Age-Related sncRNAs in Human Hippocampal Tissue Samples: Focusing on Deregulated miRNAs
Abstract
1. Introduction
2. Results
2.1. Differentially Expressed miRNAs in Human Hippocampal Tissue with Age
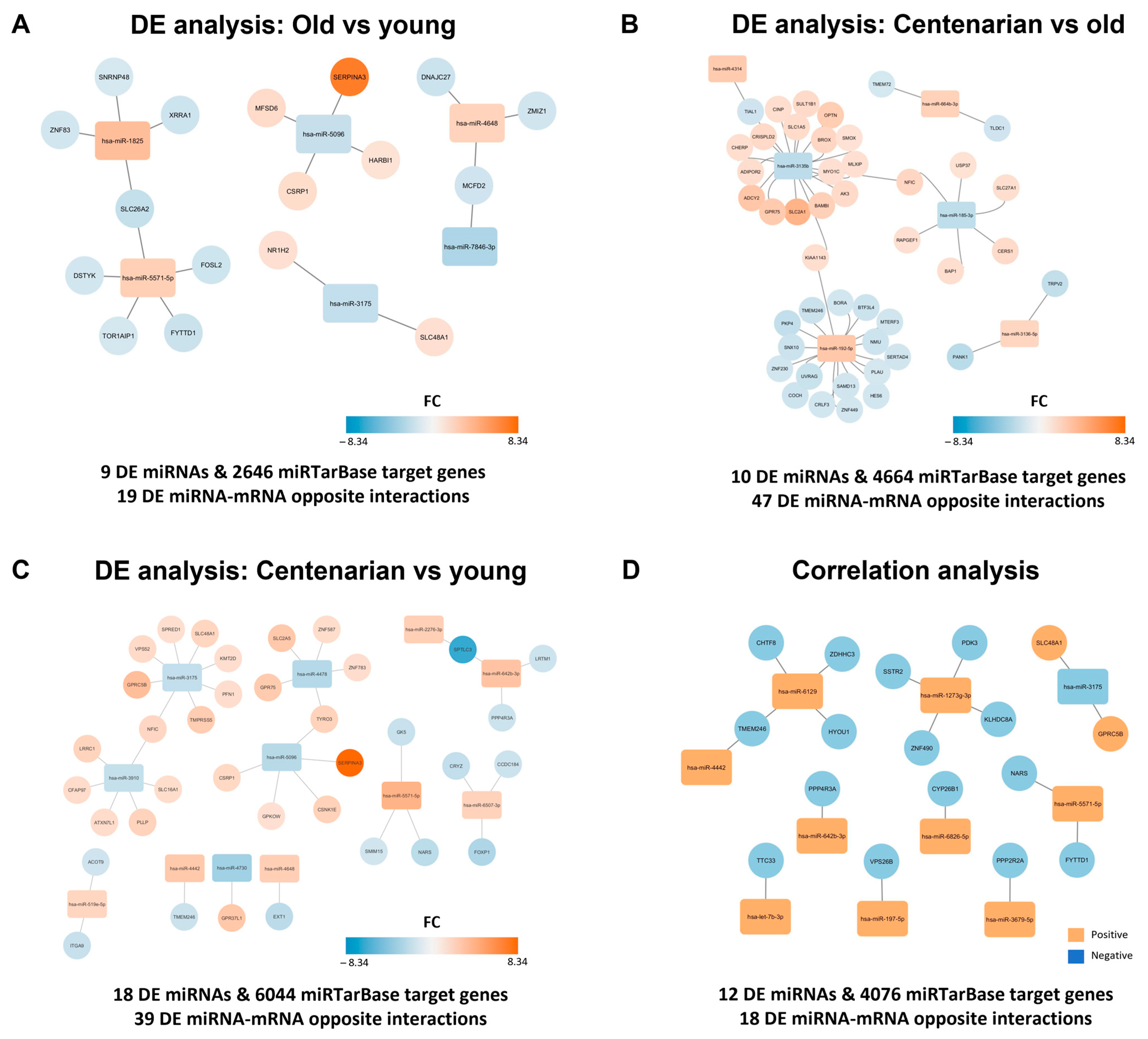
2.2. miRNA-Mediated Regulatory Networks Revealed Significant Interactions with mRNAs Analyzed in the Same Hippocampal Samples
