. Hum. Reprod. 2012, 18, 417–424. [Google Scholar] [CrossRef]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial exosomes/microvesicles in the uterine microenvironment: A new paradigm for embryo-endometrial cross talk at implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef] [PubMed]
- Rekker, K.; Altmae, S.; Suhorutshenko, M.; Peters, M.; Martinez-Blanch, J.F.; Codoner, F.M.; Vilella, F.; Simon, C.; Salumets, A.; Velthut-Meikas, A. A Two-Cohort RNA-seq Study Reveals Changes in Endometrial and Blood miRNome in Fertile and Infertile Women. Genes 2018, 9, 574. [Google Scholar] [CrossRef]
- Aplin, J.D.; Kimber, S.J. Trophoblast-uterine interactions at implantation. Reprod. Biol. Endocrinol. 2004, 2, 48. [Google Scholar] [CrossRef]
- Desrochers, L.M.; Bordeleau, F.; Reinhart-King, C.A.; Cerione, R.A.; Antonyak, M.A. Microvesicles provide a mechanism for intercellular communication by embryonic stem cells during embryo implantation. Nat. Commun. 2016, 7, 11958. [Google Scholar] [CrossRef]
- Cretoiu, D.; Xu, J.; Xiao, J.; Suciu, N.; Cretoiu, S.M. Circulating MicroRNAs as Potential Molecular Biomarkers in Pathophysiological Evolution of Pregnancy. Dis. Markers 2016, 2016, 3851054. [Google Scholar] [CrossRef]
- Liu, L.; Guo, J.; Gao, W.; Gao, M.; Ma, X. Research progress in the role of non-coding RNAs and embryo implantation. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2023, 48, 1377–1387. [Google Scholar] [PubMed]
- Gurung, S.; Greening, D.W.; Catt, S.; Salamonsen, L.; Evans, J. Exosomes and soluble secretome from hormone-treated endometrial epithelial cells direct embryo implantation. Mol. Hum. Reprod. 2020, 26, 510–520. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Kale, V. Extracellular vesicles: Mediators of embryo-maternal crosstalk during pregnancy and a new weapon to fight against infertility. Eur. J. Cell Biol. 2020, 99, 151125. [Google Scholar] [CrossRef] [PubMed]
- Atay, S.; Gercel-Taylor, C.; Suttles, J.; Mor, G.; Taylor, D.D. Trophoblast-derived exosomes mediate monocyte recruitment and differentiation. Am. J. Reprod. Immunol. 2011, 65, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Whitley, G.S.; Dash, P.R.; Ayling, L.J.; Prefumo, F.; Thilaganathan, B.; Cartwright, J.E. Increased apoptosis in first trimester extravillous trophoblasts from pregnancies at higher risk of developing preeclampsia. Am. J. Pathol. 2007, 170, 1903–1909. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.; Zhong, X.Y.; Rusterholz, C.; Hahn, S.; Holzgreve, W.; Redman, C.W.; Sargent, I.L. The effect of labour and placental separation on the shedding of syncytiotrophoblast microparticles, cell-free DNA and mRNA in normal pregnancy and pre-eclampsia. Placenta 2008, 29, 942–949. [Google Scholar] [CrossRef]
- Czernek, L.; Duchler, M. Exosomes as Messengers Between Mother and Fetus in Pregnancy. Int. J. Mol. Sci. 2020, 21, 4264. [Google Scholar] [CrossRef]
- Truong, G.; Guanzon, D.; Kinhal, V.; Elfeky, O.; Lai, A.; Longo, S.; Nuzhat, Z.; Palma, C.; Scholz-Romero, K.; Menon, R.; et al. Oxygen tension regulates the miRNA profile and bioactivity of exosomes released from extravillous trophoblast cells—Liquid biopsies for monitoring complications of pregnancy. PLoS ONE 2017, 12, e0174514. [Google Scholar] [CrossRef]
- Yang, X.W.; Shen, G.Z.; Cao, L.Q.; Jiang, X.F.; Peng, H.P.; Shen, G.; Chen, D.; Xue, P. MicroRNA-1269 promotes proliferation in human hepatocellular carcinoma via downregulation of FOXO1. BMC Cancer 2014, 14, 909. [Google Scholar] [CrossRef]
