Enhancing Antifungal Drug Discovery Through Co-Culture with Antarctic Streptomyces albidoflavus Strain CBMAI 1855
Abstract
:1. Introduction
2. Results
2.1. Bioactivity Assays
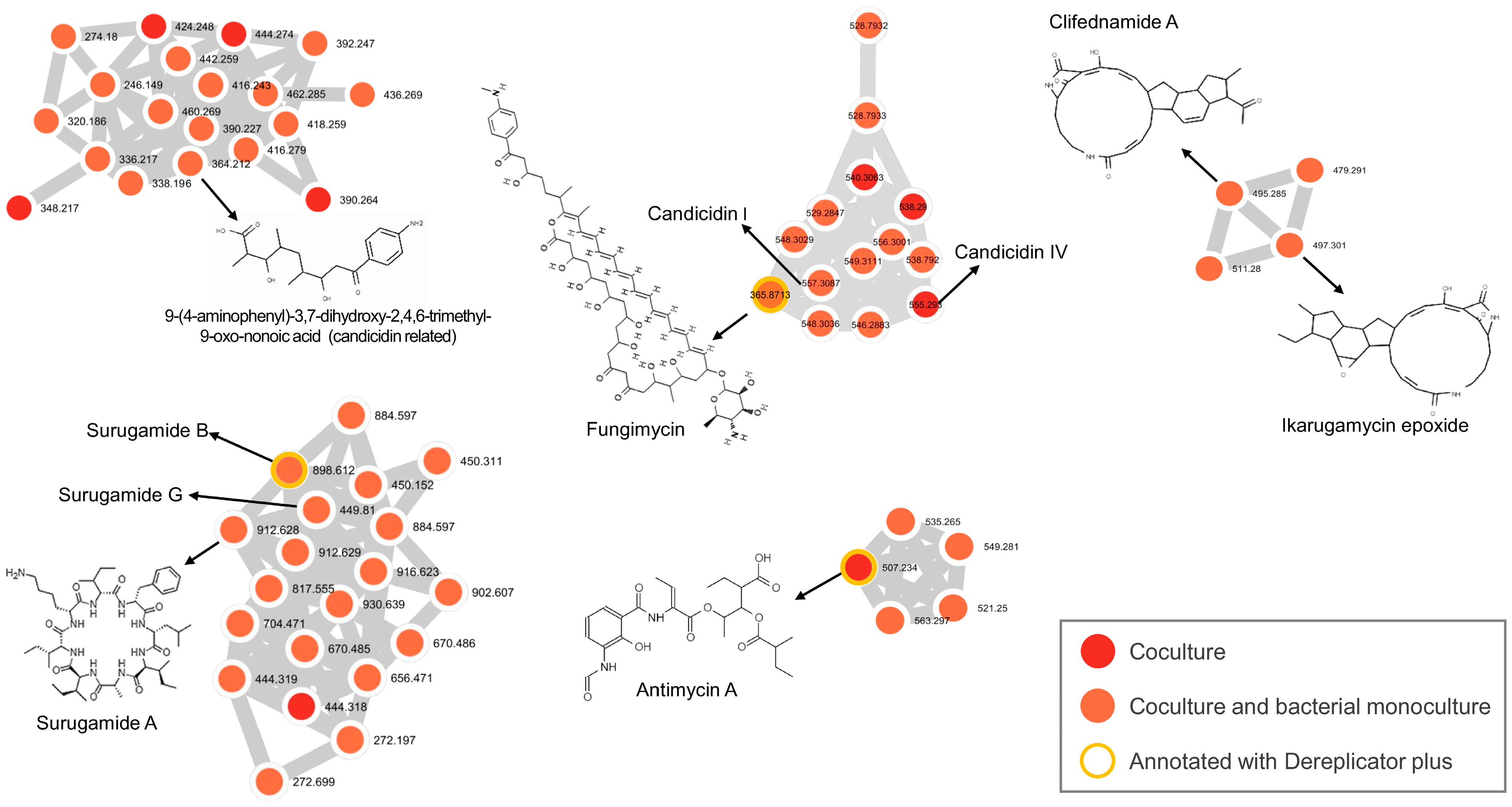
2.2. Metabolomic Analysis
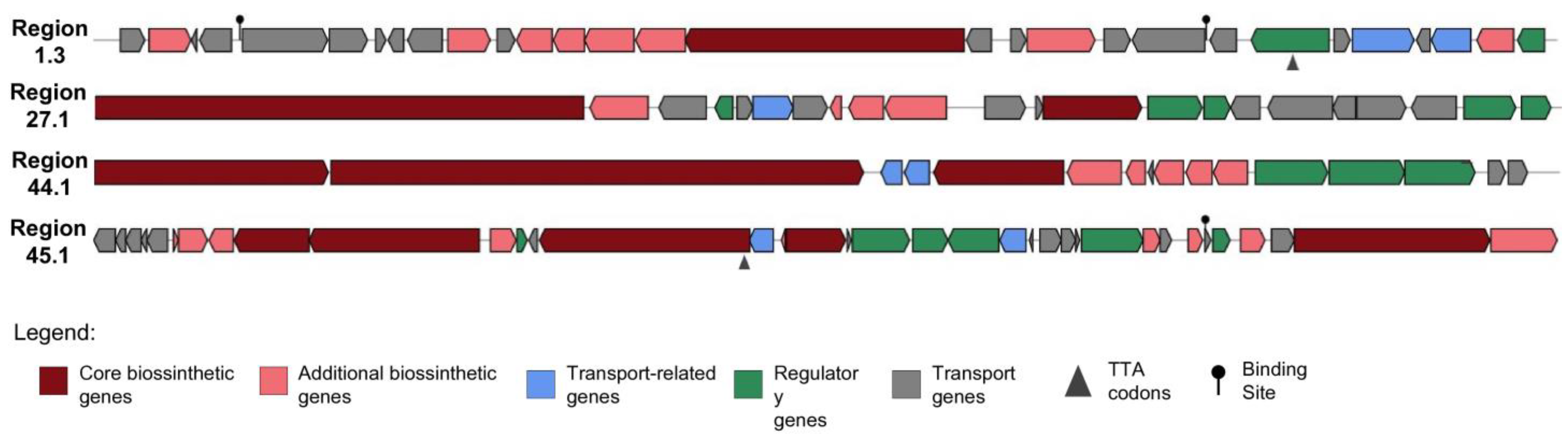
2.3. Antifungal Genome Mining
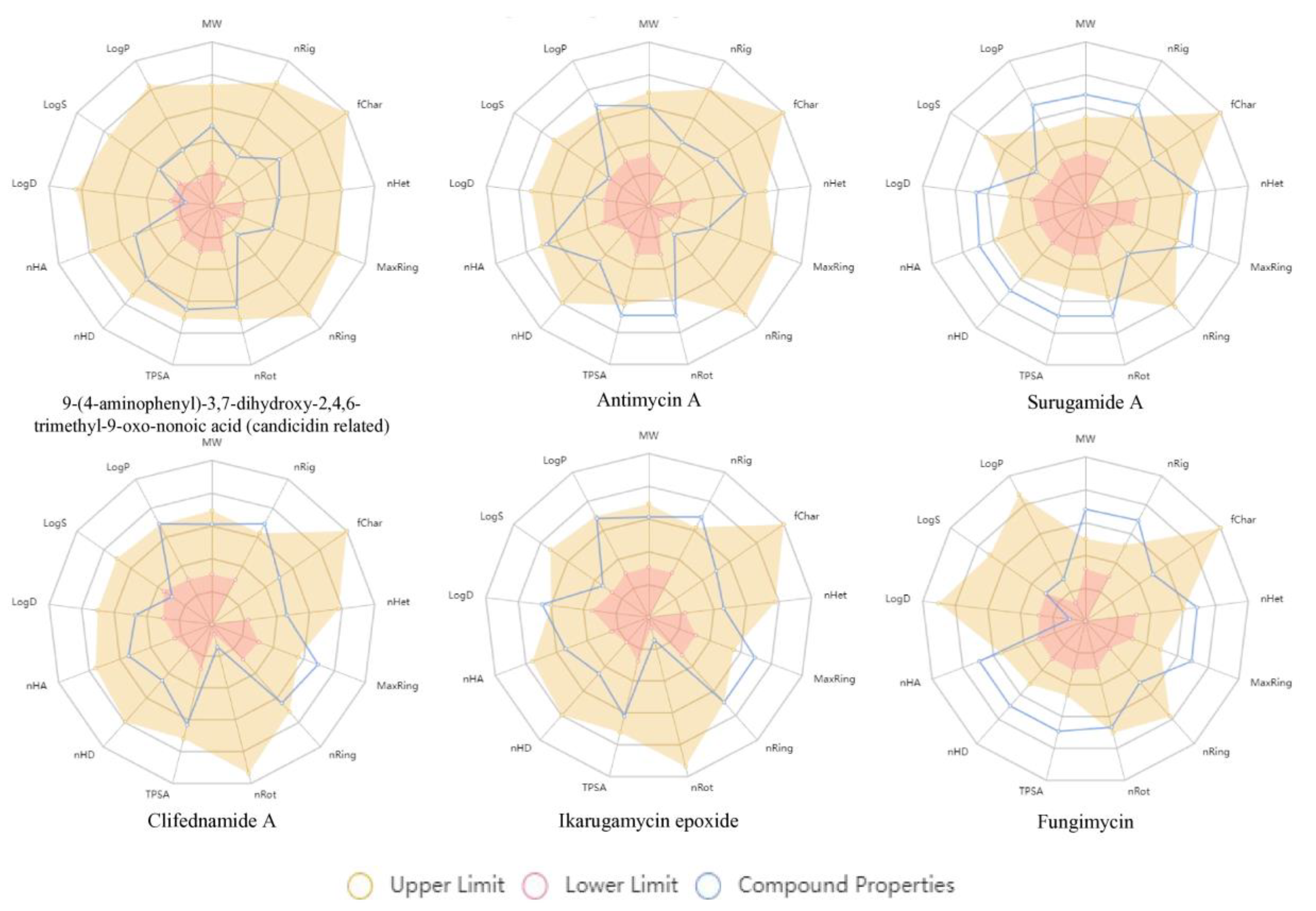
2.4. In Silico Predictions of Drug-Likeness and Toxicity
3. Discussion
4. Materials and Methods
4.1. Strains
4.1.1. Streptomyces albidoflavus CBMAI 1855
4.1.2. Fungal Pathogens
- Candida albicans (ATCC 90028);
- Candida parapsilosis (ATCC 22019);
- Candida krusei (ATCC 6258);
- Aspergillus fumigatus (ATCC 204305);
- Aspergillus flavus (ATCC 204304);
- Cryptococcus neoformans (ATCC 90113);
- Cryptococcus gattii (ATCC 56990);
- Trichophyton mentagrophytes (ATCC 9533).
- LIF 12560 (C. albicans): The resistance mechanism involves substitutions at amino acids E116D, T128K, E266D, and A298V in the ERG11 gene [82];
- LIF E-10 (C. albicans): The resistance mechanism involves substitutions G448V and G464S in the ERG11 gene [82];
- LIF 2552-4.9 (A. fumigatus): The resistance mechanism is characterized by the CYP51A TR34/L98H/S297T/F495I mutations [83];
- LIF 2444.6 (A. fumigatus): The resistance mechanism is characterized by the CYP51A TR34/L98H/S297T/F495I mutation [83];
- LIF 263-e (A. fumigatus): The resistance mechanism is characterized by the CYP51A TR46/F495I mutation [84];
- LIF 1889 (A. fumigatus): No mutations were observed in the CYP51A gene, and the resistance mechanism remains still unknown (unpublished data);
- LIF 2328 (A. fumigatus): No mutations were observed in the CYP51A gene, and the resistance mechanism remains still unknown [84];
- LIF 2486 (A. terreus);
- LIF 16607 (C. auris): Sensitive to all antifungal agents tested (unpublished data);
- LIF 16615 (C. auris): Exhibits resistance to amphotericin B (unpublished data);
- LIF 1455 (Rhizopus oryzae).