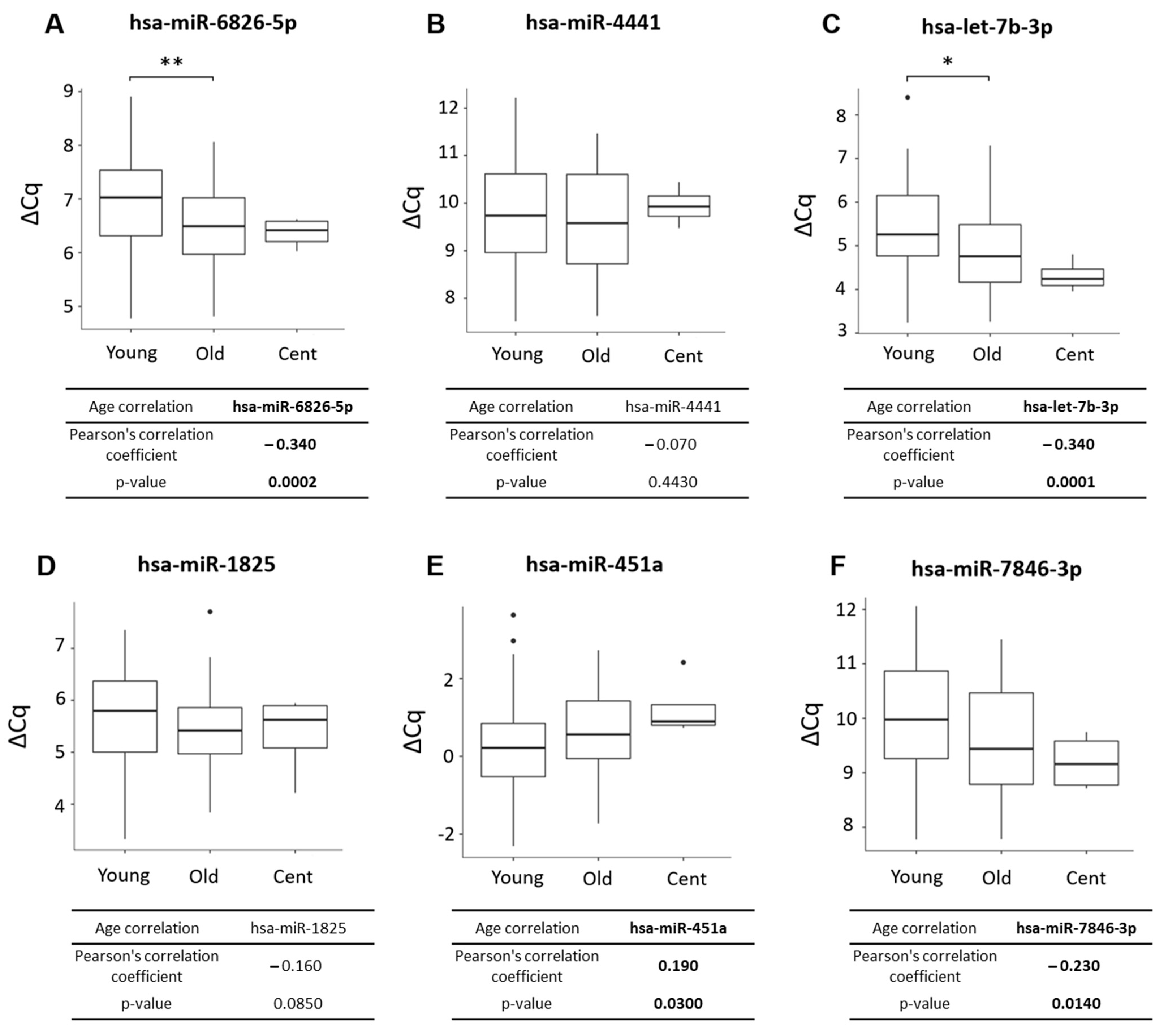
2.3. hsa-miR-6826-5p, hsa-let-7b-3p, hsa-miR-7846-3p, and hsa-miR-451a Significantly Correlated with Age
3. Discussion
4. Materials and Methods
4.1. Human Hippocampal Brain Samples
4.2. RNA Extraction and Global sncRNA Analysis
- (1)
- Old vs. young;
- (2)
- Centenarian vs. old;
- (3)
- Centenarian vs. young.
