New Insights into the Role of KLF10 in Tissue Fibrosis
Abstract
1. Introduction
2. KLF10 Characteristics
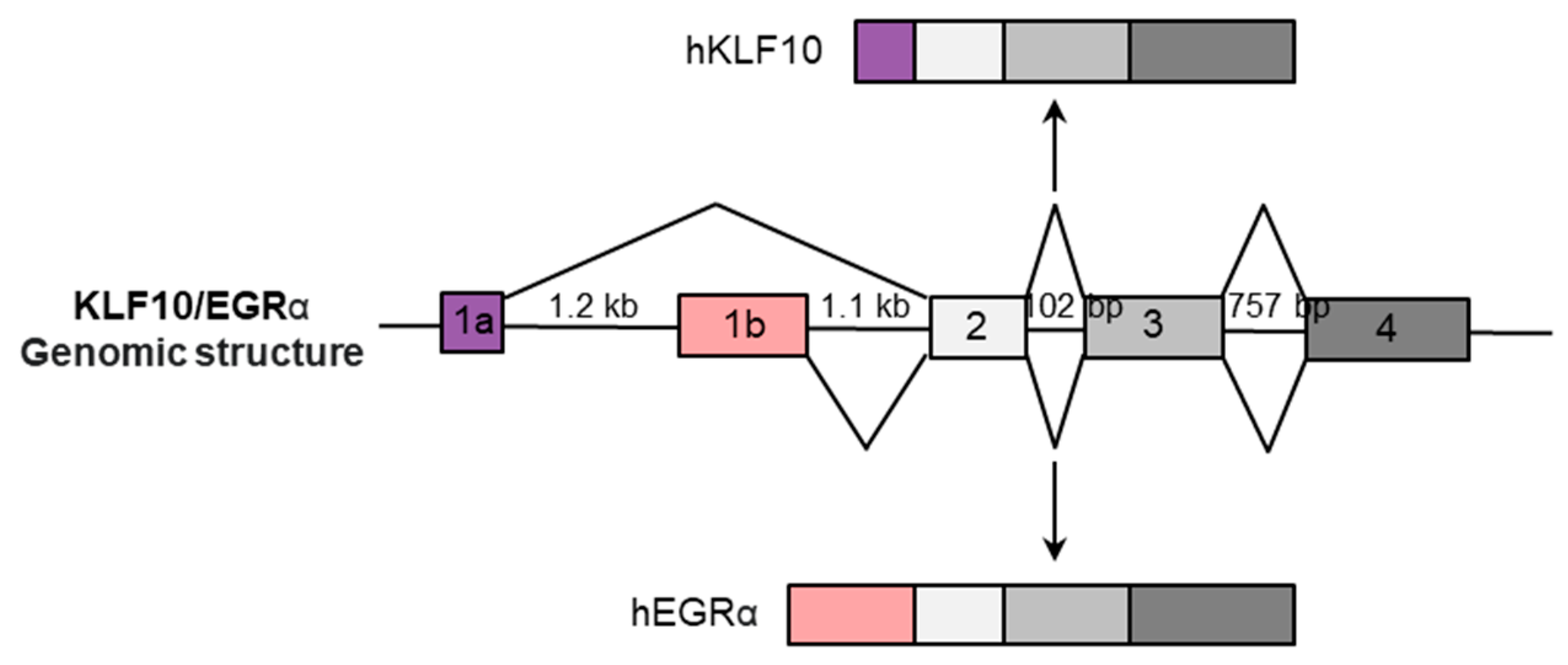
2.1. KLF10 Gene Structure and Variations
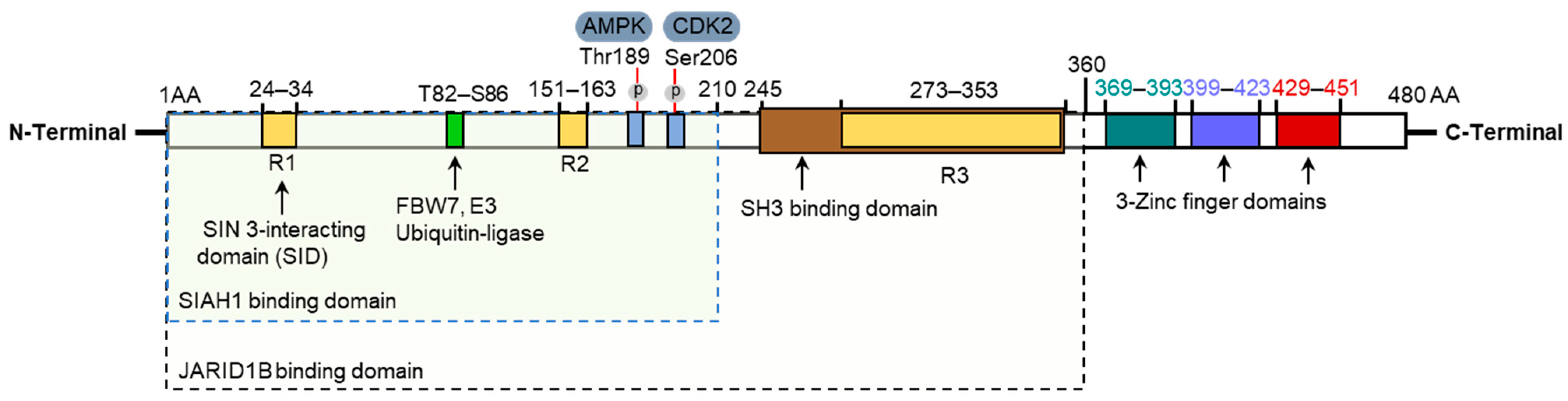
2.2. KLF10 Protein Structure
2.3. KLF10 Activation, Interactions, and Protein Stability
3. KLF10 Functional Regulation
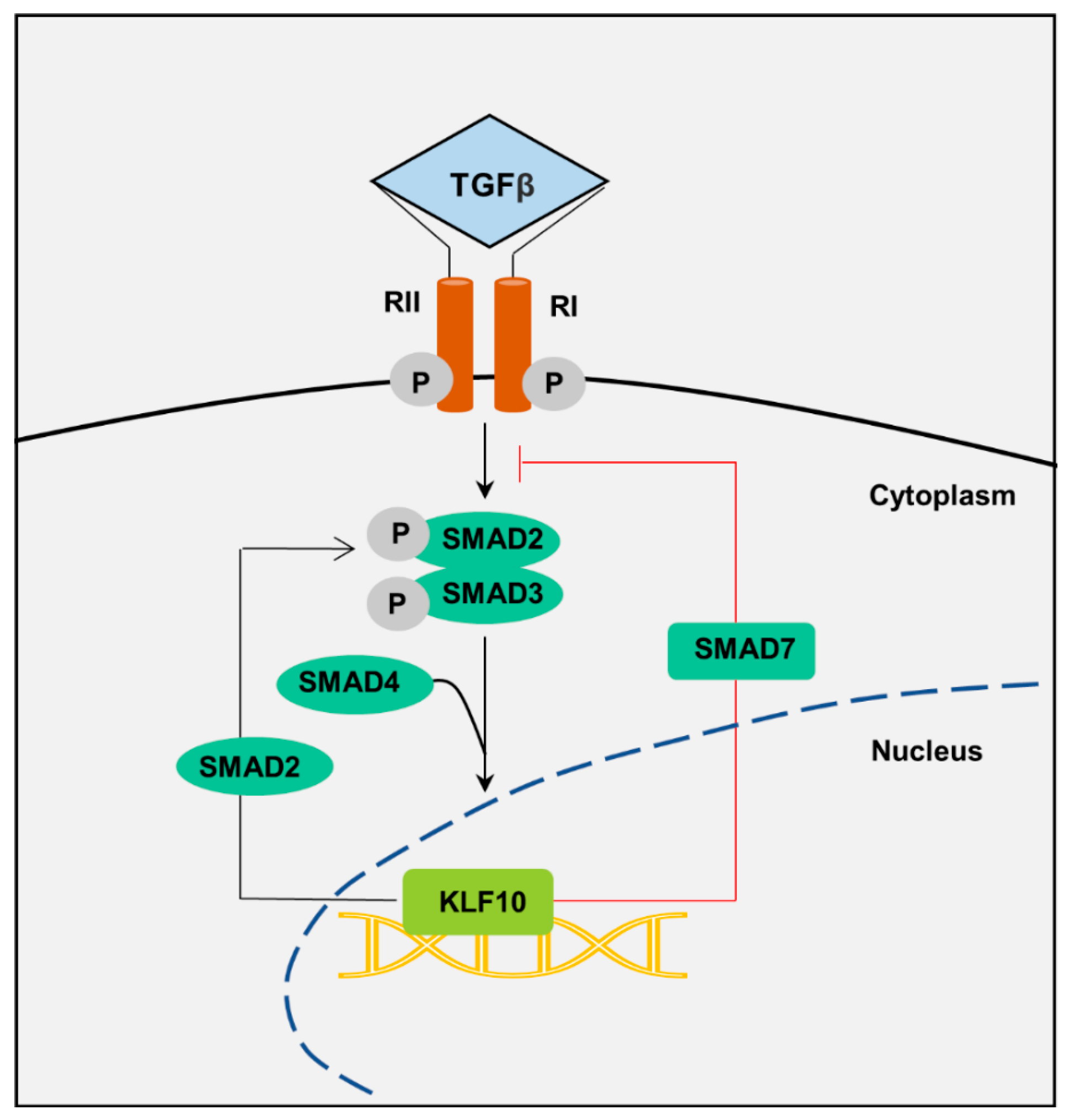
3.1. TGF-β Regulation
3.2. Nutrient and Metabolic Pathway Regulation
3.3. Inflammation and Metabolic Regulation
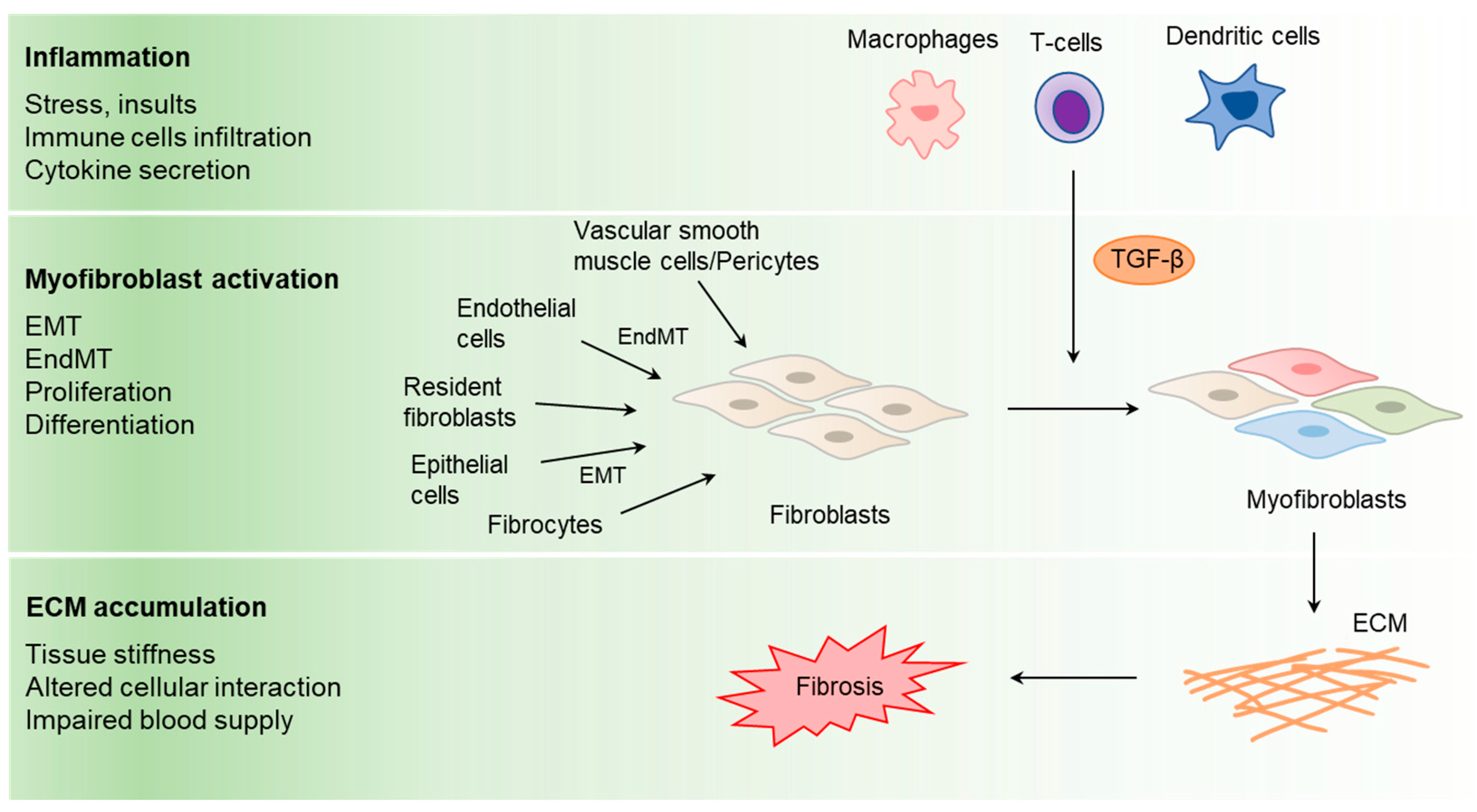
4. Effect of KLF10 in Tissue Fibrosis
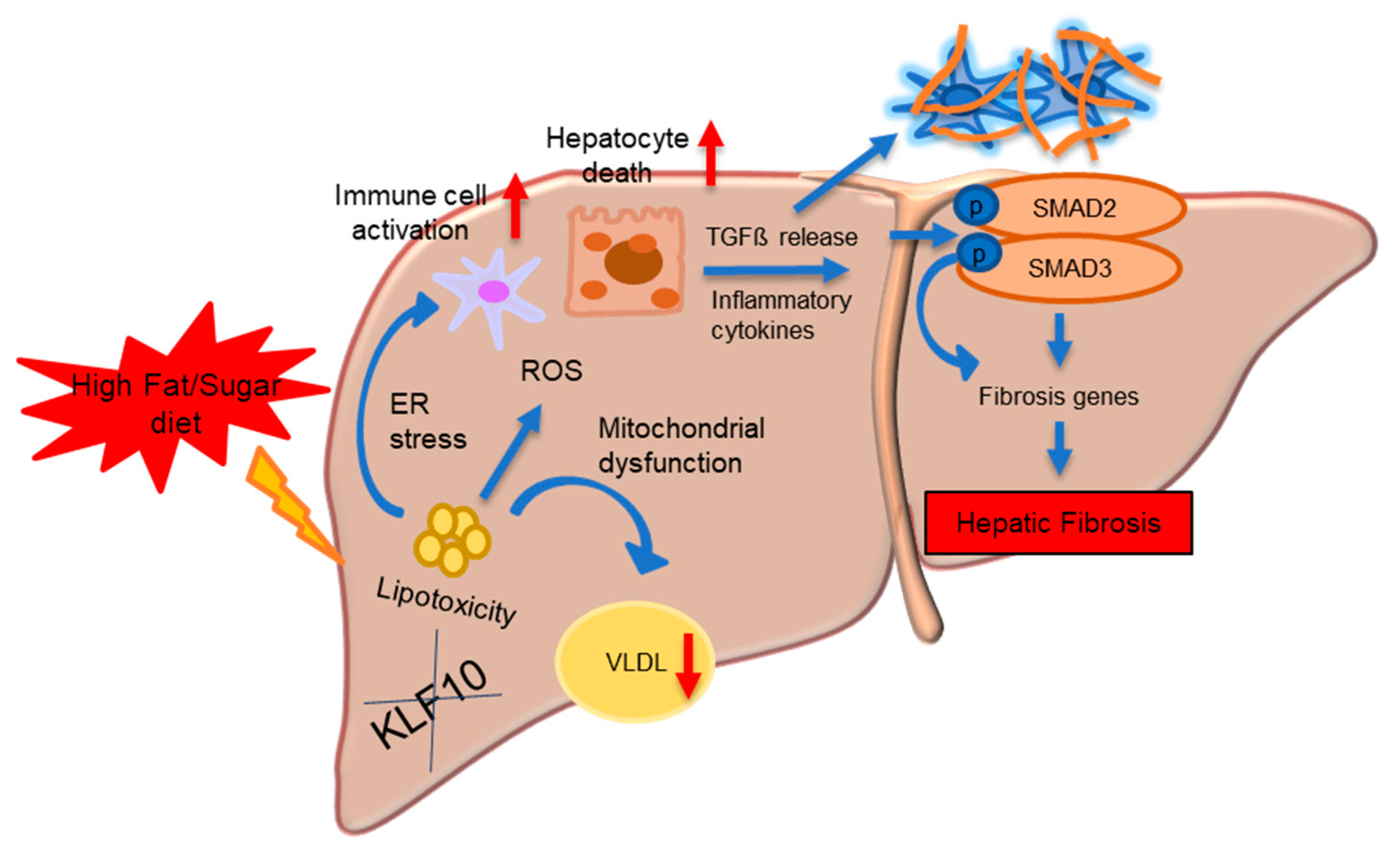
4.1. KLF10 in Hepatic Fibrosis
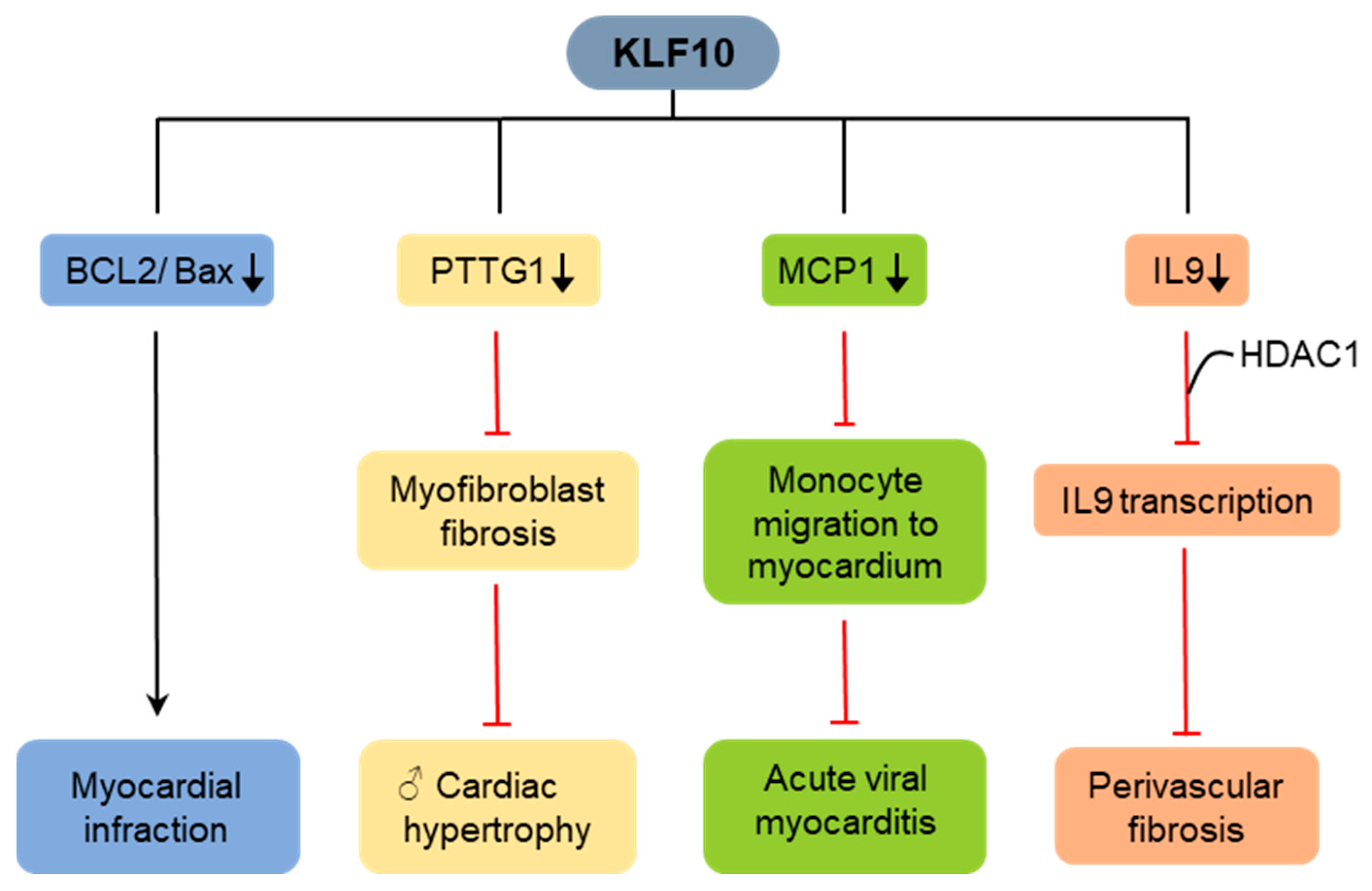
4.2. KLF10 in Cardiac Fibrosis
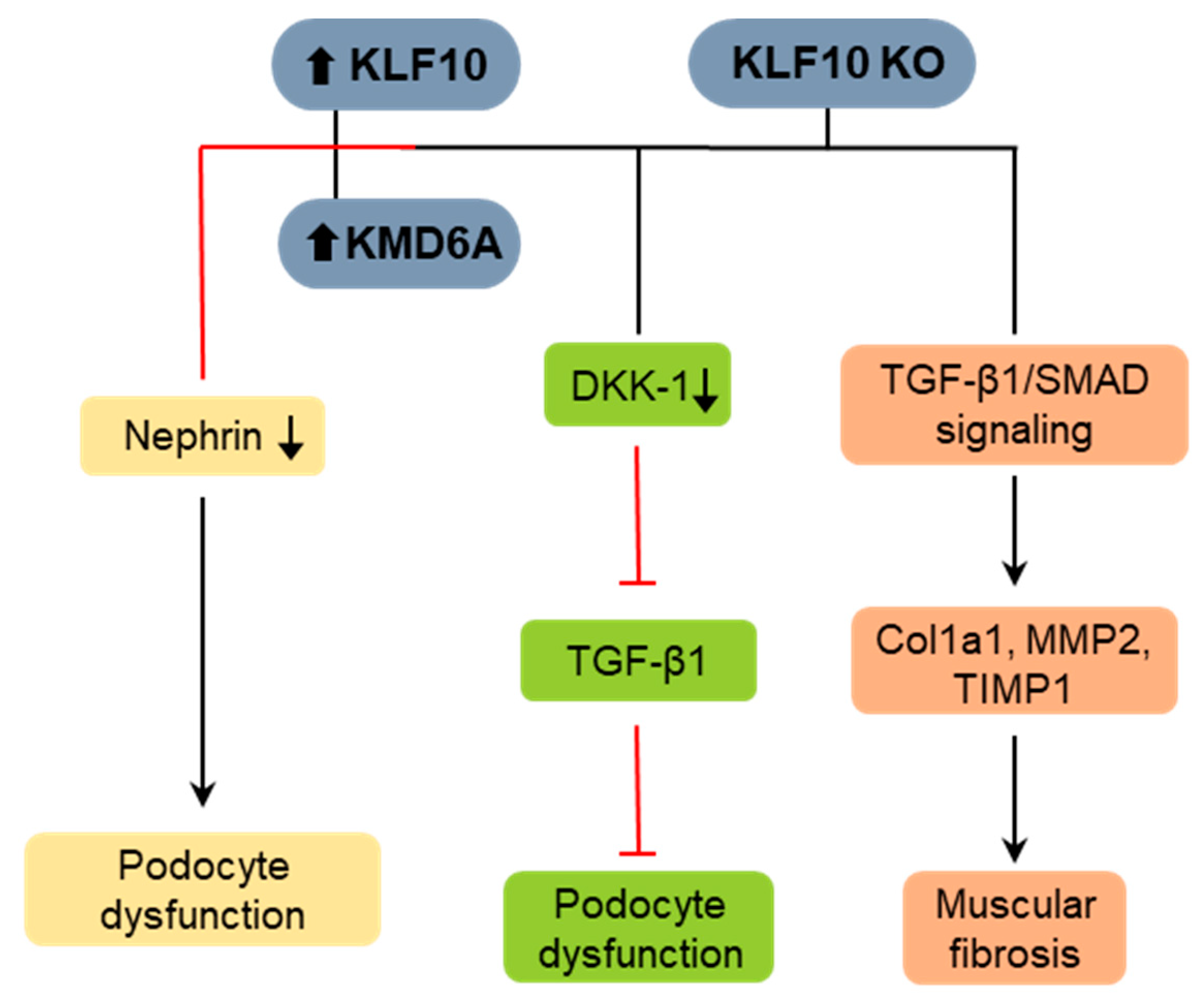
4.3. KLF10 in Renal Fibrosis
4.4. KLF10 in Muscular Fibrosis
4.5. KLF10 in Pulmonary Fibrosis
4.6. KLF10 in Skin Fibrosis
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMPK | AMP-activated protein kinase |
| CDK2 | Cyclin-dependent kinase 2 |
| ChREBP | Carbohydrate response element-binding protein |
| ECM | Extracellular matrix |
| HSC | Hepatic stellate cell |
| KLF10 | Krüppel-like factor 10 |
| KO | Knockout |
| NAFLD | Non-alcoholic fatty liver disease |
| NASH | Non-alcoholic steatohepatitis |
| SIAH1 | Seven in Absentia homologue-1 |
| SMAD | Suppressor of mothers against decapentaplegic |
| TGF-β | Transforming growth factor-beta |
| VMC | Viral myocarditis |
References
- Wynn, T.A. Cellular and molecular mechanisms of fibrosis. J. Pathol. 2008, 214, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Wynn, T.A.; Ramalingam, T.R. Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 2012, 18, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Klingberg, F.; Hinz, B.; White, E.S. The myofibroblast matrix: Implications for tissue repair and fibrosis. J. Pathol. 2013, 229, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Jun, J.I.; Lau, L.F. Resolution of organ fibrosis. J. Clin. Investig. 2018, 128, 97–107. [Google Scholar] [CrossRef]
- Rockey, D.C.; Bell, P.D.; Hill, J.A. Fibrosis—A Common Pathway to Organ Injury and Failure. N. Engl. J. Med. 2015, 372, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Frangogiannis, N. Transforming growth factor-beta in tissue fibrosis. J. Exp. Med. 2020, 217, e20190103. [Google Scholar] [CrossRef]
- Parsons, C.J.; Takashima, M.; Rippe, R.A. Molecular mechanisms of hepatic fibrogenesis. J. Gastroenterol. Hepatol. 2007, 22 (Suppl. 1), S79–S84. [Google Scholar] [CrossRef]
- Lipson, K.E.; Wong, C.; Teng, Y.; Spong, S. CTGF is a central mediator of tissue remodeling and fibrosis and its inhibition can reverse the process of fibrosis. Fibrogenesis Tissue Repair 2012, 5 (Suppl. 1), S24. [Google Scholar] [CrossRef]