4.2. Bioactivity Assays
4.2.1. Antagonism Test (Co-Culture)
4.2.2. Broth Microdilution
4.3. Untargeted Metabolomic Analysis
4.3.1. Sample Preparation
4.3.2. Data Acquisition
4.3.3. Molecular Network and Metabolite Annotation
4.3.4. Statistical Analysis
4.4. Antifungal Genome Mining
4.5. In Silico Predictions of Drug-Likeness and Toxicity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Denning, D.W. Global incidence and mortality of severe fungal disease. Lancet Infect. Dis. 2024, 24, e428–e438. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.G.; Diekema, D.J. What is new in fungal infections? Mod. Pathol. 2023, 36, 100187. [Google Scholar] [CrossRef] [PubMed]
- Friedman, D.Z.P.; Schwartz, I.S. Emerging fungal infections: New patients, new patterns, and new Pathogens. J. Fungi 2019, 5, 67. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Guarner, J. Emerging and reemerging fungal infections. Semin. Diagn. Pathol. 2019, 36, 177–181. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.C.; Hawkins, N.J.; Sanglard, D.; Gurr, S.J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 2018, 360, 739–742. [Google Scholar] [CrossRef]
- De Carolis, E.; Marchionni, F.; La Rosa, M.; Meis, J.F.; Chowdhary, A.; Posteraro, B.; Sanguinetti, M. Are We Ready for Nosocomial Candida auris infections? Front. Cell Infect. Microbiol. 2021, 11, 645049. [Google Scholar]
- Keighley, C.; Garnham, K.; Harch, S.A.J.; Robertson, M.; Chaw, K.; Teng, J.C.; Chen, S.C. Candida auris: Diagnostic challenges and emerging opportunities for the clinical microbiology laboratory. Curr. Fungal Infect. Rep. 2021, 15, 116–126. [Google Scholar] [CrossRef]
- Fisher, M.C.; Alastruey-Izquierdo, A.; Berman, J.; Bicanic, T.; Bignell, E.M.; Bowyer, P.; Bromley, M.; Brüggemann, R.; Garber, G.; Cornely, O.A.; et al. Tackling the emerging threat of antifungal resistance to human health. Nat. Rev. Microbiol. 2022, 20, 557–571. [Google Scholar] [CrossRef]
- Hui, S.T.; Gifford, H.; Rhodes, J. Emerging antifungal resistance in fungal pathogens. Curr. Clin. Microbiol. Rep. 2024, 11, 43–50. [Google Scholar] [CrossRef]
- Woods, M.; McAlister, J.A.; Geddes-McAlister, J. A One Health approach to overcoming fungal disease and antifungal resistance. WIREs Mech. Dis. 2023, 15, e1610. [Google Scholar] [CrossRef]
- Van Daele, R.; Spriet, I.; Wauters, J.; Maertens, J.; Mercier, T.; Van Hecke, S.; Brüggemann, R. Antifungal drugs: What brings the future? Med. Mycol. 2019, 57, S328–S343. [Google Scholar] [CrossRef] [PubMed]
- Ostrosky-Zeichner, L.; Casadevall, A.; Galgiani, J.N.; Odds, F.C.; Rex, J.H. An insight into the antifungal pipeline: Selected new molecules and beyond. Nat. Rev. Drug Discov. 2010, 9, 719–727. [Google Scholar] [CrossRef] [PubMed]
- Houšť, J.; Spížek, J.; Havlíček, V. Antifungal drugs. Metabolites 2020, 10, 106. [Google Scholar] [CrossRef] [PubMed]
- Day, J.N.; Chau, T.T.H.; Wolbers, M.; Mai, P.P.; Dung, N.T.; Mai, N.H.; Phu, N.H.; Nghia, H.D.; Phong, N.D.; Thai, C.Q.; et al. Combination antifungal therapy for cryptococcal meningitis. N. Engl. J. Med. 2013, 368, 1291–1302. [Google Scholar] [CrossRef] [PubMed]
- Benitez, L.L.; Carver, P.L. Adverse Effects Associated with long-term administration of azole antifungal agents. Drugs 2019, 79, 833–853. [Google Scholar] [CrossRef]
- Shafiei, M.; Peyton, L.; Hashemzadeh, M.; Foroumadi, A. History of the development of antifungal azoles: A review on structures, SAR, and mechanism of action. Bioorg. Chem. 2020, 104, 104240. [Google Scholar] [CrossRef]
- Garcia-Effron, G. Rezafungin-mechanisms of action, susceptibility and resistance: Similarities and differences with the other echinocandins. J. Fungi 2020, 6, 262. [Google Scholar] [CrossRef]
- Szymański, M.; Chmielewska, S.; Czyżewska, U.; Malinowska, M.; Tylicki, A. Echinocandins-structure, mechanism of action and use in antifungal therapy. J. Enzyme Inhib. Med. Chem. 2022, 37, 876–894. [Google Scholar]
- Nett, J.E.; Andes, D.R. Antifungal agents: Spectrum of activity, pharmacology, and clinical indications. Infect. Dis. Clin. N. Am. 2016, 30, 51–83. [Google Scholar] [CrossRef]
- Aris, P.; Wei, Y.; Mohamadzadeh, M.; Xia, X. Griseofulvin: An updated overview of old and current knowledge. Molecules 2022, 27, 7034. [Google Scholar] [CrossRef]
- Leonardelli, F.; Macedo, D.; Dudiuk, C.; Cabeza, M.S.; Gamarra, S.; Garcia-Effron, G. Aspergillus fumigatus intrinsic fluconazole resistance is due to the naturally occurring T301I substitution in Cyp51Ap. Antimicrob. Agents Chemother. 2016, 60, 5420–5426. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A.; et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Revie, N.M.; Iyer, K.R.; Robbins, N.; Cowen, L.E. Antifungal drug resistance: Evolution, mechanisms and impact. Curr. Opin. Microbiol. 2018, 45, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Roemer, T.; Krysan, D.J. Antifungal drug development: Challenges, unmet clinical needs, and new approaches. Cold Spring Harb. Perspect. Med. 2014, 4, a019703. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, H.C.; Bezerra, B.T.; Rodrigues, M.L. Antifungal development and the urgency of minimizing the impact of fungal diseases on public health. ACS Bio Med Chem Au 2022, 3, 137–146. [Google Scholar] [CrossRef]