4.3. Bioinformatic In Silico Analysis
4.4. RT-qPCR Candidates Validation
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of Aging: An Expanding Universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef] [PubMed]
- Tartiere, A.G.; Freije, J.M.P.; López-Otín, C. The Hallmarks of Aging as a Conceptual Framework for Health and Longevity Research. Front. Aging 2024, 5, 1334261. [Google Scholar] [CrossRef]
- Rivero-Segura, N.A.; Bello-Chavolla, O.Y.; Barrera-Vázquez, O.S.; Gutierrez-Robledo, L.M.; Gomez-Verjan, J.C. Promising Biomarkers of Human Aging: In Search of a Multi-Omics Panel to Understand the Aging Process from a Multidimensional Perspective. Ageing Res. Rev. 2020, 64, 101164. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Culla, M.; Irizar, H.; Gorostidi, A.; Alberro, A.; Osorio-Querejeta, I.; Ruiz-Martínez, J.; Olascoaga, J.; López de Munain, A.; Otaegui, D. Progressive Changes in Non-Coding RNA Profile in Leucocytes with Age. Aging 2017, 9, 1202–1218. [Google Scholar] [CrossRef]
- Morris, B.J.; Willcox, B.J.; Donlon, T.A. Genetic and Epigenetic Regulation of Human Aging and Longevity. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1718–1744. [Google Scholar] [CrossRef]
- Marttila, S.; Chatsirisupachai, K.; Palmer, D.; de Magalhães, J.P. Ageing-Associated Changes in the Expression of LncRNAs in Human Tissues Reflect a Transcriptional Modulation in Ageing Pathways. Mech. Ageing Dev. 2020, 185, 111177. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, H.; Wang, C.; Fu, L.; Wang, Q.; Li, S.; Cong, B. Forensic Age Estimation from Human Blood Using Age-Related MicroRNAs and Circular RNAs Markers. Front. Genet. 2022, 13, 1031806. [Google Scholar] [CrossRef]
- Niu, Y.Y.; Zhong, J.F.; Wen, H.Y.; Yan, H.Y.; Diao, Z.Q.; Li, J.X.; Bai, X.R.; Qiu, J.M.; Xu, Z.T.; Chen, L.H.; et al. Association of Combined Healthy Lifestyle Factors With Incident Dementia in Participants With and Without Multimorbidity: A Large Population-Based Prospective Cohort Study. J. Gerontol. A Biol. Sci. Med. Sci. 2024, 79, glae034. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of MiRNAs and SiRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Maier, A.B.; Kroemer, G. Gerogenes and Gerosuppression: The Pillars of Precision Geromedicine. Cell Res. 2024, 34, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Persengiev, S.P.; Kondova, I.I.; Bontrop, R.E. The Impact of MicroRNAs on Brain Aging and Neurodegeneration. Curr. Gerontol. Geriatr. Res. 2012, 2012, 359369. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.; Smith, F.; Kumar, S.; Vijayan, M.; Reddy, P.H. Are MicroRNAs True Sensors of Ageing and Cellular Senescence? Ageing Res. Rev. 2017, 35, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Harries, L.W. MicroRNAs as Mediators of the Ageing Process. Genes 2014, 5, 656–670. [Google Scholar] [CrossRef]
