Abstract
Dramatic shifts in global climate have intensified abiotic and biotic stress faced by plants. Plant microRNAs (miRNAs)—20–24 nucleotide non-coding RNA molecules—form a key regulatory system of plant gene expression; playing crucial roles in plant growth; development; and defense against abiotic and biotic stress. Moreover, they participate in cross-kingdom communication. This communication encompasses interactions with other plants, microorganisms, and insect species, collectively exerting a profound influence on the agronomic traits of crops. This article comprehensively reviews the biosynthesis of plant miRNAs and explores their impact on plant growth, development, and stress resistance through endogenous, non-transboundary mechanisms. Furthermore, this review delves into the cross-kingdom regulatory effects of plant miRNAs on plants, microorganisms, and pests. It proceeds to specifically discuss the design and modification strategies for artificial miRNAs (amiRNAs), as well as the protection and transport of miRNAs by exosome-like nanovesicles (ELNVs), expanding the potential applications of plant miRNAs in crop breeding. Finally, the current limitations associated with harnessing plant miRNAs are addressed, and the utilization of synthetic biology is proposed to facilitate the heterologous expression and large-scale production of miRNAs. This novel approach suggests a plant-based solution to address future biosafety concerns in agriculture.
1. Introduction
MicroRNAs (miRNAs) are a class of small, non-coding RNAs widely found in animals and plants. They are crucial in regulating gene expression by targeting and binding mRNAs, resulting in their subsequent silencing or degradation. Accordingly, miRNAs participate in myriad critical processes, including cellular differentiation, proliferation, and survival [1]. Although miRNAs are typically only 22–24 nucleotides (nt), they are among the most abundant gene-regulating molecules, affecting various protein-encoding genes [2]. In 1993, the first miRNA, lin-4, was identified in the nematode Caenorhabditis elegans. Specifically, lin-4 regulates the timing of postembryonic developmental events in C. elegans by negatively regulating the expression of the nuclear protein lin-14 [3]. Seven years later, a second miRNA, let-7, was discovered in C. elegans [4]. Similar to lin-4, let-7 regulates nematode development by binding to the 3′-untranslated region (UTR) of specific target mRNAs, including lin-14, lin-28, lin-41, lin-42, and daf-12. Knocking out lin-4 or let-7 in C. elegans results in severe developmental abnormalities, suggesting they are crucial in organismal development [5]. These findings have sparked widespread interest in exploring the functions of other miRNAs. Notably, the most commonly used miRNA database, miRBase version 22.1 (accessed on 26 October 2023; https://www.mirbase.org), has registered 38,589 miRNAs from 271 organisms [6], with most existing as single copies, multiple copies, or clusters [7,8,9].
miRNAs are processed from transcripts with hairpin structures produced endogenously within cells [10]. Animal miRNAs are a class of evolutionarily conserved genes that are key regulators of cellular activity [11] and serve as biomarkers for detecting the state of in vivo microenvironments, including analysis of the tumor microenvironment [12,13,14,15,16]. However, plant and animal miRNAs have evolved independently and, therefore, differ in their corresponding sequences, precursors, and maturation processes [17,18]. The regulatory effects of plant miRNAs on endogenous genes have significant impacts on crop growth and development, playing important roles in resistance to abiotic stress, such as drought, cold, and nutrient deficiencies, as well as biotic stress, such as pathogens and harmful pests.
miRNA-dependent cross-species regulatory mechanisms of plant disease resistance have been reported. For example, Zhang et al. found that endogenous miRNAs of cotton, namely miR166 and miR159, translocate into pathogenic bacteria, regulating their pathogenicity and, ultimately, inhibiting Verticillium dahliae colonization of cotton, thus improving cotton resistance to bacterial infection [19]. Furthermore, Chen et al. found that miR162a in rice can negatively regulate brown planthopper reproduction and development via transboundary silencing, representing an important component of the rice defense system against brown planthopper infestation [20]. The cross-species regulatory mechanisms of plant miRNAs can provide novel strategies for crop genetic breeding and novel pesticide development.
This article comprehensively reviews miRNA biogenesis and regulatory mechanisms. It delves into how miRNAs respond to biotic and abiotic stress, employing endogenous and cross-kingdom regulatory mechanisms. Furthermore, it presents the most recent insights into the functional characteristics of miRNAs as potential regulators in the stress responses of plants. Finally, the prospects and challenges of miRNA applications are presented, suggesting potentially viable solutions.