- Li, L. Accessing hidden microbial biosynthetic potential from underexplored sources for novel drug discovery. Biotechnol. Adv. 2023, 66, 108176. [Google Scholar] [CrossRef]
- Xu, L.; Meng, W.; Cao, C.; Wang, J.; Shan, W.; Wang, Q. Antibacterial and antifungal compounds from marine fungi. Mar. Drugs 2015, 13, 3479–3513. [Google Scholar] [CrossRef]
- Habbu, P.; Warad, V.; Shastri, R.; Madagundi, S.; Kulkarni, V.H. Antimicrobial metabolites from marine microorganisms. Chin. J. Nat. Med. 2016, 14, 101–116. [Google Scholar] [CrossRef]
- Caffrey, P.; Hogan, M.; Song, Y. New Glycosylated polyene macrolides: Refining the ore from genome mining. Antibiotics 2022, 11, 334. [Google Scholar] [CrossRef]
- Revilla-Guarinos, A.; Dürr, F.; Popp, P.F.; Döring, M.; Mascher, T. Amphotericin B Specifically Induces the Two-Component System LnrJK: Development of a novel whole-cell biosensor for the detection of amphotericin-like polyenes. Front. Microbiol. 2020, 11, 2022. [Google Scholar] [CrossRef]
- Silva, L.J.; Crevelin, E.J.; Souza, D.T.; Lacerda-Júnior, G.V.; de Oliveira, V.M.; Ruiz, A.L.T.G.; Rosa, L.H.; Moraes, L.A.B.; Melo, I.S. Actinobacteria from Antarctica as a source for anticancer discovery. Sci. Rep. 2020, 10, 13870. [Google Scholar] [CrossRef] [PubMed]
- Thompson, T.P.; Gilmore, B.F. Exploring halophilic environments as a source of new antibiotics. Crit. Rev. Microbiol. 2024, 50, 341–370. [Google Scholar] [CrossRef] [PubMed]
- de França, P.; Costa, J.H.; Fill, T.P.; Lancellotti, M.; Ruiz, A.L.T.G.; Fantinatti-Garboggini, F. Genome Mining Reveals Secondary Metabolites of Antarcticsacterium Streptomyces albidoflavus related to antimicrobial and antiproliferative activities. Arch. Microbiol. 2023, 205, 354. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Coelho, C.; Cordero, R.J.B.; Dragotakes, Q.; Jung, E.; Vij, R.; Wear, M.P. The capsule of Cryptococcus neoformans. Virulence 2019, 10, 822–831. [Google Scholar] [CrossRef]
- Melhem, M.S.C.; Leite Júnior, D.P.; Takahashi, J.P.F.; Macioni, M.B.; Oliveira, L.; de Araújo, L.S.; Fava, W.S.; Bonfietti, L.X.; Paniago, A.M.M.; Venturini, J.; et al. Antifungal resistance in cryptococcal infections. Pathogens 2024, 13, 128. [Google Scholar] [CrossRef]
- Wang, F.; Xu, M.; Li, Q.; Sattler, I.; Lin, W. p-Aminoacetophenonic acids produced by a mangrove Endophyte Streptomyces sp. (Strain HK10552). Molecules 2010, 15, 2782–2790. [Google Scholar] [CrossRef]
- Moreira, E.A.; Rezende-Teixeira, P.; Albernaz, L.C.; Bauermeister, A.; Jimenez, P.C.; Espindola, L.S.; Costa-Lotufo, L.V.; Lopes, N.P. Marine bacteria from the southeast coast of Brazil as a source of insecticidal compounds. Rev. Bras. Farmacogn. 2022, 32, 724–733. [Google Scholar] [CrossRef]
- Gil, J.A.; Naharro, G.; Villanueva, J.R.; Martín, J.F. Characterization and regulation of p-Aminobenzoic acid synthase from Streptomyces griseus. J. Gen. Microbiol. 1985, 131, 1279–1287. [Google Scholar] [CrossRef]
- Belakhov, V.V. Heptaene Macrolide antibiotic perimycin: Preparation, physicochemical properties, structure, biological activity, and application in agriculture as an eco-friendly fungicide (A Review). Russ. J. Gen. Chem. 2021, 91, 2943–2952. [Google Scholar] [CrossRef]
- Berger, S.; El Chazli, Y.; Babu, A.F.; Coste, A.T. Azole resistance in Aspergillus fumigatus: A consequence of antifungal use in agriculture? Front. Microbiol. 2017, 8, 1024. [Google Scholar] [CrossRef]
- Azevedo, M.M.; Faria-Ramos, I.; Cruz, L.C.; Pina-Vaz, C.; Rodrigues, A.G. Genesis of azole antifungal resistance from agriculture to clinical settings. J. Agric. Food Chem. 2015, 63, 7463–7468. [Google Scholar] [CrossRef] [PubMed]
- Mohimani, H.; Gurevich, A.; Shlemov, A.; Mikheenko, A.; Korobeynikov, A.; Cao, L.; Shcherbin, E.; Nothias, L.F.; Dorrestein, P.C.; Pevzner, P.A. Dereplication of microbial metabolites through database search of mass spectra. Nat. Commun. 2018, 9, 4035. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Weng, C.; Shen, X.; Zhu, X. Aptly chosen, effectively emphasizing the action and mechanism of antimycin A1. Front. Microbiol. 2024, 15, 1371850. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F.; Hutchings, M.I. The regulation and biosynthesis of antimycins. Beilstein J. Org. Chem. 2013, 9, 2556–2563. [Google Scholar] [CrossRef]
- Xu, F.; Nazari, B.; Moon, K.; Bushin, L.B.; Seyedsayamdost, M.R. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 Using High-Throughput Elicitor Screens. J. Am. Chem. Soc. 2017, 139, 9203–9212. [Google Scholar] [CrossRef]