- Smith-Vikos, T.; Slack, F.J. MicroRNAs and Their Roles in Aging. J. Cell Sci. 2012, 125, 7–17. [Google Scholar] [CrossRef]
- Saenz-Antoñanzas, A.; Muñoz-Culla, M.; Rigo, P.; Ruiz-Barreiro, L.; Moreno-Valladares, M.; Alberro, A.; Cruces-Salguero, S.; Arroyo-Izaga, M.; Arranz, A.M.; Otaegui, D.; et al. Centenarian Hippocampus Displays High Levels of Astrocytic Metallothioneins. Aging Cell 2024, 23, e14201. [Google Scholar] [CrossRef]
- Kumar Dev, P.; Gray, A.J.; Scott-Hamilton, J.; Hagstrom, A.D.; Murphy, A.; Denham, J. Co-Expression Analysis Identifies Networks of MiRNAs Implicated in Biological Ageing and Modulated by Short-Term Interval Training. Mech. Ageing Dev. 2021, 199, 111552. [Google Scholar] [CrossRef]
- Lu, J.; Li, S.; Li, X.; Zhao, W.; Duan, X.; Gu, X.; Xu, J.; Yu, B.; Sigal, L.J.; Dong, Z.; et al. Declined MiR-181a-5p Expression Is Associated with Impaired Natural Killer Cell Development and Function with Aging. Aging Cell 2021, 20, e13353. [Google Scholar] [CrossRef]
- Massaro, F.; Corrillon, F.; Stamatopoulos, B.; Dubois, N.; Ruer, A.; Meuleman, N.; Bron, D.; Lagneaux, L. Age-Related Changes in Human Bone Marrow Mesenchymal Stromal Cells: Morphology, Gene Expression Profile, Immunomodulatory Activity and MiRNA Expression. Front. Immunol. 2023, 14, 1267550. [Google Scholar] [CrossRef]
- Qi, L.; Li, X.; Liu, S.M.; Jiao, D.L.; Hu, D.; Ju, X.Y.; Zhao, S.Y.; Si, S.H.; Hu, L.; Li, G.N.; et al. Identification of a Hippocampal LncRNA-Regulating Network in a Natural Aging Rat Model. BMC Neurosci. 2022, 23, 56. [Google Scholar] [CrossRef] [PubMed]
- Krarup, J.; Araya, L.; Álvarez, F.; Bórquez, D.A.; Urrutia, P.J. A Brain Anti-Senescence Transcriptional Program Triggered by Hypothalamic-Derived Exosomal MicroRNAs. Int. J. Mol. Sci. 2024, 25, 5467. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Jiménez, E.P.; Flor-García, M.; Terreros-Roncal, J.; Rábano, A.; Cafini, F.; Pallas-Bazarra, N.; Ávila, J.; Llorens-Martín, M. Adult Hippocampal Neurogenesis Is Abundant in Neurologically Healthy Subjects and Drops Sharply in Patients with Alzheimer’s Disease. Nat. Med. 2019, 25, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Toda, T.; Parylak, S.L.; Linker, S.B.; Gage, F.H. The Role of Adult Hippocampal Neurogenesis in Brain Health and Disease. Mol. Psychiatry 2019, 24, 67–87. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.R.; Petralia, M.C.; Ciurleo, R.; Bramanti, A.; Fagone, P.; Shahjaman, M.; Wu, L.; Sun, Y.; Turanli, B.; Arga, K.Y.; et al. Comprehensive Analysis of RNA-Seq Gene Expression Profiling of Brain Transcriptomes Reveals Novel Genes, Regulators, and Pathways in Autism Spectrum Disorder. Brain Sci. 2020, 10, 747. [Google Scholar] [CrossRef]