- Fan, W.; Li, S.W.; Li, W.H.; Wang, Y.; Gong, Y.; Ma, Q.H.; Luo, S. FOXO1 expression and regulation in endometrial tissue during the menstrual cycle and in early pregnancy decidua. Gynecol. Obstet. Invest. 2012, 74, 56–63. [Google Scholar] [CrossRef]
- Kamali Simsek, N.; Benian, A.; Sevgin, K.; Ergun, Y.; Goksever Celik, H.; Karahuseyinoglu, S.; Gunel, T. Microrna analysis of human decidua mesenchymal stromal cells from preeclampsia patients. Placenta 2021, 115, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Zandvakili, I.; Lin, Y.; Morris, J.C.; Zheng, Y. Rho GTPases: Anti- or pro-neoplastic targets? Oncogene 2017, 36, 3213–3222. [Google Scholar] [CrossRef]
- Taga, S.; Hayashi, M.; Nunode, M.; Nakamura, N.; Ohmichi, M. miR-486-5p inhibits invasion and migration of HTR8/SVneo trophoblast cells by down-regulating ARHGAP5. Placenta 2022, 123, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Holcik, M.; Korneluk, R.G. XIAP, the guardian angel. Nat. Rev. Mol. Cell Biol. 2001, 2, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.; Price, M.; Straszewski-Chavez, S.; Torry, R.J.; Mor, G.; Torry, D.S. XIAP protein is induced by placenta growth factor (PLGF) and decreased during preeclampsia in trophoblast cells. Syst. Biol. Reprod. Med. 2014, 60, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Nunode, M.; Hayashi, M.; Nagayasu, Y.; Sawada, M.; Nakamura, M.; Sano, T.; Fujita, D.; Ohmichi, M. miR-515-5p suppresses trophoblast cell invasion and proliferation through XIAP regulation in preeclampsia. Mol. Cell Endocrinol. 2023, 559, 111779. [Google Scholar] [CrossRef] [PubMed]
- Jauniaux, E.; Gulbis, B.; Burton, G.J. Physiological implications of the materno-fetal oxygen gradient in human early pregnancy. Reprod. Biomed. Online 2003, 7, 250–253. [Google Scholar] [CrossRef]
- Wang, Y.; Lumbers, E.R.; Arthurs, A.L.; Corbisier de Meaultsart, C.; Mathe, A.; Avery-Kiejda, K.A.; Roberts, C.T.; Pipkin, F.B.; Marques, F.Z.; Morris, B.J.; et al. Regulation of the human placental (pro)renin receptor-prorenin-angiotensin system by microRNAs. Mol. Hum. Reprod. 2018, 24, 453–464. [Google Scholar] [CrossRef]
- Lalevee, S.; Lapaire, O.; Buhler, M. miR455 is linked to hypoxia signaling and is deregulated in preeclampsia. Cell Death Dis. 2014, 5, e1408. [Google Scholar] [CrossRef]
- Gunel, T.; Zeybek, Y.G.; Akcakaya, P.; Kalelioglu, I.; Benian, A.; Ermis, H.; Aydinli, K. Serum microRNA expression in pregnancies with preeclampsia. Genet. Mol. Res. 2011, 10, 4034–4040. [Google Scholar] [CrossRef]
- Campos, C.B.; Marques, T.M.; Pereira, R.W.; Sandrim, V.C. Reduced circulating miR-196b levels is associated with preeclampsia. Pregnancy Hypertens. 2014, 4, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Sandrim, V.C.; Eleuterio, N.; Pilan, E.; Tanus-Santos, J.E.; Fernandes, K.; Cavalli, R. Plasma levels of increased miR-195-5p correlates with the sFLT-1 levels in preeclampsia. Hypertens. Pregnancy 2016, 35, 150–158. [Google Scholar] [CrossRef]
- Sandrim, V.C.; Luizon, M.R.; Palei, A.C.; Tanus-Santos, J.E.; Cavalli, R.C. Circulating microRNA expression profiles in pre-eclampsia: Evidence of increased miR-885-5p levels. BJOG 2016, 123, 2120–2128. [Google Scholar] [CrossRef] [PubMed]
- Sheng, C.; Zhao, Y.; Zhu, L. Down-regulation of EDN1 gene expression by circulating miR-206 is associated with risk of preeclampsia. Medicine 2020, 99, e20319. [Google Scholar] [CrossRef] [PubMed]
