Protein Dynamics in Plant Immunity: Insights into Plant–Pest Interactions
Abstract
1. Introduction
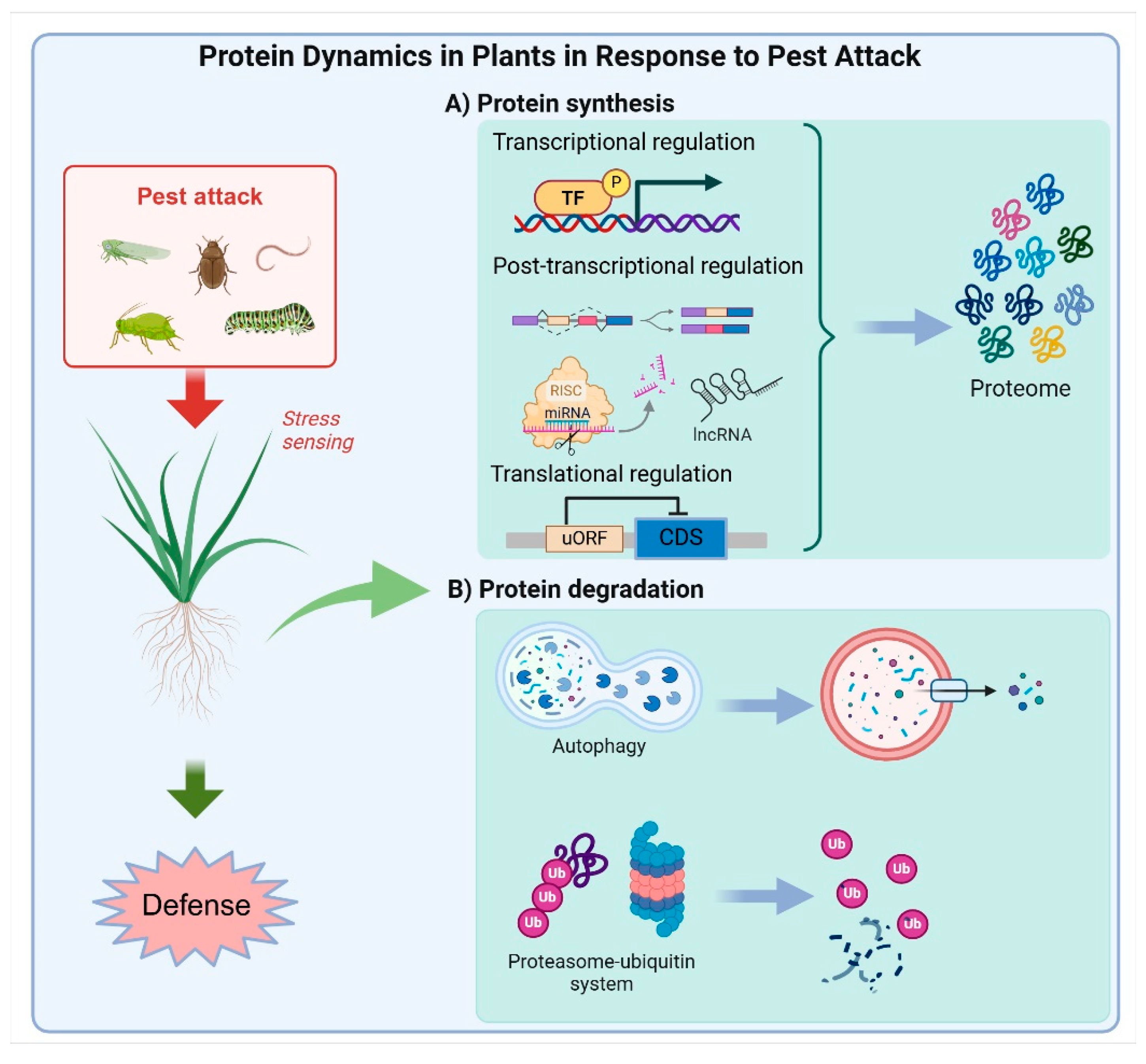
2. Plant Protein Regulations in Response to Pest Invasion
2.1. Regulation of Protein Production
2.2. Protein Degradation
2.2.1. The Ubiquitin–Proteasome System
2.2.2. Autophagy
3. Plant Proteins Targeted by Pests
3.1. UPS Components
3.2. Autophagy-Related Proteins
3.3. Phytohormone Signaling Components
3.4. Transcriptional Factors
3.5. Other Signaling Components
4. Regulation of Plant Host Proteins by Insect-Borne Microbes
5. Concluding Remarks and Future Directions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Ngou, B.; Ding, P.; Jones, J. Thirty years of resistance: Zig-zag through the plant immune system. Plant Cell 2022, 34, 1447–1478. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhu, X.; Chen, X.; Zhou, J.M. From plant immunity to crop disease resistance. J. Genet. Genomics 2022, 49, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Orosa, B.; Ustun, S.; Calderon, V.L.; Genschik, P.; Gibbs, D.; Holdsworth, M.J.; Isono, E.; Lois, M.; Trujillo, M.; Sadanandom, A. Plant proteostasis-shaping the proteome: A research community aiming to understand molecular mechanisms that control protein abundance. New Phytol. 2020, 227, 1028–1033. [Google Scholar] [CrossRef] [PubMed]
- Harper, J.W.; Bennett, E.J. Proteome complexity and the forces that drive proteome imbalance. Nature 2016, 537, 328–338. [Google Scholar] [CrossRef]
- Sels, J.; Mathys, J.; De Coninck, B.M.; Cammue, B.P.; De Bolle, M.F. Plant pathogenesis-related (PR) proteins: A focus on pr peptides. Plant Physiol. Biochem. 2008, 46, 41–950. [Google Scholar] [CrossRef]
- Hartl, M.; Giri, A.P.; Kaur, H.; Baldwin, I.T. Serine protease inhibitors specifically defend solanum nigrum against generalist herbivores but do not influence plant growth and development. Plant Cell 2010, 22, 4158–4175. [Google Scholar] [CrossRef]
- Hofius, D.; Li, L.; Hafren, A.; Coll, N.S. Autophagy as an emerging arena for plant-pathogen interactions. Curr. Opin. Plant Biol. 2017, 38, 117–123. [Google Scholar] [CrossRef]
- Tang, J.; Bassham, D.C. Autophagy in crop plants: What’s new beyond Arabidopsis? Open Biol. 2018, 8, 180162. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, C.; Kim, D.Y.; Huang, Y.; Chatt, E.; He, P.; Vierstra, R.D.; Shan, L. Ubiquitylome analysis reveals a central role for the ubiquitin-proteasome system in plant innate immunity. Plant Physiol. 2021, 185, 1943–1965. [Google Scholar] [CrossRef]
- Raffeiner, M.; Zhu, S.; Gonzalez-Fuente, M.; Ustun, S. Interplay between autophagy and proteasome during protein turnover. Trends Plant Sci. 2023, 28, 698–714. [Google Scholar] [CrossRef] [PubMed]
- Hogenhout, S.A.; Bos, J.I. Effector proteins that modulate plant-insect interactions. Curr. Opin. Plant Biol. 2011, 14, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Kaloshian, I.; Walling, L.L. Hemipteran and dipteran pests: Effectors and plant host immune regulators. J. Integr. Plant Biol. 2016, 58, 350–361. [Google Scholar] [CrossRef] [PubMed]
- van der Hoorn, R.A.; Kamoun, S. From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 2008, 20, 2009–2017. [Google Scholar] [CrossRef]