- Yang, J.; Qi, Y.; Blodgett, J.A.V.; Wencewicz, T.A. Multifunctional P450 Monooxygenase CftA diversifies the clifednamide pool through tandem C-H bond activations. J. Nat. Prod. 2022, 85, 47–55. [Google Scholar] [CrossRef]
- Lacret, R.; Oves-Costales, D.; Gómez, C.; Díaz, C.; de la Cruz, M.; Pérez-Victoria, I.; Vicente, F.; Genilloud, O.; Reyes, F. New ikarugamycin derivatives with antifungal and antibacterial properties from Streptomyces zhaozhouensis. Mar. Drugs 2014, 13, 128–140. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, S.; Li, J.; Liu, L. The biological and chemical diversity of tetramic acid compounds from marine-derived microorganisms. Mar. Drugs 2020, 18, 114. [Google Scholar] [CrossRef]
- Maimone, N.M.; de Oliveira, L.F.P.; Santos, S.N.; de Lira, S.P. Elicitation of Streptomyces lunalinharesii secondary metabolism through co-cultivation with Rhizoctonia solani. Microbiol. Res. 2021, 251, 126836. [Google Scholar] [CrossRef]
- Xu, Y.; Tan, D.S. Total Synthesis of the bacterial diisonitrile chalkophore SF2768. Org. Lett. 2019, 21, 8731–8735. [Google Scholar] [CrossRef]
- Sezaki, M.; Sasaki, T.; Nakazawa, T.; Takeda, U.; Iwata, M.; Watanabe, T.; Koyama, M.; Kai, F.; Shomura, T.; Kojima, M. A new antibiotic SF-2370 produced by Actinomadura. J. Antibiot. 1985, 38, 1437–1439. [Google Scholar] [CrossRef] [PubMed]
- Almeida, E.L.; Kaur, N.; Jennings, L.K.; Carrillo Rincón, A.F.; Jackson, S.A.; Thomas, O.P.; Dobson, A.D.W. Genome Mining Coupled with OSMAC-based cultivation reveal differential production of surugamide A by the marine sponge isolate Streptomyces sp. SM17 when compared to its terrestrial relative S. albidoflavus J1074. Microorganisms 2019, 7, 394. [Google Scholar] [CrossRef] [PubMed]
- Duda-Madej, A.; Stecko, J.; Sobieraj, J.; Szymańska, N.; Kozłowska, J. Naringenin and its derivatives-health-promoting phytobiotic against resistant bacteria and fungi in humans. Antibiotics 2022, 11, 1628. [Google Scholar] [CrossRef] [PubMed]
- McLean, T.C.; Wilkinson, B.; Hutchings, M.I.; Devine, R. Dissolution of the Disparate: Co-ordinate regulation in antibiotic biosynthesis. Antibiotics 2019, 8, 83. [Google Scholar] [CrossRef]
- Ward, A.C.; Allenby, N.E. Genome mining for the search and discovery of bioactive compounds: The Streptomyces paradigm. FEMS Microbiol. Lett. 2018, 365, fny240. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, Z.; Li, H.; Chen, X.L.; Deng, Z.; Bai, L.; Pang, X. Production of the antibiotic FR-008/candicidin in Streptomyces sp. FR-008 is co-regulated by two regulators, FscRI and FscRIV, from different transcription factor families. Microbiology 2015, 161, 539–552. [Google Scholar] [CrossRef]
- Li, Y.; Pinto-Tomás, A.A.; Rong, X.; Cheng, K.; Liu, M.; Huang, Y. Population genomics insights into adaptive evolution and ecological differentiation in Streptomycetes. Appl. Environ. Microbiol. 2019, 85, e02555-18. [Google Scholar] [CrossRef]
- Antón, N.; Santos-Aberturas, J.; Mendes, M.V.; Guerra, S.M.; Martín, J.F.; Aparicio, J.F. PimM, a PAS domain positive regulator of pimaricin biosynthesis in Streptomyces natalensis. Microbiology 2007, 153, 3174–3183. [Google Scholar] [CrossRef]
- Al-Shaibani, M.M.; Radin Mohamed, R.M.S.; Sidik, N.M.; Enshasy, H.A.E.; Al-Gheethi, A.; Noman, E.; Al-Mekhlafi, N.A.; Zin, N.M. Biodiversity of secondary metabolites compounds isolated from phylum Actinobacteria and its therapeutic applications. Molecules 2021, 26, 4504. [Google Scholar] [CrossRef]
- Hui, M.L.; Tan, L.T.; Letchumanan, V.; He, Y.W.; Fang, C.M.; Chan, K.G.; Law, J.W.; Lee, L.H. The extremophilic Actinobacteria: From microbes to medicine. Antibiotics 2021, 10, 682. [Google Scholar] [CrossRef]
- Núñez-Montero, K.; Barrientos, L. Advances in Antarctic research for antimicrobial discovery: A comprehensive narrative review of bacteria from Antarctic environments as potential sources of novel antibiotic compounds against human pathogens and microorganisms of industrial importance. Antibiotics 2018, 7, 90. [Google Scholar] [CrossRef] [PubMed]
- Flores Clavo, R.; Ruiz Quiñones, N.; Hernández-Tasco, Á.J.; José Salvador, M.; Tasca Gois Ruiz, A.L.; de Oliveira Braga, L.E.; Henrique Costa, J.; Pacheco Fill, T.; Arce Gil, Z.L.; Serquen Lopez, L.M.; et al. Evaluation of antimicrobial and antiproliferative activities of Actinobacteria isolated from the saline lagoons of northwestern Peru. PLoS ONE 2021, 16, e0240946. [Google Scholar] [CrossRef] [PubMed]
- WHO Fungal Priority Pathogens List to Guide Research, Development and Public Health Action; World Health Organization: Geneva, Switzerland, 2022; Available online: https://iris.who.int/bitstream/handle/10665/363682/9789240060241-eng.pdf?sequence=1 (accessed on 24 August 2024).