- Akgor, U.; Ayaz, L.; Cayan, F. Expression levels of maternal plasma microRNAs in preeclamptic pregnancies. J. Obstet. Gynaecol. 2021, 41, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, S.E.; Shaker, O.G.; Aboshama, R.A.; Etman, M.K.; Khalefa, A.A.; Khamiss Abd Elguaad, M.M.; Zaki, O.M.; Ali, D.Y.; Hemeda, N.F.; Amin, A.; et al. Expression profile of LncRNA ANRIL, miR-186, miR-181a, and MTMR-3 in patients with preeclampsia. Noncoding RNA Res. 2023, 8, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Sekar, D.; Lakshmanan, G.; Mani, P.; Biruntha, M. Methylation-dependent circulating microRNA 510 in preeclampsia patients. Hypertens. Res. 2019, 42, 1647–1648. [Google Scholar] [CrossRef]
- Yang, Q.; Lu, J.; Wang, S.; Li, H.; Ge, Q.; Lu, Z. Application of next-generation sequencing technology to profile the circulating microRNAs in the serum of preeclampsia versus normal pregnant women. Clin. Chim. Acta 2011, 412, 2167–2173. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Kotlabova, K.; Ondrackova, M.; Kestlerova, A.; Novotna, V.; Hympanova, L.; Doucha, J.; Krofta, L. Circulating C19MC microRNAs in preeclampsia, gestational hypertension, and fetal growth restriction. Mediat. Inflamm. 2013, 2013, 186041. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, H.; Lin, H.; Qi, J.; Zhu, C.; Gao, Z.; Wang, H. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction 2012, 143, 389–397. [Google Scholar] [CrossRef]
- Khaliq, O.P.; Murugesan, S.; Moodley, J.; Mackraj, I. Differential expression of miRNAs are associated with the insulin signaling pathway in preeclampsia and gestational hypertension. Clin. Exp. Hypertens. 2018, 40, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Eghbal-Fard, S.; Yousefi, M.; Heydarlou, H.; Ahmadi, M.; Taghavi, S.; Movasaghpour, A.; Jadidi-Niaragh, F.; Yousefi, B.; Dolati, S.; Hojjat-Farsangi, M.; et al. The imbalance of Th17/Treg axis involved in the pathogenesis of preeclampsia. J. Cell Physiol. 2019, 234, 5106–5116. [Google Scholar] [CrossRef] [PubMed]
- Witvrouwen, I.; Mannaerts, D.; Ratajczak, J.; Boeren, E.; Faes, E.; Van Craenenbroeck, A.H.; Jacquemyn, Y.; Van Craenenbroeck, E.M. MicroRNAs targeting VEGF are related to vascular dysfunction in preeclampsia. Biosci. Rep. 2021, 41, BSR20210874. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Park, M.; Kim, J.Y.; Kim, T.; Hwang, J.Y.; Ha, K.S.; Won, M.H.; Ryoo, S.; Kwon, Y.G.; Kim, Y.M. Circulating miRNAs Associated with Dysregulated Vascular and Trophoblast Function as Target-Based Diagnostic Biomarkers for Preeclampsia. Cells 2020, 9, 2003. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, D.; Dai, Y.; Li, R.; Zheng, Y.; Zhao, G.; Wang, J.; Diao, Z.; Cao, C.; Lv, H.; et al. Elevated Placental microRNA-155 Is a Biomarker of a Preeclamptic Subtype. Hypertension 2023, 80, 370–384. [Google Scholar] [CrossRef] [PubMed]
- Motawi, T.M.K.; Sabry, D.; Maurice, N.W.; Rizk, S.M. Role of mesenchymal stem cells exosomes derived microRNAs; miR-136, miR-494 and miR-495 in pre-eclampsia diagnosis and evaluation. Arch. Biochem. Biophys. 2018, 659, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Du, X.; Wang, J. Transfer of miR-15a-5p by placental exosomes promotes pre-eclampsia progression by regulating PI3K/AKT signaling pathway via CDK1. Mol. Immunol. 2020, 128, 277–286. [Google Scholar] [CrossRef]