2. Plant miRNA–Associated Biosynthesis and Plant Physiology
2.1. Biosynthesis of Plant miRNAs
miRNAs exert central roles in cell differentiation, proliferation, and survival by binding to complementary target mRNAs, resulting in their translational repression or degradation [1]. Plant miRNAs are produced through a multistep process encompassing the transcription, precursor processing, methylation, and assembly of miRNA-induced silencing complexes (miRISC) [21]. Based on sequence complementarity, miRNAs directly impact mRNA cleavage, translational repression, and DNA methylation [21]. Importantly, miRNA production is typically conserved across various species [1,22].
In animals, the processing of primary miRNA (pri-miRNA) occurs in the nucleus and cytoplasm via two distinct endonucleases (Drosha and Dicer, respectively) belonging to different subclasses of the RNase III family [23,24]. Pri-miRNAs produce 60–70 nt precursor miRNAs (pre-miRNA) after cleavage [25], which undergo cleavage mediated by the nuclear class II RNase III Drosha-DGCR8 complex and are delivered to the cytoplasm through exportin-5. In the cytoplasm, the pre-miRNA molecules undergo further cleavage by the class III RNase III enzyme Dicer, creating an ~22-nt miRNA duplex [1,26]. In contrast, both stages of pri-miRNA processing in plants are catalyzed within the nucleus by Dicer-LIKE 1 (DCL1), a Dicer homolog [21,27,28]. Plant miRNA genes are predominantly located in intergenic regions of the genome and function as independent transcriptional units. Although miRNA promoters are structurally similar to those of protein-coding genes [29], a small number of miRNAs located in the intronic sequences of protein-coding genes can be co-transcribed with their host genes [30,31,32]. Genes encoding miRNAs are transcribed in the nucleus under the action of RNA polymerase II, forming the pri-miRNA, approximately several hundred nucleotides in length (Figure 1) [33,34]. Subsequently, the Dicer-like enzyme, DCL1, produces the pre-miRNA and continues to process it, forming a double-stranded miRNA [21]. The final double-stranded miRNA undergoes methylation modification at the last nucleotide of the 3′ end on both strands [35], facilitated by miRNA methyltransferase HEN1 [22,36]. The double-stranded miRNA and its complementary strand (i.e., miRNA*) become integrated into the Argonaute protein in the RNA-induced silencing complex (RISC), enabling the complex to search for and bind to the target mRNA sequence, whereas miRNA* would normally degrade [26,37]. Mature miRNAs are subsequently transported to the cytoplasm by HASTY—an exportin5 homolog—or transported to the cytoplasm by HASTY before binding to ribosomal proteins [36]. RISC silences gene expression via one of three mechanisms: (i) interfering at the protein translation level; (ii) degrading mRNA at the transcription level; or (iii) heterochromatin formation or DNA elimination at the genome level [37].
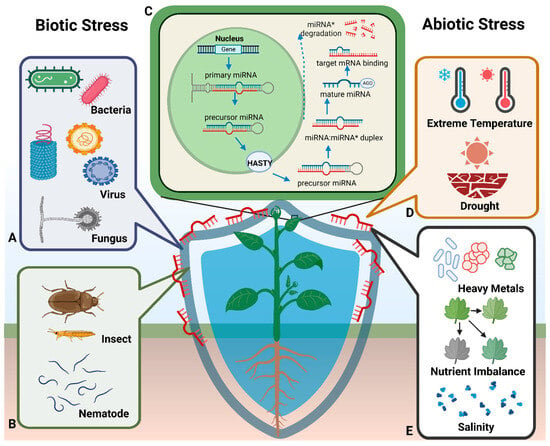
Figure 1.
miRNA synthesis in plants and associated plant responses to abiotic and biotic stress. (C) Summary of the miRNA synthesis process in plants: pri-miRNAs are translated in the nucleus, processed into pre-miRNAs by the DCL1 enzyme, and transported to the cytoplasm via HASTY, where mature miRNAs are generated. (A,B) Associated biotic stress (e.g., microorganisms and insects). (D,E) Associated abiotic stress (e.g., extreme temperature, drought, heavy metal stress, nutrient stress, and salt stress).