- Selegato, D.M.; Castro-Gamboa, I. Enhancing chemical and biological diversity by co-cultivation. Front. Microbiol. 2023, 14, 1117559. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, H.; Li, Y.; Liao, S.; Liu, Y. Exploring diverse bioactive secondary metabolites from marine microorganisms using co-culture strategy. Molecules 2023, 28, 6371. [Google Scholar] [CrossRef] [PubMed]
- Bazzano, C.F.; de Felicio, R.; Alves, L.F.G.; Costa, J.H.; Ortega, R.; Vieira, B.D.; Morais-Urano, R.P.; Furtado, L.C.; Ferreira, E.L.F.; Gubiani, J.R.; et al. NP3 MS Workflow: An open-source software system to empower natural product-based drug discovery using untargeted metabolomics. Anal. Chem. 2024, 96, 7460–7469. [Google Scholar] [CrossRef] [PubMed]
- Parra, J.; Jarmusch, S.A.; Duncan, K.R. Multi-omics analysis of antagonistic interactions among free-living Pseudonocardia from diverse ecosystems. Environ. Microbiol. 2024, 26, e16635. [Google Scholar] [CrossRef]
- Arora, D.; Gupta, P.; Jaglan, S.; Roullier, C.; Grovel, O.; Bertrand, S. Expanding the chemical diversity through microorganisms co-culture: Current status and outlook. Biotechnol. Adv. 2020, 40, 107521. [Google Scholar] [CrossRef]
- Dias, D.A.; Urban, S.; Roessner, U. A historical overview of natural products in drug discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef]
- Durán-Iturbide, N.A.; Díaz-Eufracio, B.I.; Medina-Franco, J.L. In silico ADME/Tox profiling of natural products: A focus on BIOFACQUIM. ACS Omega 2020, 5, 16076–16084. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Xiong, G.; Wu, Z.; Yi, J.; Fu, L.; Yang, Z.; Hsieh, C.; Yin, M.; Zeng, X.; Wu, C.; Lu, A.; et al. ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021, 49, W5–W14. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Wang, T.; Jiang, J.; Wang, Y.; Lv, C.; Sun, K.; Sun, J.; Yan, B.; Kang, C.; Guo, L.; et al. Biological control and plant growth promotion properties of Streptomyces albidoflavus St-220 isolated from Salvia miltiorrhiza rhizosphere. Front. Plant Sci. 2022, 13, 976813. [Google Scholar] [CrossRef] [PubMed]
- Diabankana, R.G.C.; Frolov, M.; Keremli, S.; Validov, S.Z.; Afordoanyi, D.M. Genomic insights into the microbial agent Streptomyces albidoflavus MGMM6 for various biotechnology applications. Microorganisms 2023, 11, 2872. [Google Scholar] [CrossRef] [PubMed]
- Kunova, A.; Cortesi, P.; Saracchi, M.; Migdal, G.; Pasquali, M. Draft genome sequences of two Streptomyces albidoflavus strains DEF1AK and DEF147AK with plant growth-promoting and biocontrol potential. Ann. Microbiol. 2021, 71, 1–8. [Google Scholar] [CrossRef]
- Pylro, V.S.; Dias, A.C.F.; Andreote, F.D.; Varani, A.M.; Andreote, C.C.F.; Ribeiro, I.A.F.; Kitano, I.T.; Bernardo, E.R.A. Draft Genomic Sequences of Streptomyces misionensis ACT66 and Streptomyces albidoflavus ACT77, Bacteria with Potential Application for Phytopathogen Biocontrol. Microbiol. Resour. Announc. 2019, 8, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Carlucci, A.; Raimondo, M.L.; Colucci, D.; Lops, F. Streptomyces albidoflavus Strain CARA17 as a biocontrol agent against fungal soil-borne pathogens of fennel plants. Plants 2022, 11, 1420. [Google Scholar] [CrossRef]
- Fang, W.; Wu, J.; Cheng, M.; Zhu, X.; Du, M.; Chen, C.; Liao, W.; Zhi, K.; Pan, W. Diagnosis of invasive fungal infections: Challenges and recent developments. J. Biomed. Sci. 2023, 30, 42. [Google Scholar] [CrossRef]
- Pimienta, D.A.; Cruz Mosquera, F.E.; Palacios Velasco, I.; Giraldo Rodas, M.; Oñate-Garzón, J.; Liscano, Y. Specific focus on antifungal peptides against azole-resistant Aspergillus fumigatus: Current Status, Challenges, and Future Perspectives. J. Fungi 2022, 9, 42. [Google Scholar] [CrossRef]
- Wu, Y.; Wu, M.; Gao, J.; Ying, C. Antifungal activity and mode of action of miltefosine against clinical isolates of Candida krusei. Front. Microbiol. 2020, 11, 854. [Google Scholar] [CrossRef]
- Thakur, R.; Shishodia, S.K.; Sharma, A.; Chauhan, A.; Kaur, S.; Shankar, J. Accelerating the understanding of Aspergillus terreus: Epidemiology, physiology, immunology and advances. Curr. Res. Microb. Sci. 2024, 6, 100220. [Google Scholar] [CrossRef]
- Peron, I.H.; Reichert-Lima, F.; Busso-Lopes, A.F.; Nagasako, C.K.; Lyra, L.; Moretti, M.L.; Schreiber, A.Z. Resistance surveillance in Candida albicans: A five-year antifungal susceptibility evaluation in a Brazilian university hospital. PLoS ONE 2016, 11, e01581262016. [Google Scholar] [CrossRef] [PubMed]
- Pontes, L.; Beraquet, C.A.G.; Arai, T.; Pigolli, G.L.; Lyra, L.; Watanabe, A.; Moretti, M.L.; Schreiber, A.Z. Aspergillus fumigatus clinical isolates carrying CYP51A with TR34/L98H/S297T/F495I substitutions detected after four-year retrospective azole resistance screening in Brazil. Antimicrob. Agents Chemother. 2020, 64, e02059-19. [Google Scholar] [CrossRef] [PubMed]
- Pontes, L.; Arai, T.; Gualtieri Beraquet, C.A.; Giordano, A.L.P.L.; Reichert-Lima, F.; da Luz, E.A.; Fernanda de Sá, C.; Ortolan Levy, L.; Tararam, C.A.; Watanabe, A.; et al. Uncovering a novel cyp51A Mutation and antifungal resistance in Aspergillus fumigatus through culture collection screening. J. Fungi 2024, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard Document M27-A3; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- CLSI. Reference method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi; Approved Standard M38-A2, 2nd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2008. [Google Scholar]
- EUCAST (The European Committee on Antimicrobial Susceptibility Testing). Routine and Extended Internal Quality Control for MIC Determination and Agar Dilution for Yeasts, Moulds and Dermatophytes as Recommended by EUCAST. Version 6.0. 2022. Available online: http://www.eucast.org (accessed on 24 August 2024).