- Ntsethe, A.; Mackraj, I. An Investigation of Exosome Concentration and Exosomal microRNA (miR-155 and miR-222) Expression in Pregnant Women with Gestational Hypertension and Preeclampsia. Int. J. Womens Health 2022, 14, 1681–1689. [Google Scholar] [CrossRef]
- Aharon, A.; Rebibo-Sabbah, A.; Ahmad, R.S.; Dangot, A.; Bar-Lev, T.H.; Brenner, B.; Cohen, A.H.; David, C.B.; Weiner, Z.; Solt, I. Associations of maternal and placental extracellular vesicle miRNA with preeclampsia. Front. Cell Dev. Biol. 2023, 11, 1080419. [Google Scholar] [CrossRef]
- Biro, O.; Fothi, A.; Alasztics, B.; Nagy, B.; Orban, T.I.; Rigo, J., Jr. Circulating exosomal and Argonaute-bound microRNAs in preeclampsia. Gene 2019, 692, 138–144. [Google Scholar] [CrossRef]
- Luque, A.; Farwati, A.; Crovetto, F.; Crispi, F.; Figueras, F.; Gratacos, E.; Aran, J.M. Usefulness of circulating microRNAs for the prediction of early preeclampsia at first-trimester of pregnancy. Sci. Rep. 2014, 4, 4882. [Google Scholar] [CrossRef]
- Sandrim, V.C.; Diniz, S.; Eleuterio, N.M.; Gomes, K.B.; Dusse, L.M.S.; Cavalli, R.C. Higher levels of circulating TIMP-4 in preeclampsia is strongly associated with clinical parameters and microRNA. Clin. Exp. Hypertens. 2018, 40, 609–612. [Google Scholar] [CrossRef] [PubMed]
- Gunel, T.; Kamali, N.; Hosseini, M.K.; Gumusoglu, E.; Benian, A.; Aydinli, K. Regulatory effect of miR-195 in the placental dysfunction of preeclampsia. J. Matern. Fetal Neonatal Med. 2020, 33, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Higashijima, A.; Murakami, Y.; Tsukamoto, O.; Hasegawa, Y.; Abe, S.; Fuchi, N.; Miura, S.; Kaneuchi, M.; Masuzaki, H. Circulating chromosome 19 miRNA cluster microRNAs in pregnant women with severe pre-eclampsia. J. Obstet. Gynaecol. Res. 2015, 41, 1526–1532. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Zhang, X.; Ma, L.; Gao, N.; Tang, H.; Jian, F.; Ma, Y. Downregulations of circulating miR-31 and miR-21 are associated with preeclampsia. Pregnancy Hypertens. 2019, 17, 59–63. [Google Scholar] [CrossRef] [PubMed]
- Whigham, C.A.; MacDonald, T.M.; Walker, S.P.; Pritchard, N.; Hannan, N.J.; Cannon, P.; Nguyen, T.V.; Hastie, R.; Tong, S.; Kaitu’u-Lino, T.J. Circulating GATA2 mRNA is decreased among women destined to develop preeclampsia and may be of endothelial origin. Sci. Rep. 2019, 9, 235. [Google Scholar] [CrossRef] [PubMed]
- Kolkova, Z.; Holubekova, V.; Grendar, M.; Nachajova, M.; Zubor, P.; Pribulova, T.; Loderer, D.; Zigo, I.; Biringer, K.; Hornakova, A. Association of Circulating miRNA Expression with Preeclampsia, Its Onset, and Severity. Diagnostics 2021, 11, 476. [Google Scholar] [CrossRef] [PubMed]
- Whigham, C.A.; MacDonald, T.M.; Walker, S.P.; Hiscock, R.; Hannan, N.J.; Pritchard, N.; Cannon, P.; Nguyen, T.V.; Miranda, M.; Tong, S.; et al. MicroRNAs 363 and 149 are differentially expressed in the maternal circulation preceding a diagnosis of preeclampsia. Sci. Rep. 2020, 10, 18077. [Google Scholar] [CrossRef]
- Akehurst, C.; Small, H.Y.; Sharafetdinova, L.; Forrest, R.; Beattie, W.; Brown, C.E.; Robinson, S.W.; McClure, J.D.; Work, L.M.; Carty, D.M.; et al. Differential expression of microRNA-206 and its target genes in preeclampsia. J. Hypertens. 2015, 33, 2068–2074. [Google Scholar] [CrossRef][Green Version]
- Pan, M.; Ge, Q.; Li, H.; Yang, Q.; Lu, J.; Zhang, D.; Lu, Z. Sequencing the miRNAs in maternal plasma from women before and after parturition. J. Nanosci. Nanotechnol. 2012, 12, 4035–4043. [Google Scholar] [CrossRef]