- McAnulty, R.J. Fibroblasts and myofibroblasts: Their source, function and role in disease. Int. J. Biochem. Cell Biol. 2007, 39, 666–671. [Google Scholar] [CrossRef]
- Lee, J.H.; Massague, J. TGF-beta in developmental and fibrogenic EMTs. Semin. Cancer Biol. 2022, 86 Pt 2, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Pardali, E.; Sanchez-Duffhues, G.; Gomez-Puerto, M.C.; Ten Dijke, P. TGF-beta-Induced Endothelial-Mesenchymal Transition in Fibrotic Diseases. Int. J. Mol. Sci. 2017, 18, 2157. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Yuan, W.; Mori, Y.; Levenson, A.; Trojanowska, M.; Varga, J. Stimulation of type I collagen transcription in human skin fibroblasts by TGF-beta: Involvement of Smad 3. J. Investig. Dermatol. 1999, 112, 49–57. [Google Scholar] [CrossRef]
- Roberts, A.B.; Heine, U.I.; Flanders, K.C.; Sporn, M.B. Transforming growth factor-beta. Major role in regulation of extracellular matrix. Ann. N. Y. Acad. Sci. 1990, 580, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Zakiyanov, O.; Kalousova, M.; Zima, T.; Tesar, V. Matrix metalloproteinases and tissue inhibitors of matrix metalloproteinases in kidney disease. Adv. Clin. Chem. 2021, 105, 141–212. [Google Scholar] [PubMed]
- Rohani, M.G.; Parks, W.C. Matrix remodeling by MMPs during wound repair. Matrix Biol. 2015, 44–46, 113–121. [Google Scholar] [CrossRef]
- Subramaniam, M.; Harris, S.A.; Oursler, M.J.; Rasmussen, K.; Riggs, B.L.; Spelsberg, T.C. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995, 23, 4907–4912. [Google Scholar] [CrossRef]
- Lee, J.; Oh, A.R.; Lee, H.Y.; Moon, Y.A.; Lee, H.J.; Cha, J.Y. Deletion of KLF10 Leads to Stress-Induced Liver Fibrosis upon High Sucrose Feeding. Int. J. Mol. Sci. 2020, 22, 331. [Google Scholar] [CrossRef]
- DiMario, J.X. KLF10 Gene Expression Modulates Fibrosis in Dystrophic Skeletal Muscle. Am. J. Pathol. 2018, 188, 1263–1275. [Google Scholar] [CrossRef]
- Luo, H.Y.; Zhu, J.Y.; Chen, M.; Mu, W.J.; Guo, L. Krüppel-like factor 10 (KLF10) as a critical signaling mediator: Versatile functions in physiological and pathophysiological processes. Genes Dis. 2023, 10, 915–930. [Google Scholar] [CrossRef]
- Hwang, S.; Park, S.; Yaseen, U.; Lee, H.J.; Cha, J.Y. KLF10 Inhibits TGF-β-Mediated Activation of Hepatic Stellate Cells via Suppression of ATF3 Expression. Int. J. Mol. Sci. 2023, 24, 12602. [Google Scholar] [CrossRef] [PubMed]