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef]
- van Santen, J.A.; Poynton, E.F.; Iskakova, D.; McMann, E.; Alsup, T.A.; Clark, T.N.; Fergusson, C.H.; Fewer, D.P.; Hughes, A.H.; McCadden, C.A.; et al. The Natural Products Atlas 2.0: A database of microbially-derived natural products. Nucleic Acids Res. 2022, 50, D1317–D1323. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures, and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Kumar, M.; Parveen; Raj, N.; Khatoon, S.; Fakhri, K.U.; Kumar, P.; Alamri, M.A.; Kamal, M.; Manzoor, N.; Harsha; et al. In-silico and in-vitro evaluation of antifungal bioactive compounds from Streptomyces sp. strain 130 against Aspergillus flavus. J. Biomol. Struct. Dyn. 2024, 42, 1–19. [Google Scholar] [CrossRef]
- Pokharkar, O.; Lakshmanan, H.; Zyryanov, G.; Tsurkan, M. In silico evaluation of antifungal compounds from marine sponges against COVID-19-associated mucormycosis. Mar. Drugs 2022, 20, 215. [Google Scholar] [CrossRef]
- Borba, J.V.B.; Alves, V.M.; Braga, R.C.; Korn, D.R.; Overdahl, K.; Silva, A.C.; Hall, S.U.S.; Overdahl, E.; Kleinstreuer, N.; Strickland, J.; et al. STopTox: An in silico alternative to animal testing for acute systemic and topical toxicity. Environ. Health Perspect. 2022, 130, 27012. [Google Scholar] [CrossRef] [PubMed]


| Strain | Susceptibility to Ethyl Acetate Crude Extract (mg/mL) | Susceptibility Profile to Commercial Drugs (MIC/MEC in µg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MCF | CPF | AMB | 5-FC | FCZ | VRC | ITZ | MCZ | TER | |||
| C. albicans (ATCC 90028) | MIC | 1.5 | 0.5 | 0.12 | 1 | - | 1 | 0.03 | 0.12 | 0.5 | - |
| MFC | 1.5 | ||||||||||
| C. parapsilosis (ATCC 22019) | MIC | 1.5 | 0.25 | 1 | 1 | - | 1 | 0.25 | 0.12 | 0.5 | - |
| MFC | 1.5 | ||||||||||
| C. krusei (ATCC 6258) | MIC | 1.5 | ≤0.015 | 0.25 | 1 | - | - | 0.25 | 0.12 | 0.25 | - |
| MFC | 1.5 | ||||||||||
| A. fumigatus (ATCC 204305) | MIC | 1.5 | ≤0.015 | 0.25 | 1 | - | >64 | 0.5 | 0.5 | 1 | - |
| MFC | 1.5 | ||||||||||
| A. flavus (ATCC 204304) | MIC | 1.5 | ≤0.015 | 0.25 | 1 | - | >64 | 0.25 | 0.5 | 1 | - |
| MFC | 1.5 | ||||||||||
| C. neoformans (ATCC 90113) | MIC | 1.5 | - | - | 0.5 | 1 | 2 | 0.12 | 0.06 | - | - |
| MFC | 3.0 | ||||||||||
| C. gattii (ATCC 56990) | MIC | 1.5 | - | - | 0.5 | 0.5 | 2 | 0.015 | 0.12 | - | - |
| MFC | 3.0 | ||||||||||
| T. mentagrophytes (ATCC 9533) | MIC | 1.5 | - | - | - | - | - | - | 0.125 | - | 0.03 |
| MFC | 1.5 | ||||||||||
| LIF 12560 | MIC | 1.5 | - | - | 0.25 | <0.12 | 8 | 0.25 | 0.12 | - | - |
| MFC | 1.5 | ||||||||||
| LIF E-10 | MIC | 1.5 | - | - | 0.25 | <0.12 | 64 | 0.25 | 1 | - | - |
| MFC | 1.5 | ||||||||||
| LIF 2552-4.9 | MIC | 1.5 | ≤0.015 | 0.25 | 1 | - | - | 4 | >8 | >16 | - |
| MFC | 1.5 | ||||||||||
| LIF 2444.6 | MIC | 1.5 | ≤0.015 | 0.25 | 1 | - | - | 2 | >8 | >16 | - |
| MFC | 1.5 | ||||||||||
| LIF 263-e | MIC | 1.5 | ≤0.015 | 0.125 | 1 | - | - | 4 | >8 | >16 | - |
| MFC | 1.5 | ||||||||||
| LIF 1889 | MIC | 1.5 | 0.03 | 0.25 | 1 | - | - | 8 | 4 | >16 | - |
| MFC | 1.5 | ||||||||||
| LIF 2328 | MIC | 1.5 | ≤0.015 | 0.25 | 2 | - | - | 1 | 0.5 | - | - |
| MFC | 1.5 | ||||||||||
| LIF 2486 | MIC | 1.5 | ≤0.015 | 0.25 | 1 | - | - | 2 | >8 | >16 | - |
| MFC | 1.5 | ||||||||||
| LIF 16606-1 | MIC | 1.5 | 0.12 | 0.5 | 0.5 | 0.12 | 0.5 | 0.03 | 0.03 | - | - |
| MFC | 1.5 | ||||||||||
| LIF 16615 | MIC | 1.5 | 0.07 | 0.25 | 8 | - | 0.03 | 0.07 | 0.07 | - | - |
| MFC | 1.5 | ||||||||||
| LIF 1455 | MIC | 1.5 | ≥ 16 | - | 0.5 | >64 | >64 | >8 | >8 | 1 | >16 |
| MFC | 1.5 | ||||||||||
| Compound | # | Annotation | Molecular Formula | Cosine Score | Spec MZ | MetMass | NP Pathway | NP Superclass | NP Class |
|---|---|---|---|---|---|---|---|---|---|
| 9-(4-aminophenyl)-3,7-dihydroxy-2,4,6-trimethyl-9-oxo-nonoic acid (candicidin related) | 1 | GNPS | C18H27NO5 | 0.88 | 364.21 | 338.20 | Polyketides (PKS) | Linear PKS | Open-chain PKS |
| Candicidin I | 2 | GNPS (Analog library hit) | - | 0.74 | 546.29 | 547.28 | - | - | - |
| Candicidin IV | 3 | GNPS (Analog library hit) | - | 0.71 | 557.31 | 556.28 | - | - | - |
| Antimycin A | 4 | DEREPLICATOR+ | C28H40N2O9 | - | 507.234 | 506.23 | - | - | - |
| Surugamide A | 5 | GNPS | C48H81N9O8 | 0.91 | 912.62 | 911.62 | Amino acids and Peptides | Oligopeptides | Cyclic peptides |
| Clifednamide A | 6 | GNPS | C29H36N2O5 | 0.76 | 495.28 | 493.27 | Amino acids and Peptides | Macrolides | Macrocyclic tetramic acids (PTM) |