- Li, H.; Ge, Q.; Guo, L.; Lu, Z. Maternal plasma miRNAs expression in preeclamptic pregnancies. Biomed. Res. Int. 2013, 2013, 970265. [Google Scholar] [CrossRef] [PubMed]
- Biro, O.; Alasztics, B.; Molvarec, A.; Joo, J.; Nagy, B.; Rigo, J., Jr. Various levels of circulating exosomal total-miRNA and miR-210 hypoxamiR in different forms of pregnancy hypertension. Pregnancy Hypertens. 2017, 10, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Jairajpuri, D.S.; Malalla, Z.H.; Mahmood, N.; Almawi, W.Y. Circulating microRNA expression as predictor of preeclampsia and its severity. Gene 2017, 627, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Bujold, E.; Roberge, S.; Lacasse, Y.; Bureau, M.; Audibert, F.; Marcoux, S.; Forest, J.C.; Giguere, Y. Prevention of preeclampsia and intrauterine growth restriction with aspirin started in early pregnancy: A meta-analysis. Obstet. Gynecol. 2010, 116 2 Pt 1, 402–414. [Google Scholar] [CrossRef]
- Tan, M.Y.; Wright, D.; Syngelaki, A.; Akolekar, R.; Cicero, S.; Janga, D.; Singh, M.; Greco, E.; Wright, A.; Maclagan, K.; et al. Comparison of diagnostic accuracy of early screening for pre-eclampsia by NICE guidelines and a method combining maternal factors and biomarkers: Results of SPREE. Ultrasound Obstet. Gynecol. 2018, 51, 743–750. [Google Scholar] [CrossRef]
- Ura, B.; Feriotto, G.; Monasta, L.; Bilel, S.; Zweyer, M.; Celeghini, C. Potential role of circulating microRNAs as early markers of preeclampsia. Taiwan. J. Obstet. Gynecol. 2014, 53, 232–234. [Google Scholar] [CrossRef]
- Winger, E.E.; Reed, J.L.; Ji, X. First trimester PBMC microRNA predicts adverse pregnancy outcome. Am. J. Reprod. Immunol. 2014, 72, 515–526. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Kotlabova, K.; Doucha, J.; Dlouha, K.; Krofta, L. Absolute and relative quantification of placenta-specific micrornas in maternal circulation with placental insufficiency-related complications. J. Mol. Diagn. 2012, 14, 160–167. [Google Scholar] [CrossRef]
- Winger, E.E.; Reed, J.L.; Ji, X. First-trimester maternal cell microRNA is a superior pregnancy marker to immunological testing for predicting adverse pregnancy outcome. J. Reprod. Immunol. 2015, 110, 22–35. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, G.; Zhang, Y.; Yang, H.; Long, Y.; Liang, Q.; Zheng, Z. MiR-942 decreased before 20 weeks gestation in women with preeclampsia and was associated with the pathophysiology of preeclampsia in vitro. Clin. Exp. Hypertens. 2017, 39, 108–113. [Google Scholar] [CrossRef]
- Nakashima, A.; Yamanaka-Tatematsu, M.; Fujita, N.; Koizumi, K.; Shima, T.; Yoshida, T.; Nikaido, T.; Okamoto, A.; Yoshimori, T.; Saito, S. Impaired autophagy by soluble endoglin, under physiological hypoxia in early pregnant period, is involved in poor placentation in preeclampsia. Autophagy 2013, 9, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Mavreli, D.; Lykoudi, A.; Lambrou, G.; Papaioannou, G.; Vrachnis, N.; Kalantaridou, S.; Papantoniou, N.; Kolialexi, A. Deep Sequencing Identified Dysregulated Circulating MicroRNAs in Late Onset Preeclampsia. In Vivo 2020, 34, 2317–2324. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Han, Y.; Xu, P.; Yin, L.; Si, Y.; Zhang, C.; Meng, Y.; Feng, W.; Pan, Z.; Gao, Z.; et al. Elevated microRNA-125b inhibits cytotrophoblast invasion and impairs endothelial cell function in preeclampsia. Cell Death Discov. 2020, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhao, Y.; Liu, M.; Wang, Y.; Wang, H.; Li, Y.X.; Zhu, X.; Yao, Y.; Wang, H.; Qiao, J.; et al. Variations of microRNAs in human placentas and plasma from preeclamptic pregnancy. Hypertension 2014, 63, 1276–1284. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Fierro, M.L.; Garza-Veloz, I. Analysis of Circulating microRNA Signatures and Preeclampsia Development. Cells 2021, 10, 1003. [Google Scholar] [CrossRef] [PubMed]