- Ma, Z.C.; Lin, Z.; Song, T.Q.; Wang, Y.; Zhang, Q.; Xia, Y.Q.; Qiu, M.; Lin, Y.C.; Li, H.Y.; Kong, L.; et al. A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 2017, 355, 710–714. [Google Scholar] [CrossRef]
- Sonawala, U.; Beasley, H.; Thorpe, P.; Varypatakis, K.; Senatori, B.T.; Jones, J.; Derevnina, L.; Sebastian. A gene with a thousand alleles: The hyper-variable effectors of plant-parasitic nematodes. Cell Genom. 2024, 4, 100580. [Google Scholar] [CrossRef]
- Erb, M.; Reymond, P. Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 2019, 70, 527–557. [Google Scholar] [CrossRef]
- Divekar, P.A.; Narayana, S.; Divekar, B.A.; Kumar, R.; Gadratagi, B.G.; Ray, A.; Singh, A.K.; Rani, V.; Singh, V.; Singh, A.K.; et al. Plant secondary metabolites as defense tools against herbivores for sustainable crop protection. Int. J. Mol. Sci. 2022, 23, 2690. [Google Scholar] [CrossRef]
- Kehr, J. Phloem sap proteins: Their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 2006, 57, 767–774. [Google Scholar] [CrossRef]
- Absmanner, B.; Stadler, R.; Hammes, U.Z. Phloem development in nematode-induced feeding sites: The implications of auxin and cytokinin. Front. Plant Sci. 2013, 4, 241. [Google Scholar] [CrossRef]
- Ali, M.A.; Anjam, M.S.; Nawaz, M.A.; Lam, H.M.; Chung, G. Signal transduction in plant-nematode interactions. Int. J. Mol. Sci. 2018, 19, 1648. [Google Scholar]
- Gatehouse, J.A. Plant resistance towards insect herbivores: A dynamic interaction. New Phytol. 2002, 156, 145–169. [Google Scholar] [CrossRef] [PubMed]
- Appel, H.M.; Fescemyer, H.; Ehlting, J.; Weston, D.; Rehrig, E.; Joshi, T.; Xu, D.; Bohlmann, J.; Schultz, J. Transcriptional responses of Arabidopsis thaliana to chewing and sucking insect herbivores. Front. Plant Sci. 2014, 5, 565. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, L.; Hull, J.J.; Liang, S.; Daniell, H.; Jin, S.; Zhang, X. Transcriptome analysis reveals a comprehensive insect resistance response mechanism in cotton to infestation by the phloem feeding insect Bemisia tabaci (Whitefly). Plant Bio. J. 2016, 14, 1956–1975. [Google Scholar] [CrossRef] [PubMed]
- Ji, M.; Bui, H.; Ramirez, R.A.; Clark, R.M. Concerted cis and trans effects underpin heightened defense gene expression in multi-herbivore-resistant maize lines. Plant J. 2022, 111, 508–528. [Google Scholar] [CrossRef] [PubMed]
- Zha, W.; Li, C.; Wu, Y.; Chen, J.; Li, S.; Sun, M.; Wu, B.; Shi, S.; Liu, K.; Xu, H.; et al. Single-cell RNA sequencing of leaf sheath cells reveals the mechanism of rice resistance to brown planthopper (Nilaparvata lugens). Front. Plant Sci. 2023, 14, 1200014. [Google Scholar] [CrossRef]
- Lee, M.J.; Yaffe, M.B. Protein regulation in signal transduction. Cold Spring Harb. Perspect. Biol. 2016, 8, a005918. [Google Scholar] [CrossRef]
- He, J.; Liu, Y.; Yuan, D.; Duan, M.; Liu, Y.; Shen, Z.; Yang, C.; Qiu, Z.; Liu, D.; Wen, P.; et al. An R2R3 MYB transcription factor confers brown planthopper resistance by regulating the phenylalanine ammonia-lyase pathway in rice. Proc. Natl. Acad. Sci. USA 2020, 117, 271–277. [Google Scholar] [CrossRef]
- Kumar, A.; Sichov, N.; Bucki, P.; Miyara, S.B. SlWRKY16 and SlWRKY31 of tomato, negative regulators of plant defense, involved in susceptibility activation following root-knot nematode Meloidogyne javanica infection. Sci. Rep. 2023, 13, 14592. [Google Scholar] [CrossRef]