- Ren, F.-J.; Yao, Y.; Cai, X.-Y.; Fang, G.-Y. Emerging Role of MiR-192-5p in Human Diseases. Front. Pharmacol. 2021, 12, 614068. [Google Scholar] [CrossRef]
- Zhao, B.; Jiang, X. Hsa-MiR-518-5p/Hsa-MiR-3135b Regulates the REL/SOD2 Pathway in Ischemic Cerebral Infarction. Front. Neurol. 2022, 13, 852013. [Google Scholar] [CrossRef]
- Shi, J.; Ren, Y.; Liu, Y.; Cheng, Y.; Liu, Y. Circulating MiR-3135b and MiR-107 Are Potential Biomarkers for Severe Hypertension. J. Hum. Hypertens. 2021, 35, 343–350. [Google Scholar] [CrossRef]
- Chen, F.; Yang, J.; Li, Y.; Wang, H. Circulating MicroRNAs as Novel Biomarkers for Heart Failure. Hellenic J. Cardiol. 2018, 59, 209–214. [Google Scholar] [CrossRef]
- Ramaswamy, P.; Athira, S.V.; Misra, P.; Chauhan, V.S.; Adhvaryu, A.; Gupta, A.; Ankita, G.; Sibin, M.K. Circulating MicroRNA Profiling Identifies MicroRNAs Linked to Prediabetes Associated with Alcohol Dependence Syndrome. Alcohol 2024, 42, 109–118. [Google Scholar] [CrossRef]
- Wang, F.; Li, Z.; Zhao, M.; Ye, W.; Wu, H.; Liao, Q.; Bu, S.; Zhang, Y. Circulating MiRNAs MiR-574-5p and MiR-3135b Are Potential Metabolic Regulators for Serum Lipids and Blood Glucose in Gestational Diabetes Mellitus. Gynecol. Endocrinol. 2021, 37, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Wagner, V.; Kern, F.; Hahn, O.; Schaum, N.; Ludwig, N.; Fehlmann, T.; Engel, A.; Henn, D.; Rishik, S.; Isakova, A.; et al. Characterizing Expression Changes in Noncoding RNAs during Aging and Heterochronic Parabiosis across Mouse Tissues. Nat. Biotechnol. 2024, 42, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Demirel, G.; Tanoglu, E.G.; Aslıyuksek, H. Evaluation of MicroRNA Let-7b-3p Expression Levels in Methamphetamine Abuse. Rev. Assoc. Med. Bras. (1992) 2023, 69, e20221391. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.D.; van Vliet, E.A.; Chen, B.J.; Janitz, M.; Anink, J.J.; Baayen, J.C.; Idema, S.; Devore, S.; Friedman, D.; Diehl, B.; et al. Coding and Non-Coding Transcriptome of Mesial Temporal Lobe Epilepsy: Critical Role of Small Non-Coding RNAs. Neurobiol. Dis. 2020, 134, 104612. [Google Scholar] [CrossRef] [PubMed]
- Moreau, M.P.; Bruse, S.E.; Jornsten, R.; Liu, Y.; Brzustowicz, L.M. Chronological Changes in MicroRNA Expression in the Developing Human Brain. PLoS ONE 2013, 8, e60480. [Google Scholar] [CrossRef]