- Hromadnikova, I.; Kotlabova, K.; Ivankova, K.; Krofta, L. First trimester screening of circulating C19MC microRNAs and the evaluation of their potential to predict the onset of preeclampsia and IUGR. PLoS ONE 2017, 12, e0171756. [Google Scholar] [CrossRef]
- Hromadnikova, I.; Dvorakova, L.; Kotlabova, K.; Krofta, L. The Prediction of Gestational Hypertension, Preeclampsia and Fetal Growth Restriction via the First Trimester Screening of Plasma Exosomal C19MC microRNAs. Int. J. Mol. Sci. 2019, 20, 2972. [Google Scholar] [CrossRef]
- Kondracka, A.; Kondracki, B.; Jaszczuk, I.; Staniczek, J.; Kwasniewski, W.; Filip, A.; Kwasniewska, A. Diagnostic potential of microRNAs Mi 517 and Mi 526 as biomarkers in the detection of hypertension and preeclampsia in the first trimester. Ginekol. Pol. 2023, 12. [Google Scholar] [CrossRef]
- Martinez-Fierro, M.L.; Garza-Veloz, I.; Gutierrez-Arteaga, C.; Delgado-Enciso, I.; Barbosa-Cisneros, O.Y.; Flores-Morales, V.; Hernandez-Delgadillo, G.P.; Rocha-Pizana, M.R.; Rodriguez-Sanchez, I.P.; Badillo-Almaraz, J.I.; et al. Circulating levels of specific members of chromosome 19 microRNA cluster are associated with preeclampsia development. Arch. Gynecol. Obstet. 2018, 297, 365–371. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, M.; Yu, H.; Zhang, J.; Zhou, R. Circulating microRNAs as biomarkers for diagnosis and prediction of preeclampsia: A systematic review and meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 253, 121–132. [Google Scholar] [CrossRef]
- Su, S.; Yang, F.; Zhong, L.; Pang, L. Circulating noncoding RNAs as early predictive biomarkers in preeclampsia: A diagnostic meta-analysis. Reprod. Biol. Endocrinol. 2021, 19, 177. [Google Scholar] [CrossRef] [PubMed]
- Stojanovska, V.; Scherjon, S.A.; Plosch, T. Preeclampsia as Modulator of Offspring Health. Biol. Reprod. 2016, 94, 53. [Google Scholar] [CrossRef] [PubMed]
- Fraser, A.; Nelson, S.M.; Macdonald-Wallis, C.; Sattar, N.; Lawlor, D.A. Hypertensive disorders of pregnancy and cardiometabolic health in adolescent offspring. Hypertension 2013, 62, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Apicella, C.; Ruano, C.S.M.; Mehats, C.; Miralles, F.; Vaiman, D. The Role of Epigenetics in Placental Development and the Etiology of Preeclampsia. Int. J. Mol. Sci. 2019, 20, 2837. [Google Scholar] [CrossRef]
- Ji, L.; Brkic, J.; Liu, M.; Fu, G.; Peng, C.; Wang, Y.L. Placental trophoblast cell differentiation: Physiological regulation and pathological relevance to preeclampsia. Mol. Aspects Med. 2013, 34, 981–1023. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, R. miRNA and methylation: A multifaceted liaison. Chembiochem 2015, 16, 195–203. [Google Scholar] [CrossRef]
- Morales, S.; Monzo, M.; Navarro, A. Epigenetic regulation mechanisms of microRNA expression. Biomol. Concepts 2017, 8, 203–212. [Google Scholar] [CrossRef]
- Wu, D.; Shi, L.; Chen, F.; Lin, Q.; Kong, J. Methylation Status of the miR-141-3p Promoter Regulates miR-141-3p Expression, Inflammasome Formation, and the Invasiveness of HTR-8/SVneo Cells. Cytogenet. Genome Res. 2021, 161, 501–513. [Google Scholar] [CrossRef]