- Memon, A.; Lee, W.K. KLF10 as a Tumor Suppressor Gene and Its TGF-β Signaling. Cancers 2018, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Grishin, N.V. A new family of predicted Krüppel-like factor genes and pseudogenes in placental mammals. PLoS ONE 2013, 8, e81109. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, Q.; Sun, L.; Zhang, H.; Yao, L.; Cui, X.; Gao, Y.; Fang, F.; Chang, Y. KLF10 transcription factor regulates hepatic glucose metabolism in mice. Diabetologia 2017, 60, 2443–2452. [Google Scholar] [CrossRef]
- Subramaniam, M.; Hawse, J.R.; Rajamannan, N.M.; Ingle, J.N.; Spelsberg, T.C. Functional role of KLF10 in multiple disease processes. Biofactors 2010, 36, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Spittau, B.; Krieglstein, K. Klf10 and Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue Res. 2012, 347, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Res. 2022, 51, D933–D941. [Google Scholar] [CrossRef]
- Fautsch, M.P.; Vrabel, A.; Subramaniam, M.; Hefferen, T.E.; Spelsberg, T.C.; Wieben, E.D. TGFbeta-inducible early gene (TIEG) also codes for early growth response alpha (EGRalpha): Evidence of multiple transcripts from alternate promoters. Genomics 1998, 51, 408–416. [Google Scholar] [CrossRef]
- Fautsch, M.P.; Vrabel, A.; Rickard, D.; Subramaniam, M.; Spelsberg, T.C.; Wieben, E.D. Characterization of the mouse TGFbeta-inducible early gene (TIEG): Conservation of exon and transcriptional regulatory sequences with evidence of additional transcripts. Mamm. Genome 1998, 9, 838–842. [Google Scholar] [CrossRef]
- Memon, A.; Pyao, Y.; Jung, Y.; Choi, H.S.; Song, K.D.; Lee, W.K. The basal transcriptional activity of the murine Klf10 gene is regulated by the transcriptional factor JunB. Genes Genom. 2021, 43, 343–349. [Google Scholar] [CrossRef]
- Shields, J.M.; Christy, R.J.; Yang, V.W. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J. Biol. Chem. 1996, 271, 20009–20017. [Google Scholar] [CrossRef] [PubMed]
- Cook, T.; Gebelein, B.; Belal, M.; Mesa, K.; Urrutia, R. Three conserved transcriptional repressor domains are a defining feature of the TIEG subfamily of Sp1-like zinc finger proteins. J. Biol. Chem. 1999, 274, 29500–29504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.S.; Moncrieffe, M.C.; Kaczynski, J.; Ellenrieder, V.; Prendergast, F.G.; Urrutia, R. A conserved alpha-helical motif mediates the interaction of Sp1-like transcriptional repressors with the corepressor mSin3A. Mol. Cell Biol. 2001, 21, 5041–5049. [Google Scholar] [CrossRef]