- Tatara, Y.; Yamazaki, H.; Katsuoka, F.; Chiba, M.; Saigusa, D.; Kasai, S.; Nakamura, T.; Inoue, J.; Aoki, Y.; Shoji, M.; et al. Multiomics and Artificial Intelligence Enabled Peripheral Blood-Based Prediction of Amnestic Mild Cognitive Impairment. Curr. Res. Transl. Med. 2023, 71, 103367. [Google Scholar] [CrossRef]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A Compendium of RNA, Proteins, Lipids and Metabolites in Extracellular Vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, X.; Hong, T.; Zhang, M.; Cai, Y.; Cui, L. TTC3-Mediated Protein Quality Control, A Potential Mechanism for Cognitive Impairment. Cell. Mol. Neurobiol. 2021, 42, 1659–1669. [Google Scholar] [CrossRef]
- Mor, M.; Nardone, S.; Sams, D.S.; Elliott, E. Hypomethylation of MiR-142 Promoter and Upregulation of MicroRNAs That Target the Oxytocin Receptor Gene in the Autism Prefrontal Cortex. Mol. Autism 2015, 6, 46. [Google Scholar] [CrossRef]
- Huang, Z.X.; Chen, Y.; Guo, H.R.; Chen, G.F. Systematic Review and Bioinformatic Analysis of MicroRNA Expression in Autism Spectrum Disorder Identifies Pathways Associated With Cancer, Metabolism, Cell Signaling, and Cell Adhesion. Front. Psychiatry 2021, 12, 630876. [Google Scholar] [CrossRef]
- Wan, Y.; Liu, Y.; Wang, X.; Wu, J.; Liu, K.; Zhou, J.; Liu, L.; Zhang, C. Identification of Differential MicroRNAs in Cerebrospinal Fluid and Serum of Patients with Major Depressive Disorder. PLoS ONE 2015, 10, e0121975. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, P.; Chandra, S.R.; Christopher, R. Circulating MicroRNAs as Potential Biomarkers for the Identification of Vascular Dementia Due to Cerebral Small Vessel Disease. Age Ageing 2017, 46, 861–864. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.; Santagati, M.; Mirabella, F.; Lauretta, G.; Cirnigliaro, M.; Brex, D.; Barbagallo, C.; Domini, C.N.; Gulisano, M.; Barone, R.; et al. Potential Associations Among Alteration of Salivary MiRNAs, Saliva Microbiome Structure, and Cognitive Impairments in Autistic Children. Int. J. Mol. Sci. 2020, 21, 6203. [Google Scholar] [CrossRef] [PubMed]
- Agostini, S.; Mancuso, R.; Costa, A.S.; Guerini, F.R.; Trecate, F.; Miglioli, R.; Menna, E.; Arosio, B.; Clerici, M. Sarcopenia Associates with SNAP-25 SNPs and a MiRNAs Profile Which Is Modulated by Structured Rehabilitation Treatment. J. Transl. Med. 2021, 19, 315. [Google Scholar] [CrossRef] [PubMed]
- McKeever, P.M.; Schneider, R.; Taghdiri, F.; Weichert, A.; Multani, N.; Brown, R.A.; Boxer, A.L.; Karydas, A.; Miller, B.; Robertson, J.; et al. MicroRNA Expression Levels Are Altered in the Cerebrospinal Fluid of Patients with Young-Onset Alzheimer’s Disease. Mol. Neurobiol. 2018, 55, 8826. [Google Scholar] [CrossRef]