- Brodowski, L.; Schroder-Heurich, B.; von Hardenberg, S.; Richter, K.; von Kaisenberg, C.S.; Dittrich-Breiholz, O.; Meyer, N.; Dork, T.; von Versen-Hoynck, F. MicroRNA Profiles of Maternal and Neonatal Endothelial Progenitor Cells in Preeclampsia. Int. J. Mol. Sci. 2021, 22, 5320. [Google Scholar] [CrossRef]
- Lin, C.; Rajakumar, A.; Plymire, D.A.; Verma, V.; Markovic, N.; Hubel, C.A. Maternal endothelial progenitor colony-forming units with macrophage characteristics are reduced in preeclampsia. Am. J. Hypertens. 2009, 22, 1014–1019. [Google Scholar] [CrossRef]
- Suarez, Y.; Fernandez-Hernando, C.; Pober, J.S.; Sessa, W.C. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007, 100, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Suarez, Y.; Fernandez-Hernando, C.; Yu, J.; Gerber, S.A.; Harrison, K.D.; Pober, J.S.; Iruela-Arispe, M.L.; Merkenschlager, M.; Sessa, W.C. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 14082–14087. [Google Scholar] [CrossRef] [PubMed]
- Vitale, S.G.; Fulghesu, A.M.; Mikuš, M.; Watrowski, R.; D’Alterio, M.N.; Lin, L.T.; Shah, M.; Reyes-Muñoz, E.; Sathyapalan, T.; Angioni, S. The Translational Role of miRNA in Polycystic Ovary Syndrome: From Bench to Bedside-A Systematic Literature Review. Biomedicines 2022, 10, 1816. [Google Scholar] [CrossRef] [PubMed]
| miRNAs | Diagnosed PE | Early Onset PE | Late Onset PE | Mild PE | Severe PE | Prediction of PE | Ref. |
|---|---|---|---|---|---|---|---|
| miR-15a-5p |  | [111] | |||||
 | [112] | ||||||
| miR-15b |  | [113] | |||||
| miR-16 |  | [114] | |||||
 | [115] | ||||||
| miR-16-5p |  | [116] | |||||
| miR-17-5p |  | [117] | |||||
| miR-18a |  | [118] | |||||
 | [113] | ||||||
 | [115] | ||||||
| miR-19b1 |  | [118] | |||||
 | [113] | ||||||
| miR-21 |  | [119] | |||||
 | [113] | ||||||
| miR-21-5p |  | [120] | |||||
| miR-22-5p |  | [111] | |||||
| miR-24-3p |  | ||||||
| miR-23b-5p |  | [121] | |||||
| miR-24 |  | [122] | |||||
| miR-26a |  | [122] | |||||
| miR-29a |  | [123] | |||||
 | [124] | ||||||
 | [113] | ||||||
 | [125] | ||||||
| miR-29a-3p |  | [117] | |||||
| miR-31 |  | [119] | |||||
| miR-31-5p |  | [126] | |||||
| miR-92a-1 |  | [118] | |||||
| miR-92-a-1-3p |  | [118] | |||||
| miR-93-5p |  | [111] | |||||
| miR-99-5p |  | [121] | |||||
| miR-103 |  | [122] | |||||
| miR-106a |  | [111] | |||||
| miR-106b |  | [127] | |||||
| miR-125b |  | [111] | |||||
 | [44] | ||||||
 | [125] | ||||||
| miR-125a-5p |  | [125] | |||||
| miR-126 |  | [128] | |||||
| miR-126-3p |  | [111] | |||||
| miR-130a-3p |  | [111] | |||||
| miR-130b |  | [122] | |||||
| miR-132 |  | [129] | |||||
| miR-132-3p |  | [117] | |||||
| miR-133b |  | [129] | |||||
| miR-136 |  | [130] | |||||
 | [125] | ||||||
| miR-141 |  | [131] | |||||
 | [123] | ||||||
| miR-144 |  |  | [123] | ||||
 | [113] | ||||||
| miR-146a |  | [129] | |||||
| miR-149 |  | [115] | |||||
| miR-151a-3p |  | [132] | |||||
| miR-152 |  | [133] | |||||
| miR-155 |  | [113] | |||||
 | [134] | ||||||
 | [135] | ||||||
| miR-155-5p |  | [126] | |||||
 | [120] | ||||||
| miR-181a |  | [122] | |||||
 | [122] | ||||||
 | [136] | ||||||
| miR-185 |  | [125] | |||||
| miR-186 |  | [136] | |||||
| miR-191-5p |  | [111] | |||||
 | [117] | ||||||
| miR-195-5p |  | [137] | |||||
| miR-196b |  | [138] | |||||
| miR197-3p |  | [117] | |||||
| miR-200c |  | [114] | |||||
| miR-204-3p |  | [111] | |||||