- Ling, Z.; Zhou, W.; Baldwin, I.T.; Xu, S. Insect herbivory elicits genome-wide alternative splicing responses in Nicotiana attenuata. Plant J. 2015, 824, 28–43. [Google Scholar] [CrossRef]
- Chen, D.Y.; Chen, Q.Y.; Wang, D.D.; Mu, Y.P.; Wang, M.Y.; Huang, J.R.; Mao, Y.B. Differential transcription and alternative splicing in cotton underly specialized defense responses against pests. Front. Plant Sci. 2020, 11, 573131. [Google Scholar] [CrossRef]
- Pandey, S.P.; Shahi, P.; Gase, K.; Baldwin, I.T. Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana Attenuata. Proc. Natl. Acad. Sci. USA 2008, 105, 4559–4564. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Jin, J.; Xu, J.; Wang, L.; Li, J.; Lou, Y.; Baldwin, I.T. Long non-coding RNAs associate with jasmonate-mediated plant defence against herbivores. Plant Cell Environ. 2021, 44, 982–994. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zha, W.; Qiu, D.; Guo, J.; Liu, G.; Li, C.; Wu, B.; Li, S.; Chen, J.; Hu, L.; et al. Comprehensive identification and characterization of lncRNAs and circRNAs reveal potential brown planthopper-responsive ceRNA networks in rice. Front. Plant Sci. 2023, 14, 1242089. [Google Scholar] [CrossRef]
- Fan, D.; Ran, L.; Hu, J.; Ye, X.; Xu, D.; Li, J.; Su, H.; Wang, X.; Ren, S.; Luo, K. miR319a/TCP module and DELLA protein regulate trichome initiation synergistically and improve insect defenses in Populus tomentosa. New Phytol. 2020, 227, 867–883. [Google Scholar] [CrossRef]
- Sun, B.; Shen, Y.; Zhu, L.; Yang, X.; Liu, X.; Li, D.; Zhu, M.; Miao, X.; Shi, Z. OsmiR319-OsPCF5 modulate resistance to brown planthopper in rice through association with MYB proteins. BMC Biol. 2024, 22, 68. [Google Scholar] [CrossRef]
- Zhao, W.; Li, Z.; Fan, J.; Hu, C.; Yang, R.; Qi, X.; Chen, H.; Zhao, F.; Wang, S. Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J. Exp. Bot. 2015, 66, 4653–4667. [Google Scholar] [CrossRef]
- Withers, J.; Dong, X. Post-translational regulation of plant immunity. Curr. Opin. Plant Biol. 2017, 38, 124–132. [Google Scholar] [CrossRef]
- Xu, G.; Greene, G.H.; Yoo, H.; Liu, L.; Marques, J.; Motley, J.; Dong, X. Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature 2017, 545, 487–490. [Google Scholar] [CrossRef]
- Yoo, H.; Greene, G.H.; Yuan, M.; Xu, G.; Burton, D.; Liu, L.; Marques, J.; Dong, X. Translational regulation of metabolic dynamics during effector-triggered immunity. Mol. Plant 2020, 13, 88–98. [Google Scholar] [CrossRef]
- von Arnim, A.G.; Jia, Q.; Vaughn, J.N. Vaughn, Regulation of plant translation by upstream open reading frames. Plant Sci. 2014, 214, 1–12. [Google Scholar] [CrossRef]
- Xu, G.; Yuan, M.; Ai, C.; Liu, L.; Zhuang, E.; Karapetyan, S.; Wang, S.; Dong, X. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature 2017, 545, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Ai, G.; Liu, J.; Fu, X.; Li, T.; Zhu, H.; Zhai, Y.; Xia, C.; Pan, W.; Li, J.; Jing, M.; et al. Making use of plant uORFs to control transgene translation in response to pathogen attack. BioDesign Res. 2022, 2022, 9820540. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; J, L.; QH, S. Regulation of NLR stability in plant immunity. Front. Agr. Sci. Eng. 2019, 2, 97–104. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, L. Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity. Plant Commun. 2020, 1, 100041. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. The logic of the 26S proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef]