- Wang, L.; Zhen, H.; Sun, Y.; Rong, S.; Li, B.; Song, Z.; Liu, Z.; Li, Z.; Ding, J.; Yang, H.; et al. Plasma Exo-MiRNAs Correlated with AD-Related Factors of Chinese Individuals Involved in Aβ Accumulation and Cognition Decline. Mol. Neurobiol. 2022, 59, 6790–6804. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Exploratory Study on MicroRNA Profiles from Plasma-Derived Extracellular Vesicles in Alzheimer’s Disease and Dementia with Lewy Bodies. Transl. Neurodegener. 2019, 8, 31. [Google Scholar] [CrossRef]
- Seršić, L.V.; Alić, V.K.; Biberić, M.; Zrna, S.; Jagoić, T.; Tarčuković, J.; Grabušić, K. Real-Time PCR Quantification of 87 MiRNAs from Cerebrospinal Fluid: MiRNA Dynamics and Association with Extracellular Vesicles after Severe Traumatic Brain Injury. Int. J. Mol. Sci. 2023, 24, 4751. [Google Scholar] [CrossRef]
- Feng, H.; Hu, P.; Chen, Y.; Sun, H.; Cai, J.; He, X.; Cao, Q.; Yin, M.; Zhang, Y.; Li, Q.; et al. Decreased MiR-451a in Cerebrospinal Fluid, a Marker for Both Cognitive Impairment and Depressive Symptoms in Alzheimer’s Disease. Theranostics 2023, 13, 3021–3040. [Google Scholar] [CrossRef]
- Agostini, S.; Mancuso, R.; Citterio, L.A.; Mihali, G.A.; Arosio, B.; Clerici, M. Evaluation of Serum MiRNAs Expression in Frail and Robust Subjects Undergoing Multicomponent Exercise Protocol (VIVIFRAIL). J. Transl. Med. 2023, 21, 67. [Google Scholar] [CrossRef]
- Trattnig, C.; Üçal, M.; Tam-Amersdorfer, C.; Bucko, A.; Zefferer, U.; Grünbacher, G.; Absenger-Novak, M.; Öhlinger, K.A.; Kraitsy, K.; Hamberger, D.; et al. MicroRNA-451a Overexpression Induces Accelerated Neuronal Differentiation of Ntera2/D1 Cells and Ablation Affects Neurogenesis in MicroRNA-451a−/− Mice. PLoS ONE 2018, 13, e0207575. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Oh, S.J.; Gim, J.A.; Shin, O.S. MiR-10a, MiR-30c, and MiR-451a Encapsulated in Small Extracellular Vesicles Are Prosenescence Factors in Human Dermal Fibroblasts. J. Invest. Dermatol. 2022, 142, 2570–2579.e6. [Google Scholar] [CrossRef] [PubMed]
- Chenery, A.; Burrows, K.; Antignano, F.; Underhill, T.M.; Petkovich, M.; Zaph, C. The Retinoic Acid-Metabolizing Enzyme Cyp26b1 Regulates CD4 T Cell Differentiation and Function. PLoS ONE 2013, 8, e72308. [Google Scholar] [CrossRef] [PubMed]
- Spoorendonk, K.M.; Peterson-Maduro, J.; Renn, J.; Trowe, T.; Kranenbarg, S.; Winkler, C.; Schulte-Merker, S. Retinoic Acid and Cyp26b1 Are Critical Regulators of Osteogenesis in the Axial Skeleton. Development 2008, 135, 3765–3774. [Google Scholar] [CrossRef]
- Laue, K.; Jänicke, M.; Plaster, N.; Sonntag, C.; Hammerschmidt, M. Restriction of Retinoic Acid Activity by Cyp26b1 Is Required for Proper Timing and Patterning of Osteogenesis during Zebrafish Development. Development 2008, 135, 3775–3787. [Google Scholar] [CrossRef]