2.2. Role of Plant miRNAs in Plant Physiology
Plant miRNAs play important regulatory roles in various biological processes, including plant development, metabolism, responses to abiotic stress, and defense against pathogens. These miRNAs can effectively recalibrate internal plant signaling and metabolic pathways, regulate gene expression, and modulate gene responses [38]. Additionally, plant miRNAs can inhibit gene expression in pathogens, most notably regarding resistance traits, thus increasing the ability of plants to resist infection. Plant miRNAs can also control the adaptation of cellular biological responses to environmental stress and low nutrient supplies [39,40]. Moreover, as important environmental response regulators, miRNAs regulate gene expression and mediate plant growth and development under low-temperatures, heat stress, salt stress, and fungal stress [38].
As the study of miRNAs advances, an increasing body of evidence has demonstrated their roles in regulating plant cell differentiation, embryo formation, fertility, plant flowering, and fruiting, along with regulating plant type and seed yield. For example, Luo et al. reported that miR397 is highly expressed in undifferentiated rice embryonic healing tissues, leading to the downregulation of laccase genes; this causes the formation of thin-walled meristematic cells [41]. Alternatively, Hou et al. found that overexpressing miR171 in tomatoes results in larger tomato plants by suppressing the target gene family GRAS (SLGRAS24) [42]. Similarly, rice miR397 increases seed size and promotes spike branching by downregulating OsLAC expression, improving rice yield [43]. Moreover, Arabidopsis plants overexpressing miR397 can produce more inflorescence buds, increasing seed yield [44]. Meanwhile, miR528 participates in regulating the tillering number and regeneration ability of switchgrass through the miR528-SOD module, which enriches the regulatory network of miRNA-target genes [45].
3. Plant miRNAs Mediate Plant Resistance via Endogenous Regulation
Plant-derived miRNAs not only participate in regulating the host plant’s endogenous genes related to growth and development but also contribute to responses to abiotic (Table 1) and biotic (Table 2) stress by modulating the host’s endogenous target genes, thus impacting the agronomic traits of crop varieties.
3.1. miRNA Responses to Abiotic Stress
3.1.1. miRNAs and Heavy Metal Stress
Heavy metals, such as cadmium, aluminum, arsenic, lead, and mercury, are typically absorbed by plant root systems and can significantly inhibit plant growth [46]. Cadmium is a particularly persistent trace metal and a hazardous environmental pollutant [47] that is not essential for key plant metabolic or growth processes [48]. Therefore, Ali et al. constructed Arabidopsis thaliana miR397 mutant lines to investigate the molecular mechanisms responsible for the potential relationship between cadmium tolerance and miR397 expression [49]. They found that overexpressing miR397 alters lignin content and reduces cadmium tolerance by regulating laccase 2, 4, and 17 expressions. Similarly, knocking down miR535 improves cadmium tolerance in rice plants under cadmium stress, while overexpressing miR35 elicits the opposite effect compared to wild-type (WT) plants [50]. Additionally, Yan et al. found that growth inhibition, oxidative damage, and antioxidant enzyme disorders are more pronounced in tomato plants overexpressing miR398 under cadmium stress than in WT plants. Conversely, miR398 down-regulation exerts a protective effect against cadmium stress in tomato plants by regulating antioxidant enzyme activities alongside cadmium uptake and translocation [51].
3.1.2. miRNAs and Drought Stress
Drought is a major stressor that negatively impacts plant growth and development, leading to significant crop losses globally [52]. Notably, Jiang et al. reported that the loss of miR159 function in A. thaliana increases drought tolerance, attributed to fewer open stomata and lower water loss rates [53]. Meanwhile, Li et al. found that Arabidopsis plants overexpressing miR169a exhibit enhanced leaf water loss and greater sensitivity to drought stress than WT plants [54]. Additionally, reducing miR1119 expression in wheat roots increases MYC2 levels, upregulating the activity of certain enzymes, including catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD), effectively improving drought resistance [55].
3.1.3. miRNAs and Temperature Stress
Extreme temperatures can disrupt normal cellular and biochemical functions in plants, inhibiting their normal growth. High-temperature stress leads to oxidative damage through the generation of reactive oxygen species (ROS), which damage organelles by disrupting membranes via lipid peroxidation, ultimately reducing chlorophyll content and diminishing photosynthetic capacity [56,57]. Alternatively, low temperatures adversely affect plant growth and yield, significantly reducing growth efficiency [58]. Indeed, some plants can be irreversibly damaged by low-temperature stress [59]. To address these challenges, Matthews et al. found that miR156 can help alleviate high-temperature stress in alfalfa; miR156-overexpressing plants exhibit notably enhanced antioxidant scavenging capacity, reducing ROS-associated damage [60]. Moreover, using high-throughput screening, Zeng et al. reported that miR166e, miR319, and Bra-novel-miR3936-5p may have important roles in plant responses to cold stress [61]. This was further demonstrated by Shi et al., who constructed sha-miR319d-overexpressing tomato plants with enhanced cold and heat stress tolerance [62].