- Berndsen, C.E.; Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014, 21, 301–307. [Google Scholar] [CrossRef]
- Dielen, A.S.; Badaoui, S.; Candresse, T.; German-Retana, S. The ubiquitin/26S proteasome system in plant-pathogen interactions: A never-ending hide-and-seek game. Mol. Plant Pathol. 2010, 11, 293–308. [Google Scholar] [CrossRef]
- Langin, G.; Gonzalez-Fuente, M.; Ustun, S. The plant ubiquitin-proteasome system as a target for microbial manipulation. Annu. Rev. Phytopathol. 2023, 61, 351–375. [Google Scholar] [CrossRef]
- Shirasu, K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu. Rev. Plant Biol. 2009, 60, 139–164. [Google Scholar] [CrossRef]
- Kitagawa, K.; Skowyra, D.; Elledge, S.J.; Harper, J.W.; Hieter, P. SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 1999, 4, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Catlett, M.G.; Kaplan, K.B. Sgt1p is a unique co-chaperone that acts as a client adaptor to link Hsp90 to Skp1p. J. Biol. Chem. 2006, 281, 33739–33748. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.K.; Li, Q.; Liu, Y.; Dinesh-Kumar, S.P.; Kaloshian, I. The MI-1-mediated pest resistance requires HSP90 and SGT1. Plant Physiol. 2007, 144, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Erb, M.; Meldau, S.; Howe, G.A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012, 17, 250–259. [Google Scholar] [CrossRef]
- Yan, C.; Fan, M.; Yang, M.; Zhao, J.; Zhang, W.; Su, Y.; Xiao, L.; Deng, H.; Xie, D. Injury Activates Ca2+/Calmodulin-Dependent Phosphorylation of JAV1-JAZ8-WRKY51 Complex for Jasmonate Biosynthesis. Mol. Cell 2018, 70, 136–149. [Google Scholar] [CrossRef]
- Chen, C.Y.; Liu, Y.Q.; Song, W.M.; Chen, D.Y.; Chen, F.Y.; Chen, X.Y.; Chen, Z.W.; Ge, S.X.; Wang, C.Z.; Zhan, S.; et al. An effector from cotton bollworm oral secretion impairs host plant defense signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 14331–14338. [Google Scholar] [CrossRef]
- Zou, J.; Chen, X.; Liu, C.; Guo, M.; Kanwar, M.K.; Qi, Z.; Yang, P.; Wang, G.; Bao, Y.; Bassham, D.C.; et al. Autophagy promotes jasmonate-mediated defense against nematodes. Nat. Commun. 2023, 14, 4769. [Google Scholar] [CrossRef]
- Meur, G.; Budatha, M.; Srinivasan, T.; Rajesh, K.K.; Dutta, G.A.; Kirti, P.B. Constitutive expression of Arabidopsis NPR1 confers enhanced resistance to the early instars of Spodoptera litura in transgenic tobacco. Physiol. Plant 2008, 133, 765–775. [Google Scholar] [CrossRef]
- Zarate, S.I.; Kempema, L.A.; Walling, L.L. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 2007, 143, 866–875. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhong, S.; Li, Q.; Zhu, Z.; Lou, Y.; Wang, L.; Wang, J.; Wang, M.; Li, Q.; Yang, D.; et al. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 2007, 5, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Afsheen, S.; Xin, Z.; Han, X.; Lou, Y. OsNPR1 negatively regulates herbivore-induced JA and ethylene signaling and plant resistance to a chewing herbivore in rice. Physiol. Plant. 2013, 147, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Mou, Z.; Tada, Y.; Spivey, N.W.; Genschik, P.; Dong, X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 2009, 137, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Skelly, M.J.; Furniss, J.J.; Grey, H.; Wong, K.W.; Spoel, S.H. Dynamic ubiquitination determines transcriptional activity of the plant immune coactivator NPR1. eLife 2019, 8, e47005. [Google Scholar] [CrossRef]