- Gunther, M.; Laithier, M.; Brison, O. A set of proteins interacting with transcription factor Sp1 identified in a two-hybrid screening. Mol. Cell Biochem. 2000, 210, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Shin, S.; Subramaniam, M.; Bruinsma, E.; Kim, T.D.; Hawse, J.R.; Spelsberg, T.C.; Janknecht, R. Histone demethylase JARID1B/KDM5B is a corepressor of TIEG1/KLF10. Biochem. Biophys. Res. Commun. 2010, 401, 412–416. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, S.A.; Subramaniam, M.; Monroe, D.G.; Janknecht, R.; Spelsberg, T.C. Modulation of transforming growth factor beta (TGFbeta)/Smad transcriptional responses through targeted degradation of TGFbeta-inducible early gene-1 by human seven in absentia homologue. J. Biol. Chem. 2002, 277, 30754–30759. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Wang, F.; Tan, X.; Gao, G.L.; Pan, W.J.; Luan, Y.; Ge, X. FBW7 targets KLF10 for ubiquitin-dependent degradation. Biochem. Biophys. Res. Commun. 2018, 495, 2092–2097. [Google Scholar] [CrossRef]
- Lin, C.H.; Lin, S.Y.; Chang, H.W.; Ko, L.J.; Tseng, Y.S.; Chang, V.H.; Yu, W.C. CDK2 phosphorylation regulates the protein stability of KLF10 by interfering with binding of the E3 ligase SIAH1. Biochim. Biophys. Acta 2015, 1853, 1174–1181. [Google Scholar] [CrossRef]
- Chen, Y.C.; Chen, R.J.; Peng, S.Y.; Yu, W.C.Y.; Chang, V.H. Therapeutic Targeting of Nonalcoholic Fatty Liver Disease by Downregulating SREBP-1C Expression via AMPK-KLF10 Axis. Front. Mol. Biosci. 2021, 8, 751938. [Google Scholar] [CrossRef]
- Yajima, S.; Lammers, C.H.; Lee, S.H.; Hara, Y.; Mizuno, K.; Mouradian, M.M. Cloning and characterization of murine glial cell-derived neurotrophic factor inducible transcription factor (MGIF). J. Neurosci. 1997, 17, 8657–8666. [Google Scholar] [CrossRef]
- Hu, Z.C.; Shi, F.; Liu, P.; Zhang, J.; Guo, D.; Cao, X.L.; Chen, C.F.; Qu, S.Q.; Zhu, J.Y.; Tang, B. TIEG1 Represses Smad7-Mediated Activation of TGF-beta1/Smad Signaling in Keloid Pathogenesis. J. Investig. Dermatol. 2017, 137, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.E. Non-Smad Signaling Pathways of the TGF-β Family. Cold Spring Harb. Perspect. Biol. 2017, 9, a022129. [Google Scholar] [CrossRef]
- Jin, W.; Di, G.; Li, J.; Chen, Y.; Li, W.; Wu, J.; Cheng, T.; Yao, M.; Shao, Z. TIEG1 induces apoptosis through mitochondrial apoptotic pathway and promotes apoptosis induced by homoharringtonine and velcade. FEBS Lett. 2007, 581, 3826–3832. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Rodríguez, R.; Barzi, M.; Berenguer, J.; Pons, S. Bone morphogenetic protein 2 opposes Shh-mediated proliferation in cerebellar granule cells through a TIEG-1-based regulation of Nmyc. J. Biol. Chem. 2007, 282, 37170–37180. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.; Bronk, S.F.; Roberts, P.J.; Urrutia, R.; Gores, G.J. The transforming growth factor beta(1)-inducible transcription factor TIEG1, mediates apoptosis through oxidative stress. Hepatology 1999, 30, 1490–1497. [Google Scholar] [CrossRef] [PubMed]