| Ikarugamycin epoxide | 7 | GNPS | C29H38N2O5 | 0.79 | 497.30 | 495.29 | Amino acids and Peptides | Macrolides | PTM |
| Fungimycin | 8 | DEREPLICATOR+ | C59H86N2O17 | - | 1095.6 | 1094.59 | - | - | - |
| Region | Type | From | To | Most Similar Known Cluster | Similarity | |
|---|---|---|---|---|---|---|
| Region 1.1 | NRPS | 164.68 | 209.1 | diisonitrile antibiotic SF2768 | NRP | 66% |
| Region 1.3 | T1PKS, NRPS | 423.35 | 472.76 | SGR PTMs | NRP + PKS | 100% |
| Region 24.1 | NRPS | 5.317 | 35.206 | surugamide A/surugamide D | NRP | 42% |
| Region 25.1 | NRPS | 1 | 41.377 | surugamide A/surugamide D | NRP | 19% |
| Region 27.1 | NRPS, LAP | 1 | 34.45 | surugamide A/surugamide D | NRP | 61% |
| Region 40.1 | T3PKS | 1.86 | 42.957 | naringenin | T3PKS | 100% |
| Region 40.2 | T1PKS | 47.893 | 101.94 | candicidin | NRP + PKS | 33% |
| Region 41.1 | T1PKS | 1 | 31.408 | rosamicin | PKS | 26% |
| Region 44.1 | T1PKS, NRPS-like | 1 | 59.019 | candicidin | NRP + PKS | 90% |
| Region 45.1 | NRPS, T1PKS, LAPclass-II, NRPS-like | 1 | 74.671 | antimycin | NRP: Cyclic depsipeptide + T3PKS | 80% |
| Compound Annotated | Physicochemical Properties | Pharmacokinetics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Weight | N° Rotatable Bonds | N° H-Bond Acceptors | Nº H-Bond Donors | Aqueous Solubility (LogS) | Octanol/Water Partition Coeffifient (LogP) | Gastrointestinal Absorption | Blood-Brain Barrier Permeability | Metabolism (Cytochrome P450 Inhibition) | Skin Permeation (Log Kp) [cm/s] | Bioavailability Score | |
| 9-(4-aminophenyl)-3,7-dihydroxy-2,4,6-trimethyl-9-oxo-nonoic acid (candicidin related) | 337.190 | 9 | 5 | 4 | −2.702 | 0.967 | High | No | No | −7.19 | 0.56 |
| Antimycin A | 492.210 | 16 | 9 | 4 | −3.984 | 3.337 | Low | No | No | −6.76 | 0.11 |
| Fungimycin | 1094.590 | 10 | 19 | 13 | −3.953 | 0.649 | Low | No | No information | No information | No information |
| Surugamide A | 911.620 | 16 | 9 | 9 | −3.175 | 5.136 | Low | No | Yes (CYP3A4) | −7.65 | 0.17 |
| Clifednamide A | 492.260 | 1 | 5 | 3 | −4.779 | 3.053 | High | No | Yes (CYP3A4) | −6.95 | 0.56 |
| Ikarugamycin epoxide | 496.290 | 1 | 5 | 3 | −3.338 | 2.893 | High | No | Yes (CYP3A4) | −6.91 | 0.55 |
| Compound Annotated | Toxicity | |||||||
|---|---|---|---|---|---|---|---|---|
| Human Hepatotoxicity | Drug-Induced Liver Injury | Rat Oral Acute Toxicity | Skin Sensitization | Carcinogenicity | Eye Corrosion | Eye Irritation | Respiratory Toxicity | |
| 9-(4-aminophenyl)-3,7-dihydroxy-2,4,6-trimethyl-9-oxo-nonoic acid (candicidin related) | -- | +++ | - | --- | --- | --- | --- | - |
| Antimycin A | ++ | +++ | --- | -- | --- | --- | --- | --- |
| Fungimycin | +++ | --- | --- | +++ | + | --- | --- | +++ |
| Surugamide A | +++ | --- | ++ | --- | --- | --- | --- | --- |
| Clifednamide A | - | +++ | - | -- | -- | --- | --- | ++ |
| Ikarugamycin epoxide | - | + | ++ | -- | --- | --- | --- | +++ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giordano, A.L.P.L.; Rodrigues, M.V.N.; dos Santos, K.G.A.; Legabão, B.C.; Pontes, L.; de Angelis, D.A.; Garboggini, F.F.; Schreiber, A.Z. Enhancing Antifungal Drug Discovery Through Co-Culture with Antarctic Streptomyces albidoflavus Strain CBMAI 1855. Int. J. Mol. Sci. 2024, 25, 12744. https://doi.org/10.3390/ijms252312744
Giordano ALPL, Rodrigues MVN, dos Santos KGA, Legabão BC, Pontes L, de Angelis DA, Garboggini FF, Schreiber AZ. Enhancing Antifungal Drug Discovery Through Co-Culture with Antarctic Streptomyces albidoflavus Strain CBMAI 1855. International Journal of Molecular Sciences. 2024; 25(23):12744. https://doi.org/10.3390/ijms252312744
Chicago/Turabian StyleGiordano, Ana Luisa Perini Leme, Marili Villa Nova Rodrigues, Karen Gabriela Araujo dos Santos, Barbara Cipulo Legabão, Lais Pontes, Derlene Attili de Angelis, Fabiana Fantinatti Garboggini, and Angelica Zaninelli Schreiber. 2024. "Enhancing Antifungal Drug Discovery Through Co-Culture with Antarctic Streptomyces albidoflavus Strain CBMAI 1855" International Journal of Molecular Sciences 25, no. 23: 12744. https://doi.org/10.3390/ijms252312744
APA StyleGiordano, A. L. P. L., Rodrigues, M. V. N., dos Santos, K. G. A., Legabão, B. C., Pontes, L., de Angelis, D. A., Garboggini, F. F., & Schreiber, A. Z. (2024). Enhancing Antifungal Drug Discovery Through Co-Culture with Antarctic Streptomyces albidoflavus Strain CBMAI 1855. International Journal of Molecular Sciences, 25(23), 12744. https://doi.org/10.3390/ijms252312744