- Lamb, C.A.; Yoshimori, T.; Tooze, S.A. The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef]
- Sertsuvalkul, N.; DeMell, A.; Dinesh-Kumar, S.P. The complex roles of autophagy in plant immunity. Febs Lett. 2022, 596, 2163–2171. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef]
- Hao, L.K.; Dangol, A.; Shavit, R.; Pitt, W.J.; Nalam, V.; Brotman, Y.; Michaeli, S.; Peled-Zehavi, H.; Tzin, V. Self-eating while being eaten: Elucidating the relationship between aphid feeding and the plant autophagy machinery in Arabidopsis leaves. bioRxiv, 2023; preprint. [Google Scholar]
- Nguyen, H.T.; Mantelin, S.; Ha, C.V.; Lorieux, M.; Jones, J.T.; Mai, C.D.; Bellafiore, S. Insights into the genetics of the Zhonghua 11 resistance to Meloidogyne graminicola and its molecular determinism in rice. Front. Plant Sci. 2022, 13, 854961. [Google Scholar] [CrossRef]
- Liao, C.Y.; Wang, P.; Yin, Y.; Bassham, D.C. Interactions between autophagy and phytohormone signaling pathways in plants. FEBS Lett. 2022, 596, 2198–2214. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Jin, W.; Zou, J.; Chen, X.; Zhao, Q.; Yu, J.; Zhou, J. NBR1a mediates root-knot nematode resistance by modulating antioxidant system, jasmonic acid and selective autophagy in Solanum lycopersicum. Plant Stress 2024, 11, 100390. [Google Scholar] [CrossRef]
- Guo, J.; Wang, H.; Guan, W.; Guo, Q.; Wang, J.; Yang, J.; Peng, Y.; Shan, J.; Gao, M.; Shi, S.; et al. A tripartite rheostat controls self-regulated host plant resistance to insects. Nature 2023, 618, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Sadanandom, A.; Bailey, M.; Ewan, R.; Lee, J.; Nelis, S. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef]
- Zientara-Rytter, K.; Sirko, A. To deliver or to degrade—An interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J. 2016, 283, 3534–3555. [Google Scholar] [CrossRef]
- Yagyu, M.; Yoshimoto, K. New insights into plant autophagy: Molecular mechanisms and roles in development and stress responses. J. Exp. Bot. 2024, 75, 1234–1251. [Google Scholar] [CrossRef]
- Su, W.; Bao, Y.; Yu, X.; Xia, X.; Liu, C.; Yin, W. Autophagy and its regulators in response to stress in plants. Int. J. Mol. Sci. 2020, 21, 8889. [Google Scholar] [CrossRef]
- Wang, H.; Shi, S.; Hua, W. Advances of herbivore-secreted elicitors and effectors in plant-insect interactions. Front. Plant Sci. 2023, 14, 1176048. [Google Scholar] [CrossRef]
- Bali, S.; Gleason, C. Unveiling the diversity: Plant parasitic nematode effectors and their plant interaction partners. Mol. Plant Microbe Interact. 2024, 37, 179–189. [Google Scholar] [CrossRef]
- Bleau, J.R.; Gaur, N.; Fu, Y.; Bos, J. Unveiling the slippery secrets of saliva: Effector proteins of phloem-feeding insects. Mol. Plant Microbe Interact. 2024, 37, 211–219. [Google Scholar] [CrossRef]