- Hefferan, T.E.; Reinholz, G.G.; Rickard, D.J.; Johnsen, S.A.; Waters, K.M.; Subramaniam, M.; Spelsberg, T.C. Overexpression of a nuclear protein, TIEG, mimics transforming growth factor-beta action in human osteoblast cells. J. Biol. Chem. 2000, 275, 20255–20259. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, S.A.; Subramaniam, M.; Katagiri, T.; Janknecht, R.; Spelsberg, T.C. Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. J. Cell Biochem. 2002, 87, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Papadakis, K.A.; Krempski, J.; Reiter, J.; Svingen, P.; Xiong, Y.; Sarmento, O.F.; Huseby, A.; Johnson, A.J.; Lomberk, G.A.; Urrutia, R.A.; et al. Kruppel-like factor KLF10 regulates transforming growth factor receptor II expression and TGF-beta signaling in CD8+ T lymphocytes. Am. J. Physiol. Cell Physiol. 2015, 308, C362–C371. [Google Scholar] [CrossRef]
- Papadakis, K.A.; Krempski, J.; Svingen, P.; Xiong, Y.; Sarmento, O.F.; Lomberk, G.A.; Urrutia, R.A.; Faubion, W.A. Krüppel-like factor KLF10 deficiency predisposes to colitis through colonic macrophage dysregulation. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G900–G909. [Google Scholar] [CrossRef]
- Vollmers, C.; Gill, S.; DiTacchio, L.; Pulivarthy, S.R.; Le, H.D.; Panda, S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 21453–21458. [Google Scholar] [CrossRef]
- Guillaumond, F.; Gréchez-Cassiau, A.; Subramaniam, M.; Brangolo, S.; Peteri-Brünback, B.; Staels, B.; Fiévet, C.; Spelsberg, T.C.; Delaunay, F.; Teboul, M. Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol. Cell Biol. 2010, 30, 3059–3070. [Google Scholar] [CrossRef]
- Ruberto, A.A.; Gréchez-Cassiau, A.; Guérin, S.; Martin, L.; Revel, J.S.; Mehiri, M.; Subramaniam, M.; Delaunay, F.; Teboul, M. KLF10 integrates circadian timing and sugar signaling to coordinate hepatic metabolism. eLife 2021, 10, e65574. [Google Scholar] [CrossRef] [PubMed]
- Uyeda, K.; Repa, J.J. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006, 4, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Filhoulaud, G.; Guilmeau, S.; Dentin, R.; Girard, J.; Postic, C. Novel insights into ChREBP regulation and function. Trends Endocrinol. Metab. 2013, 24, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.S.; Kim, D.; Lee, Y.S.; Kim, H.J.; Han, J.Y.; Im, S.S.; Chong, H.K.; Kwon, J.K.; Cho, Y.H.; Kim, W.K.; et al. Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PLoS ONE 2011, 6, e22544. [Google Scholar] [CrossRef] [PubMed]