- Snyder, J.M.; Zhong, G.; Hogarth, C.; Huang, W.; Topping, T.; LaFrance, J.; Palau, L.; Czuba, L.C.; Griswold, M.; Ghiaur, G.; et al. Knockout of Cyp26a1 and Cyp26b1 during Postnatal Life Causes Reduced Lifespan, Dermatitis, Splenomegaly, and Systemic Inflammation in Mice. FASEB J. 2020, 34, 15788–15804. [Google Scholar] [CrossRef]
- Vogel, J.W.; La Joie, R.; Grothe, M.J.; Diaz-Papkovich, A.; Doyle, A.; Vachon-Presseau, E.; Lepage, C.; Vos de Wael, R.; Thomas, R.A.; Iturria-Medina, Y.; et al. A Molecular Gradient along the Longitudinal Axis of the Human Hippocampus Informs Large-Scale Behavioral Systems. Nat. Commun. 2020, 11, 960. [Google Scholar] [CrossRef]
- Huang, H.Y.; Lin, Y.C.D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. MiRTarBase Update 2022: An Informative Resource for Experimentally Validated MiRNA–Target Interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
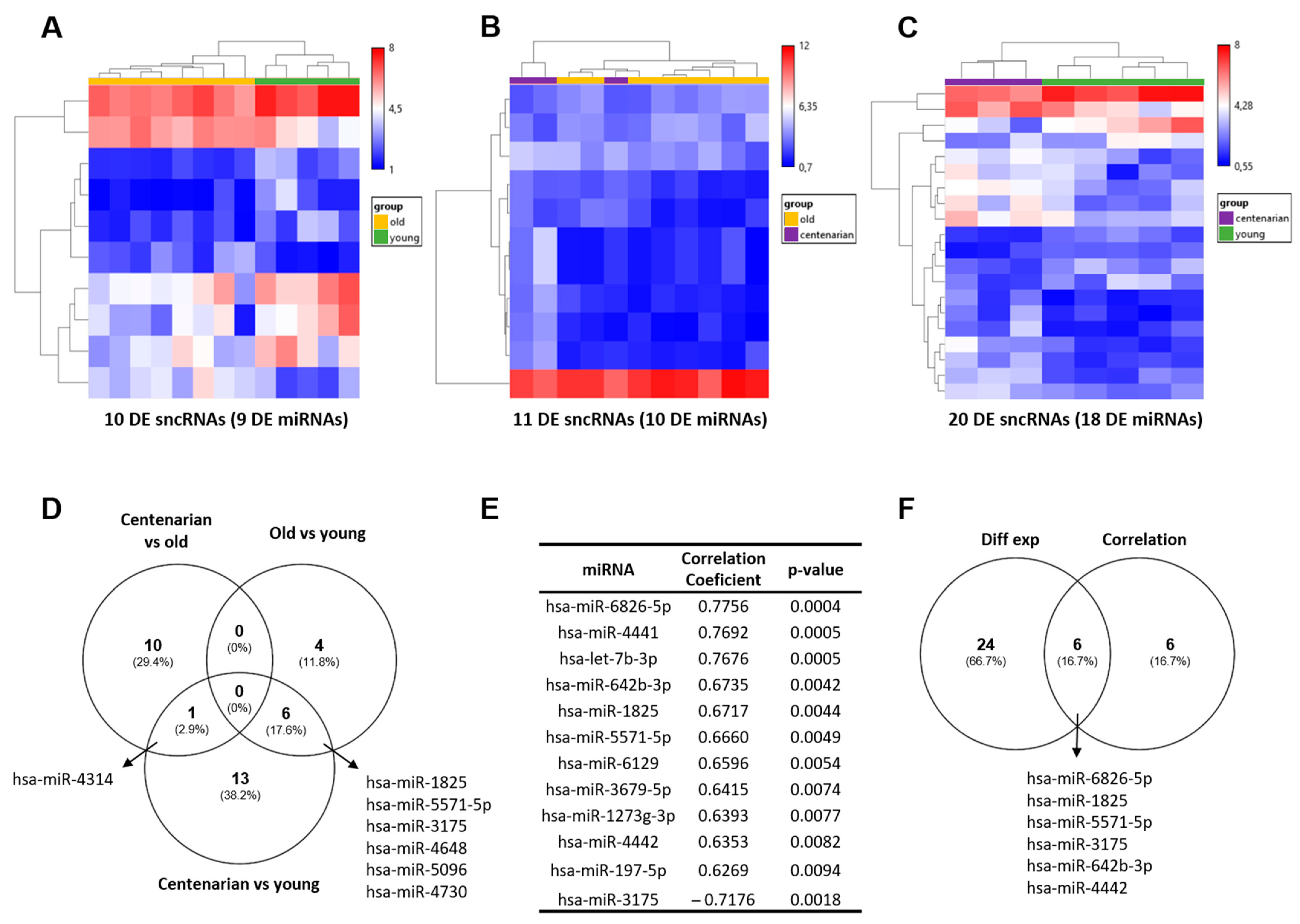
| Discovery Cohort | ||
| Age Range (Years) | Sex (Female/Male) | n |
| 27–49 | 1/4 | 5 |
| 58–88 | 1/7 | 8 |
| 97–100 | 2/1 | 3 |
| Validation Cohort | ||
| Age Range (Years) | Sex (Female/Male) | n |
| 20–50 | 12/65 | 77 |
| 65–82 | 9/28 | 37 |
| 92–96 | 3/1 | 4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alberro, A.; Bravo-Miana, R.D.C.; Iñiguez, S.G.; Iribarren-López, A.; Arroyo-Izaga, M.; Matheu, A.; Muñoz-Culla, M.; Otaegui, D. Age-Related sncRNAs in Human Hippocampal Tissue Samples: Focusing on Deregulated miRNAs. Int. J. Mol. Sci. 2024, 25, 12872. https://doi.org/10.3390/ijms252312872
Alberro A, Bravo-Miana RDC, Iñiguez SG, Iribarren-López A, Arroyo-Izaga M, Matheu A, Muñoz-Culla M, Otaegui D. Age-Related sncRNAs in Human Hippocampal Tissue Samples: Focusing on Deregulated miRNAs. International Journal of Molecular Sciences. 2024; 25(23):12872. https://doi.org/10.3390/ijms252312872
Chicago/Turabian StyleAlberro, Ainhoa, Rocío Del Carmen Bravo-Miana, Saioa GS Iñiguez, Andrea Iribarren-López, Marta Arroyo-Izaga, Ander Matheu, Maider Muñoz-Culla, and David Otaegui. 2024. "Age-Related sncRNAs in Human Hippocampal Tissue Samples: Focusing on Deregulated miRNAs" International Journal of Molecular Sciences 25, no. 23: 12872. https://doi.org/10.3390/ijms252312872
APA StyleAlberro, A., Bravo-Miana, R. D. C., Iñiguez, S. G., Iribarren-López, A., Arroyo-Izaga, M., Matheu, A., Muñoz-Culla, M., & Otaegui, D. (2024). Age-Related sncRNAs in Human Hippocampal Tissue Samples: Focusing on Deregulated miRNAs. International Journal of Molecular Sciences, 25(23), 12872. https://doi.org/10.3390/ijms252312872