- Liu, S.; Lenoir, C.; Amaro, T.; Rodriguez, P.A.; Huitema, E.; Bos, J. Virulence strategies of an insect herbivore and oomycete plant pathogen converge on host E3 SUMO ligase SIZ1. New Phytol. 2022, 235, 1599–1614. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Granados, A.; Sterken, M.G.; Overmars, H.; Ariaans, R.; Holterman, M.; Pokhare, S.S.; Yuan, Y.; Pomp, R.; Finkers-Tomczak, A.; Roosien, J.; et al. The effector GpRbp-1 of Globodera pallida targets a nuclear HECT E3 ubiquitin ligase to modulate gene expression in the host. Mol. Plant Pathol. 2020, 21, 66–82. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Zhou, J.; Yang, Y.; Lu, W.; Jin, Y.; Huang, X.; Zhang, W.; Li, J.; Ai, G.; Yin, Z.; et al. Cyclophilin effector Al106 of mirid bug Apolygus lucorum inhibits plant immunity and promotes insect feeding by targeting PUB33. New Phytol. 2023, 237, 2388–2403. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Escalante, L.N.; Chen, H.; Benatti, T.R.; Qu, J.; Chellapilla, S.; Waterhouse, R.M.; Wheeler, D.; Andersson, M.N.; Bao, R.; et al. A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor. Curr. Biol. 2015, 25, 613–620. [Google Scholar] [CrossRef]
- Lal, N.K.; Thanasuwat, B.; Huang, P.J.; Cavanaugh, K.A.; Carter, A.; Michelmore, R.W.; Dinesh-Kumar, S.P. Phytopathogen effectors use multiple mechanisms to manipulate plant autophagy. Cell Host Microbe 2020, 28, 558–571. [Google Scholar] [CrossRef]
- Chen, J.; Chen, S.; Xu, C.; Yang, H.; Achom, M.; Wang, X. A key virulence effector from cyst nematodes targets host autophagy to promote nematode parasitism. New Phytol. 2023, 237, 1374–1390. [Google Scholar] [CrossRef]
- Verhoeven, A.; Finkers-Tomczak, A.; Prins, P.; Valkenburg-Van, R.D.; van Schaik, C.C.; Overmars, H.; van Steenbrugge, J.; Tacken, W.; Varossieau, K.; Slootweg, E.J.; et al. The root-knot nematode effector MiMSP32 targets host 12-oxophytodienoate reductase 2 to regulate plant susceptibility. New Phytol. 2023, 237, 2360–2374. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, P.; Ma, Y.; Yao, X.; Sun, Y.; Huang, X.; Jin, J.; Zhang, Y.; Zhu, C.; Fang, R.; et al. A whitefly effector Bsp9 targets host immunity regulator WRKY33 to promote performance. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2019, 374, 20180313. [Google Scholar] [CrossRef]
- Xu, H.X.; Qian, L.X.; Wang, X.W.; Shao, R.X.; Hong, Y.; Liu, S.S.; Wang, X.W. A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway. Proc. Natl. Acad. Sci. USA 2019, 116, 490–495. [Google Scholar] [CrossRef]
- Ji, R.; Fu, J.; Shi, Y.; Li, J.; Jing, M.; Wang, L.; Yang, S.; Tian, T.; Wang, L.; Ju, J.; et al. Vitellogenin from planthopper oral secretion acts as a novel effector to impair plant defenses. New Phytol. 2021, 232, 802–817. [Google Scholar] [CrossRef]
- Hewezi, T.; Juvale, P.S.; Piya, S.; Maier, T.R.; Rambani, A.; Rice, J.H.; Mitchum, M.G.; Davis, E.L.; Hussey, R.S.; Baum, T.J. The cyst nematode effector protein 10A07 targets and recruits host posttranslational machinery to mediate its nuclear trafficking and to promote parasitism in Arabidopsis. Plant Cell 2015, 27, 891–907. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, P.A.; Escudero-Martinez, C.; Bos, J.I. An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol. 2017, 173, 1892–1903. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ye, Y.; Lu, J.; Wang, X.; Lu, H.; Zhang, Z.; Ye, Z.; Lu, Y.; Sun, Z.; Chen, J.; et al. Horizontally transferred salivary protein promotes insect feeding by suppressing ferredoxin-mediated plant defenses. Mol. Biol. Evol. 2023, 40, msad221. [Google Scholar] [CrossRef] [PubMed]