3.1.4. miRNAs and Nutrient Stress
A disparity exists between the nutritional requirements of plants in agricultural settings and those in their natural environments; nonetheless, irrespective of the environment, deficiencies in essential elements can induce nutritional stress, which can negatively affect normal plant growth and development [63]. miR156 expression becomes upregulated in response to nitrogen deficiency, promoting plant height development by regulating SQUAMOSA promoter-binding protein-like (SPL) genes [64] and primary and lateral root growth by regulating the expression of NAC4, ARF2, and AFB3 [65]. Alternatively, Xu et al. analyzed the transcriptome of soybean roots and leaves under phosphorus sufficiency and deficiency. They established that miR169, miR395, miR397, and miR399 are regulated to alleviate phosphorus deficiency-induced stress [66]. Moreover, under potassium deficiency, miR171 and miR166 expression levels become markedly altered in wheat; wheat seedlings synthesize more miRNAs, resulting in increased potassium absorption during the pre-growth phase to promote root differentiation and development [67].
3.1.5. miRNAs and Salt Stress
Extended exposure to saline conditions can lead to ROS production [68], which can cause oxidative damage to cellular components, including proteins, lipids, and DNA, disrupting critical plant biochemical processes [69,70]. The resultant tissue damage can lead to a reversible slowing of metabolism and growth and, in more severe cases, irreversible cell death [71]. Under salt stress, the expression levels of nine representative miRNAs, including miR159, miR168, miR169, miR172, miR393, miR395, miR396, miR399, and miR408, vary in different plant species [72]. Interestingly, the same miRNAs may have opposite expression patterns in different plant species and fulfill differing roles under salt stress. For example, in tobacco, miR396 overexpression can improve water retention and proline content, helping reduce ROS production and improve salt resistance [73]. In contrast, overexpression of miR396 reduces salinity stress tolerance in rice and A. thaliana [74].


Table 1.
Summary of endogenous miRNA regulation in plants under abiotic stress.
Table 1.
Summary of endogenous miRNA regulation in plants under abiotic stress.
| Plant | miRNA Regulation | Function | Target Gene(s) | Reference(s) |
|---|---|---|---|---|
| Arabidopsis thaliana | miR397 | Cadmium tolerance | LAC2/4/17 | [49] |
| Rice | mir535 | SPL7 | [50] | |
| Tomato | miR398 | IRT1, IRT2NRAMP2, HMA3 | [51] | |
| Barley | miR156 | HvNAT2 | [75] | |
| Arabidopsis thaliana | miR159 | Drought | ABI5 | [53] |
| Arabidopsis thaliana | miR169a | NFYA5 | [54] | |
| Wheat | miR1119 | MYC2 | [55] | |
| Arabidopsis and rice | miR393 | AsAFB2, AsTIR1 | [76] | |
| Alfalfa | miR156 | Temperature stress | SPL | [60] |
| Rapeseed | miR166e, miR319, miR3936-5p | – | [61] | |
| Tomato | sha-miR319d | MYB83, CBF1, HSFA1A, HSFA1B, HSP90, ZAT12, ZAT10 | [62] | |
| Tea plants | miR156 | Nutritional stress | SPL, DFR | [64] |
| Peanut | miR156 | NAC4, ARF2, AFB3 | [65] | |
| Soybean | miR169, miR395, miR397, miR399 | – | [66] | |
| Wheat | miR171, miR166 | ARF | [67] | |
| Tomato | miR167a | Salt stress | DREB2A | [77] |
| Tobacco | miR396a | NtGRF1, NtGRF3, NtGRF7, NtGRF8 | [73] | |
| Rice | miR396c | ABRE | [74] |
3.2. miRNA Responses to Biotic Stress
In natural environments, plants are frequently exposed to biotic stress from various sources, including bacteria, fungi, viruses, and pests. Extensive research has shown that miRNAs are effective regulators of plant resistance to pathogen infections [40,78]. When a host plant is attacked by pathogens, miRNA expression responds rapidly, altering the expression of its downstream target genes; ultimately, this regulates the plant’s resistance to pathogenic infection by affecting processes such as hormone transduction, disease resistance gene expression, and secondary metabolism (Table 2).