- Iizuka, K.; Takeda, J.; Horikawa, Y. Krüppel-like factor-10 is directly regulated by carbohydrate response element-binding protein in rat primary hepatocytes. Biochem. Biophys. Res. Commun. 2011, 412, 638–643. [Google Scholar] [CrossRef]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 251–262. [Google Scholar] [CrossRef]
- Wara, A.K.; Wang, S.; Wu, C.; Fang, F.; Haemmig, S.; Weber, B.N.; Aydogan, C.O.; Tesmenitsky, Y.; Aliakbarian, H.; Hawse, J.R.; et al. KLF10 Deficiency in CD4+ T Cells Triggers Obesity, Insulin Resistance, and Fatty Liver. Cell Rep. 2020, 33, 108550. [Google Scholar] [CrossRef]
- Hernandez-Gea, V.; Friedman, S.L. Pathogenesis of liver fibrosis. Annu. Rev. Pathol. 2011, 6, 425–456. [Google Scholar] [CrossRef]
- Loomba, R.; Sanyal, A.J. The global NAFLD epidemic. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 686–690. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, K.S.; Chang, H.Y.; Lee, W.K.; Lee, J.I. Progression of diet induced nonalcoholic steatohepatitis is accompanied by increased expression of Kruppel-like-factor 10 in mice. J. Transl. Med. 2014, 12, 186. [Google Scholar] [CrossRef]
- Woods, A.; Williams, J.R.; Muckett, P.J.; Mayer, F.V.; Liljevald, M.; Bohlooly, Y.M.; Carling, D. Liver-Specific Activation of AMPK Prevents Steatosis on a High-Fructose Diet. Cell Rep. 2017, 18, 3043–3051. [Google Scholar] [CrossRef]
- Raza, G.S.; Sodum, N.; Kaya, Y.; Herzig, K.H. Role of Circadian Transcription Factor Rev-Erb in Metabolism and Tissue Fibrosis. Int. J. Mol. Sci. 2022, 23, 12954. [Google Scholar] [CrossRef]
- Leclère, P.S.; Rousseau, D.; Patouraux, S.; Guérin, S.; Bonnafous, S.; Gréchez-Cassiau, A.; Ruberto, A.A.; Luci, C.; Subramaniam, M.; Tran, A.; et al. MCD diet-induced steatohepatitis generates a diurnal rhythm of associated biomarkers and worsens liver injury in Klf10 deficient mice. Sci. Rep. 2020, 10, 12139. [Google Scholar] [CrossRef]
- Hirota, T.; Kon, N.; Itagaki, T.; Hoshina, N.; Okano, T.; Fukada, Y. Transcriptional repressor TIEG1 regulates Bmal1 gene through GC box and controls circadian clockwork. Genes Cells 2010, 15, 111–121. [Google Scholar] [CrossRef]
- Rudic, R.D.; McNamara, P.; Curtis, A.M.; Boston, R.C.; Panda, S.; Hogenesch, J.B.; Fitzgerald, G.A. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004, 2, e377. [Google Scholar] [CrossRef]
- Zhang, D.; Tong, X.; Nelson, B.B.; Jin, E.; Sit, J.; Charney, N.; Yang, M.; Omary, M.B.; Yin, L. The hepatic BMAL1/AKT/lipogenesis axis protects against alcoholic liver disease in mice via promoting PPARalpha pathway. Hepatology 2018, 68, 883–896. [Google Scholar] [CrossRef]