- Banfield, M.J. Perturbation of host ubiquitin systems by plant pathogen/pest effector proteins. Cell Microbiol. 2015, 17, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Ashida, H.; Sasakawa, C. Bacterial E3 ligase effectors exploit host ubiquitin systems. Curr. Opin. Microbiol. 2017, 35, 16–22. [Google Scholar] [CrossRef]
- Kandoth, P.K.; Ithal, N.; Recknor, J.; Maier, T.; Nettleton, D.; Baum, T.J.; Mitchum, M.G. The Soybean Rhg1 locus for resistance to the soybean cyst nematode Heterodera glycines regulates the expression of a large number of stress- and defense-related genes in degenerating feeding cells. Plant Physiol. 2011, 155, 1960–1975. [Google Scholar] [CrossRef]
- Chronis, D.; Chen, S.; Lu, S.; Hewezi, T.; Carpenter, S.C.; Loria, R.; Baum, T.J.; Wang, X. A ubiquitin carboxyl extension protein secreted from a plant-parasitic nematode Globodera rostochiensis is cleaved in planta to promote plant parasitism. Plant J. 2013, 74, 185–196. [Google Scholar] [CrossRef]
- Kazan, K.; Lyons, R. Intervention of phytohormone pathways by pathogen effectors. Plant Cell 2014, 26, 2285–2309. [Google Scholar] [CrossRef]
- Ma, K.W.; Ma, W. Phytohormone pathways as targets of pathogens to facilitate infection. Plant Mol. Biol. 2016, 91, 713–725. [Google Scholar] [CrossRef]
- Iida, J.; Desaki, Y.; Hata, K.; Uemura, T.; Yasuno, A.; Islam, M.; Maffei, M.E.; Ozawa, R.; Nakajima, T.; Galis, I.; et al. Tetranins: New putative spider mite elicitors of host plant defense. New Phytol. 2019, 224, 875–885. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, C.X.; Chen, R.; He, S.Y. Challenging battles of plants with phloem-feeding insects and prokaryotic pathogens. Proc. Natl. Acad. Sci. USA 2019, 116, 23390–23397. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Casteel, C.L. Effector-mediated plant-virus-vector interactions. Plant Cell 2022, 34, 1514–1531. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; Kingdom, H.N.; MacLean, A.M.; Grieve, V.M.; Hogenhout, S.A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, e1254–e1263. [Google Scholar] [CrossRef] [PubMed]
- MacLean, A.M.; Orlovskis, Z.; Kowitwanich, K.; Zdziarska, A.M.; Angenent, G.C.; Immink, R.G.; Hogenhout, S.A. Phytoplasma effector SAP54 hijacks plant reproduction by degrading MADS-box proteins and promotes insect colonization in a RAD23-dependent manner. PLoS Biol. 2014, 12, e1001835. [Google Scholar] [CrossRef]
- Zhang, X.F.; Li, Z.; Lin, H.; Cheng, Y.; Wang, H.; Jiang, Z.; Ji, Z.; Huang, Z.; Chen, H.; Wei, T. A phytoplasma effector destabilizes chloroplastic glutamine synthetase inducing chlorotic leaves that attract leafhopper vectors. Proc. Natl. Acad. Sci. USA 2024, 121, e2402911121. [Google Scholar] [CrossRef]
- Li, P.; Liu, C.; Deng, W.H.; Yao, D.M.; Pan, L.L.; Li, Y.Q.; Liu, Y.Q.; Liang, Y.; Zhou, X.P.; Wang, X.W. Plant begomoviruses subvert ubiquitination to suppress plant defenses against insect vectors. PLoS Pathog. 2019, 15, e1007607. [Google Scholar] [CrossRef]