3.2.1. miRNAs Mediate Pathogen Resistance in Plants through Hormonal Signaling Pathways
Zhao et al. observed that tomato miR319-TCP4 plays a key role in influencing plant resistance to root-knot nematodes by regulating the expression of genes related to jasmonic acid (JA) synthesis and altering the levels of endogenous JA in leaves [79]. Moreover, Pinweha et al. found that, when cassava is infested with Colletotrichum gloeosporioides f. sp., the expression of miR393 and miR160 promotes a defense response against anthracnose by down-regulating the target genes TIR1 and ARF, thus suppressing growth hormone signaling [80]. Similarly, when Arabidopsis is exposed to Pseudomonas syringae flagellin, miR393 expression is induced, leading to the negative regulation of the F-box growth hormone receptor TIR1. This inhibition suppresses growth hormone signaling, promoting antimicrobial resistance in A. thaliana [81]. Furthermore, infestation of Malus hupehensis with Botryosphaeria dothidea promotes the upregulation of host miR168, targeting MhAGO1 inhibition and facilitating salicylic acid (SA)-mediated defense responses. These defense responses can delay the symptom progression of pathogen-inoculated leaves and positively regulate resistance to pathogenic bacteria [82]. Additionally, Zhang et al. reported the involvement of miR394 in the negative regulation of biotic stress. Notably, miR394 overexpression in tomato plants inhibits the expression of its target gene, LCR; this, in turn, inhibits the expression of JA synthesis–related genes, reducing resistance to disease-causing mildew [83]. Finally, Yang et al. reported that, compared to WT poplar trees, miR159a overexpression in transgenic poplars can enhance disease resistance following inoculation with the necrotrophic fungus C. chrysosperma. These findings demonstrate that JA/ethylene (Et)-inducible signaling is involved in the dynamic response to biotic stress in WT and miR159a-overexpressing lines [84].
Citrus can resist Candidatus Liberibacter asiaticus (CLas) by activating the miR2977–SAMT cascade reaction, which further utilizes methyl salicylate (MeSA) to regulate phytosome defense [85]. Interestingly, CLas hijacks key host processes by manipulating critical miRNAs in citrus, while host miRNAs also coordinate the regulation of defense-related genes. Liu et al. performed differential gene analysis of resistant Cladosporium fulvum plants and identified 54 miRNAs that may regulate hormone signaling [86]. The downregulation of miR9472 potentially upregulates the expression of SAR deficient 1 (SARD1), a key regulator of ICS1 (isochorismate synthase 1) induction and SA synthesis, to improve the level of SA in resistant plants. These findings provide a more comprehensive gene circuit and valuable targets for modulating resistance to pathogens. Additionally, silencing miR530 enhances plant defense against Verticillium dahliae, whereas its overexpression weakens plant resistance [87]. This is related to the function of miR530, which degrades GhSAP6 mRNA. Knockdown of GhSAP6 also reduces plant resistance due to the down-regulation of SA-related gene expression, including GhNPR1 and GhPR1.
3.2.2. miRNAs Mediate Pathogen Resistance in Plants by Regulating Plant Disease Resistance Proteins
The miRNA response to plant disease resistance primarily functions as a regulatory mechanism that targets plant disease resistance proteins, such as NB-LRR, chitin, and lectin. Plant NB-LRR proteins can initiate plant defense responses following microbial infection. In tomato plants, negative regulation of NB-LRR proteins by miR482 and miR5300 in vivo reduces resistance to Fusarium oxysporum f. sp. lycopersici infection [88]. In A. thaliana, the in vivo downregulation of miR393 and upregulation of its target genes, lectin receptor-like kinases (LecRLKs), occurs after treatment with bacterial lipopolysaccharide (LPS); ultimately, this response enhances the basal resistance of A. thaliana to LPS [89]. Zhang et al. reported that, in apple leaves inoculated with leaf spot fungus, MdmiR395 expression increases while that of its target genes, MdMRKY26 and MdWRKYN1, decreases; moreover, overexpression of MdWRKY or inhibition of MdmiR395 increases the expression of pathogenesis-related genes, enhancing the apple tree’s resistance [90]. Furthermore, using small RNA sequencing, Li et al. identified 58 miRNAs associated with potato virus A (PVA) infection [91]. miR482d-3p, miR397-5p, and other miRNAs become down-regulated, increasing PR gene expression. This study provides transcriptome-wide insights into the molecular basis of PVA resistance in potato leaves.