| miR-206 |  | [139] | |||||
 | [140] | ||||||
| miR-210 |  | [133] | |||||
 | [117] | ||||||
 | [118] | ||||||
 |  | [141] | |||||
 | [113] | ||||||
 | [142] | ||||||
 | [116] | ||||||
 | [129] | ||||||
| miR-214-3p |  | [126] | |||||
| miR-215 |  | [113] | |||||
| miR-218-5p |  | [117] | |||||
| miR-221 |  | [131] | |||||
| miR-218-5p |  | [117] | |||||
| miR-223 |  | [125] | |||||
| miR-302b-3p |  | [117] | |||||
| miR-320c |  | [125] | |||||
| miR-326 |  | [127] | |||||
| miR-328 |  | [117] | |||||
| miR-342-3p |  | [122] | |||||
| miR-363 |  | [115] | |||||
| miR-365a-3p |  | [111] | |||||
| miR-374a-5p |  | [111] | |||||
| miR-375 |  | [117] | |||||
| miR-424 |  | [115] | |||||
| miR-494 |  | [130] | |||||
| miR-495 |  | [130] | |||||
| miR-510 |  | [143] | |||||
| miR-512-3p |  | [144] | |||||
| miR-515-5p |  | [145] | |||||
 | [107] | ||||||
| miR-516-5p |  | [146] | |||||
| miR-516a-5p |  | [146] | |||||
| miR-516b |  | [145] | |||||
| miR-517 |  | [146] | |||||
 | [147] | ||||||
| miR-517-5p |  | [148] | |||||
 | [149] | ||||||
| mirR-517b |  | [125] | |||||
| miR-517c |  | [125] | |||||
| miR-518b |  | [113] | |||||
 | [115] | ||||||
| miR-518e |  | [125] | |||||
| miR-518f3p |  | [144] | |||||
| miR-519a |  | [125] | |||||
| miR-519d |  | [125] | |||||
| miR-520-5p |  | [145] | |||||
| miR-520a |  | [146] | |||||
| miR-520a-5p |  | [149] | |||||
| miR-520c-3p |  | [144] | |||||
| miR-520d-3p |  | [144] | |||||
| miR-520g |  | [125] | |||||
| miR-520h |  | [145] | |||||
 | [125] | ||||||
| miR-521 |  | [125] | |||||
| miR-525 |  | [146] | |||||
| miR-525-5p |  | [145] | |||||
 | [149] | ||||||
| miR-526 |  | [145] | |||||
 | [147] | ||||||
| miR-559-5p |  | [111] | |||||
| miR-526a |  | [146] | |||||
| miR-542-3p |  | [125] | |||||
| miR-573 |  | [132] | |||||
| miR-574-5p |  | [122] | |||||
 | [111] | ||||||
| miR-628-3p |  | [132] | |||||
| miR-650 |  | [113] | |||||
| miR-885-5p |  | [150] | |||||
| miR-942 |  | [151] | |||||
| miR-1229p |  | [129] | |||||
| miR-1233 |  | [152] | |||||
| miR-1244 |  | [129] | |||||
| miR-1260 |  | [125] | |||||
| miR-1272 |  | [125] | |||||
| miR-1283 |  | [115] | |||||
| miR-1290-3p |  | [126] | |||||
| miR-1323 |  | [145] | |||||
| miR-4264-5p |  | [111] | |||||
| Let-7a-5p |  | [111] | |||||
| Let-7a |  | [125] | |||||
| Let-7d |  | [125] | |||||
| Let-7f |  | [125] | |||||
| Let-7f-1 |  | [125] |
 Up-regulated;
Up-regulated;  Down-regulated.
Down-regulated.Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giannubilo, S.R.; Cecati, M.; Marzioni, D.; Ciavattini, A. Circulating miRNAs and Preeclampsia: From Implantation to Epigenetics. Int. J. Mol. Sci. 2024, 25, 1418. https://doi.org/10.3390/ijms25031418
Giannubilo SR, Cecati M, Marzioni D, Ciavattini A. Circulating miRNAs and Preeclampsia: From Implantation to Epigenetics. International Journal of Molecular Sciences. 2024; 25(3):1418. https://doi.org/10.3390/ijms25031418
Chicago/Turabian StyleGiannubilo, Stefano Raffaele, Monia Cecati, Daniela Marzioni, and Andrea Ciavattini. 2024. "Circulating miRNAs and Preeclampsia: From Implantation to Epigenetics" International Journal of Molecular Sciences 25, no. 3: 1418. https://doi.org/10.3390/ijms25031418
APA StyleGiannubilo, S. R., Cecati, M., Marzioni, D., & Ciavattini, A. (2024). Circulating miRNAs and Preeclampsia: From Implantation to Epigenetics. International Journal of Molecular Sciences, 25(3), 1418. https://doi.org/10.3390/ijms25031418