- Wu, D.; Qi, T.; Li, W.X.; Tian, H.; Gao, H.; Wang, J.; Ge, J.; Yao, R.; Ren, C.; Wang, X.B.; et al. Viral effector protein manipulates host hormone signaling to attract insect vectors. Cell Res. 2017, 27, 402–415. [Google Scholar] [CrossRef]
- Waksman, T.; Astin, E.; Fisher, S.R.; Hunter, W.N.; Bos, J. Computational prediction of structure, function, and interaction of Myzus persicae (green peach aphid) salivary effector proteins. Mol. Plant Microbe Interact. 2024, 37, 338–346. [Google Scholar] [CrossRef]
- Homma, F.; Huang, J.; van der Hoorn, R. AlphaFold-Multimer predicts cross-kingdom interactions at the plant-pathogen interface. Nat. Commun. 2023, 14, 6040. [Google Scholar] [CrossRef]
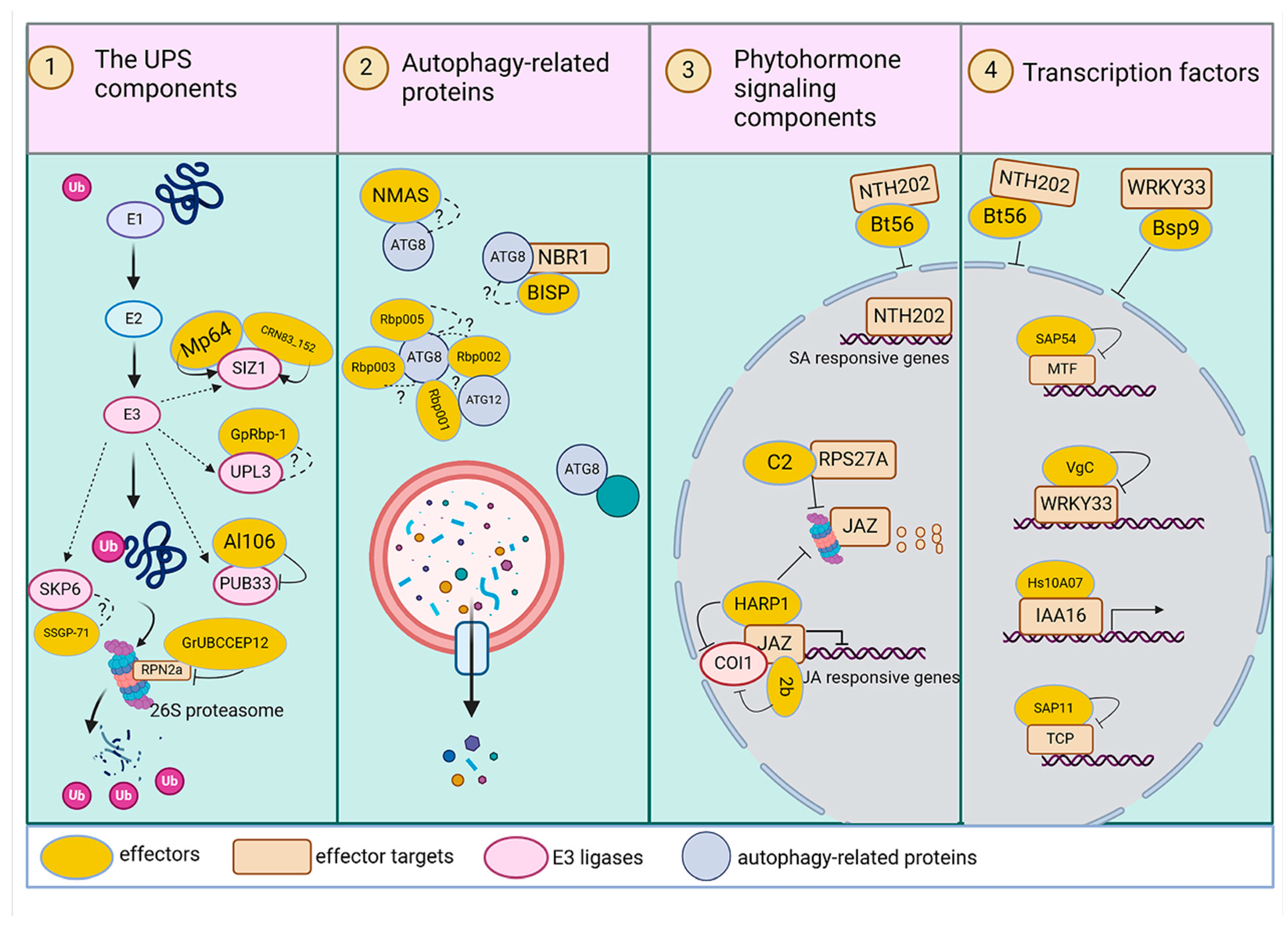
| Effector | Pest Species | Plant Protein | Plant Species | Interaction Outcome | Reference | |
|---|---|---|---|---|---|---|
| UPS components | Mp64 | Myzus persicae | SIZ1 | Arabidopsis | increase SIZ1 protein levels | [82] |
| CRN83_152 | Phytophthora capsici | SIZ1 | Nicotiana benthamiana | enhance SIZ1-E3 SUMO ligase activity | [82] | |
| GpRbp | Globodera pallida | UPL3 | Solanum tuberosum | unknown | [83] | |
| Al106 | Apolygus lucorum | PUB33 | Nicotiana benthamiana | inhibit PUB33 ubiquitination | [84] | |
| SSGP-71 | Mayetiola destructor | SKP6 | wheat | unknown | [85] | |
| Autophagy-related proteins | BISP | Nilaparvata lugens | NBR1 | Oryza sativa | degradation of BISP | [74] |
| Rbp001/2/3/5 | Globodera pallida | ATG1/8/12/20 | Arabidopsis | unknown | [86] | |
| NMAS1 | Heterodera and Globodera spp. | ATG8 | Solanum tuberosum | supress ROS | [87] | |
| Phytohormone signaling components | HARP1 | Helicoverpa armigera | JAZ proteins | Arabidopsis, Cotton, and Tobacco | stabilize JAZs | [58] |
| MiMSP32 | Meloidogyne incognita | OPR2 | Solanum lycopersicum | increase in the concentration of 12-OPDA | [88] | |
| Transcriptional factors | Bsp9 | Bemisia tabaci | WRKY33 | Arabidopsis | decrease WRKY33 in nuclear | [89] |
| Bt56 | Bemisia tabaci | NTH202 | Nicotiana benthamiana | decrease NTH202 in nuclear | [90] | |
| VgC | Laodelphax striatellus | OsWRKY71 | Oryza sativa | repress OsWRKY71 activity | [91] | |
| Hs10A07 | Heterodera schachtii | IAA16 | Arabidopsis | modulate ARF expression | [92] | |
| Other signaling components | Mp1 | Myzus persicae | VPS52 | Arabidopsis | decrease VPS52 protein level | [93] |
| BtFTSP1 | Bemisia tabaci | NtFD1 | Nicotiana tabacum | destabilize NtFD1 | [94] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Wang, Y. Protein Dynamics in Plant Immunity: Insights into Plant–Pest Interactions. Int. J. Mol. Sci. 2024, 25, 12951. https://doi.org/10.3390/ijms252312951
Zhao Y, Wang Y. Protein Dynamics in Plant Immunity: Insights into Plant–Pest Interactions. International Journal of Molecular Sciences. 2024; 25(23):12951. https://doi.org/10.3390/ijms252312951
Chicago/Turabian StyleZhao, Yan, and Yanru Wang. 2024. "Protein Dynamics in Plant Immunity: Insights into Plant–Pest Interactions" International Journal of Molecular Sciences 25, no. 23: 12951. https://doi.org/10.3390/ijms252312951
APA StyleZhao, Y., & Wang, Y. (2024). Protein Dynamics in Plant Immunity: Insights into Plant–Pest Interactions. International Journal of Molecular Sciences, 25(23), 12951. https://doi.org/10.3390/ijms252312951