- Xu, L.; Yang, T.Y.; Zhou, Y.W.; Wu, M.F.; Shen, J.; Cheng, J.L.; Liu, Q.X.; Cao, S.Y.; Wang, J.Q.; Zhang, L. Bmal1 inhibits phenotypic transformation of hepatic stellate cells in liver fibrosis via IDH1/alpha-KG-mediated glycolysis. Acta Pharmacol. Sin. 2022, 43, 316–329. [Google Scholar] [CrossRef]
- Rajamannan, N.M.; Subramaniam, M.; Abraham, T.P.; Vasile, V.C.; Ackerman, M.J.; Monroe, D.G.; Chew, T.L.; Spelsberg, T.C. TGFbeta inducible early gene-1 (TIEG1) and cardiac hypertrophy: Discovery and characterization of a novel signaling pathway. J. Cell Biochem. 2007, 100, 315–325. [Google Scholar] [CrossRef]
- Cen, M.; Hu, P.; Cai, Z.; Fang, T.; Zhang, J.; Lu, M. TIEG1 deficiency confers enhanced myocardial protection in the infarcted heart by mediating the Pten/Akt signalling pathway. Int. J. Mol. Med. 2017, 39, 569–578. [Google Scholar] [CrossRef][Green Version]
- Yang, J.; Zhang, H.; Wang, X.; Guo, J.; Wei, L.; Song, Y.; Luo, Y.; Zhao, Y.; Subramaniam, M.; Spelsberg, T.C.; et al. Kruppel-like factor 10 protects against acute viral myocarditis by negatively regulating cardiac MCP-1 expression. Cell Mol. Immunol. 2021, 18, 2236–2248. [Google Scholar] [CrossRef]
- Zhuang, R.; Chen, J.; Cheng, H.S.; Assa, C.; Jamaiyar, A.; Pandey, A.K.; Pérez-Cremades, D.; Zhang, B.; Tzani, A.; Khyrul Wara, A.; et al. Perivascular Fibrosis Is Mediated by a KLF10-IL-9 Signaling Axis in CD4+ T Cells. Circ. Res. 2022, 130, 1662–1681. [Google Scholar] [CrossRef]
- Wahab, N.A.; Weston, B.S.; Mason, R.M. Modulation of the TGFbeta/Smad signaling pathway in mesangial cells by CTGF/CCN2. Exp. Cell Res. 2005, 307, 305–314. [Google Scholar] [CrossRef]
- Chung, A.C.; Dong, Y.; Yang, W.; Zhong, X.; Li, R.; Lan, H.Y. Smad7 suppresses renal fibrosis via altering expression of TGF-beta/Smad3-regulated microRNAs. Mol. Ther. 2013, 21, 388–398. [Google Scholar] [CrossRef]
- Hsu, Y.C.; Ho, C.; Shih, Y.H.; Ni, W.C.; Li, Y.C.; Chang, H.C.; Lin, C.L. Knockout of KLF10 Ameliorated Diabetic Renal Fibrosis via Downregulation of DKK-1. Molecules 2022, 27, 2644. [Google Scholar] [CrossRef]
- Killick, R.; Ribe, E.M.; Al-Shawi, R.; Malik, B.; Hooper, C.; Fernandes, C.; Dobson, R.; Nolan, P.M.; Lourdusamy, A.; Furney, S.; et al. Clusterin regulates β-amyloid toxicity via Dickkopf-1-driven induction of the wnt-PCP-JNK pathway. Mol. Psychiatry 2014, 19, 88–98. [Google Scholar] [CrossRef]
- Mallipattu, S.K.; Estrada, C.C.; He, J.C. The critical role of Kruppel-like factors in kidney disease. Am. J. Physiol. Ren. Physiol. 2017, 312, F259–F265. [Google Scholar] [CrossRef]
- Lin, C.L.; Hsu, Y.C.; Huang, Y.T.; Shih, Y.H.; Wang, C.J.; Chiang, W.C.; Chang, P.J. A KDM6A-KLF10 reinforcing feedback mechanism aggravates diabetic podocyte dysfunction. EMBO Mol. Med. 2019, 11, e9828. [Google Scholar] [CrossRef]
- Kammoun, M.; Pouletaut, P.; Morandat, S.; Subramaniam, M.; Hawse, J.R.; Bensamoun, S.F. Kruppel-like factor 10 regulates the contractile properties of skeletal muscle fibers in mice. Muscle Nerve 2021, 64, 765–769. [Google Scholar] [CrossRef]
- Tran, S.; Ksajikian, A.; Overbey, J.; Li, P.; Li, Y. Pathophysiology of Pulmonary Fibrosis in the Context of COVID-19 and Implications for Treatment: A Narrative Review. Cells 2022, 11, 2489. [Google Scholar] [CrossRef]
- Mishra, V.K.; Subramaniam, M.; Kari, V.; Pitel, K.S.; Baumgart, S.J.; Naylor, R.M.; Nagarajan, S.; Wegwitz, F.; Ellenrieder, V.; Hawse, J.R.; et al. Kruppel-like Transcription Factor KLF10 Suppresses TGFbeta-Induced Epithelial-to-Mesenchymal Transition via a Negative Feedback Mechanism. Cancer Res. 2017, 77, 2387–2400. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.T.; Chang, H.W.; Wu, M.J.; Lai, Y.T.; Wu, W.C.; Yu, W.C.Y.; Chang, V.H.S. Klf10 deficiency in mice exacerbates pulmonary inflammation by increasing expression of the proinflammatory molecule NPRA. Int. J. Biochem. Cell Biol. 2016, 79, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Zhu, B.; Liang, Y.; Bi, L.; Hu, Z.; Chen, B.; Zhang, K.; Zhu, J. Asiaticoside suppresses collagen expression and TGF-beta/Smad signaling through inducing Smad7 and inhibiting TGF-betaRI and TGF-betaRII in keloid fibroblasts. Arch. Dermatol. Res. 2011, 303, 563–572. [Google Scholar] [CrossRef]
- Yu, H.; Bock, O.; Bayat, A.; Ferguson, M.W.; Mrowietz, U. Decreased expression of inhibitory SMAD6 and SMAD7 in keloid scarring. J. Plast. Reconstr. Aesthet. Surg. 2006, 59, 221–229. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yaseen, U.; Hwang, S.; Park, S.; Kim, S.-B.; Lee, H.-J.; Cha, J.-Y. New Insights into the Role of KLF10 in Tissue Fibrosis. Int. J. Mol. Sci. 2024, 25, 1276. https://doi.org/10.3390/ijms25021276
Yaseen U, Hwang S, Park S, Kim S-B, Lee H-J, Cha J-Y. New Insights into the Role of KLF10 in Tissue Fibrosis. International Journal of Molecular Sciences. 2024; 25(2):1276. https://doi.org/10.3390/ijms25021276
Chicago/Turabian StyleYaseen, Uzma, Soonjae Hwang, Sangbin Park, Soo-Bin Kim, Ho-Jae Lee, and Ji-Young Cha. 2024. "New Insights into the Role of KLF10 in Tissue Fibrosis" International Journal of Molecular Sciences 25, no. 2: 1276. https://doi.org/10.3390/ijms25021276
APA StyleYaseen, U., Hwang, S., Park, S., Kim, S.-B., Lee, H.-J., & Cha, J.-Y. (2024). New Insights into the Role of KLF10 in Tissue Fibrosis. International Journal of Molecular Sciences, 25(2), 1276. https://doi.org/10.3390/ijms25021276