Leucine-rich repeat proteins (NLRs) are critical to the plant–pathogen defense response, while 22 nt miRNAs are essential for NLR regulation, improving the response rate of plants to most pathogens [92]. NLRs are the largest class of disease-resistance (R) proteins in plants [93]. miR2118 in rice negatively regulates Xanthomonas oryzae pv. oryzae resistance via negative regulation of three nucleotide-binding NLR genes (LOC_Os08g42700.1, LOC_Os01g05600.1, and LOC_Os12g37290.1) [94].
3.2.3. miRNAs Regulate Reactive Oxygen Species Accumulation and Plant Secondary Metabolism Responses to Abiotic Stress
When rice plants are subjected to viral damage, miR528 expression is suppressed, promoting the expression of its target gene, L-ascorbate oxidase (AO), and, subsequently, reducing AO-mediated ROS accumulation [95]. Moreover, the miR528-AO defense module is regulated by SPL9, which binds to specific motifs within the miR528 promoter region, activating miR528 gene expression [96]. Similarly, reducing miR398b expression in tomato plants reduces ROS accumulation and up-regulates MeJA-responsive defense genes, increasing plant resistance to Botrytis cinerea [97]. Downregulation of miR397 expression during the plant response to pathogenic microbial infestation enhances laccase activity, induces the synthesis of lignin and other phenolic polymers, and causes rapid deposition in the cell wall; ultimately, this limits the growth and invasion of pathogenic microorganisms [98,99]. For example, infection of potatoes with PVA and maize with sugarcane mosaic virus (SCMV) is inhibited by the downregulation of miR397 expression, enhancing laccase activity and increasing lignin formation [91,100].
Alternatively, Chandran et al. demonstrated that transgenic rice lines overexpressing miR396 are highly susceptible to Aspergillus oryzae infection, whereas those overexpressing its target genes (OsGRF6, OsGRF7, OsGRF8, and OsGRF9) exhibit enhanced A. oryzae resistance [101]. Furthermore, miR396 and GRF8-OE plants exhibit increased flavonoid content, which is associated with enhanced pathogen resistance, revealing a novel pathogen resistance mechanism mediated by the miR396–GRF8–F3H–flavonoid pathway [102]. In contrast, overexpression of miR396 in alfalfa enhances resistance to Spodoptera litura larvae; this may be attributed to the increased lignin content and enhanced biosynthesis of low-molecular-weight flavonoids and glucosinolates [103].


Table 2.
Summary of endogenous miRNA regulation in plants under biotic stress.
Table 2.
Summary of endogenous miRNA regulation in plants under biotic stress.
| Plant | miRNA Regulation | Pathogen | Pathogen Resistance Function | Target Gene | Reference(s) | |
|---|---|---|---|---|---|---|
| Cassava | miR393, miR160 | Colletotrichum gloeosporioides f. sp. | Hormonal signaling | IAA | TIR1 ARF | [80] |
| Arabidopsis thaliana | miR393 | Pseudomonas syringae | TIR1 | [81] | ||
| Malus hupehensis | miR168 | Botrytis cinerea | SA | MhAGO1 | [82] | |
| Tomato | miR319 | Root-knot nematode | JA | TCP4 | [79] | |
| Tomato | miR394 | Disease-causing mildew | LCR | [83] | ||
| Poplar tree | miR159a | Necrotrophic fungus C. chrysosperma | JA\ET | ERF, PR3, MPK3, MPK6 | [84] | |
| Tomato | miR482, miR5300 | Fusarium oxysporum f. sp. lycopersici | Plant disease resistance proteins | NB-LRR | [88] | |
| Arabidopsis thaliana | miR393 | Bacterial lipopolysaccharide | LecRLKs | [89] | ||
| Apple | miR395 | Leaf spot fungus | PR | [90] | ||
| Rice | miR528 | Rice stripe virus | Reactive oxygen and plant secondary metabolism | AO, SPL9 | [95,96] | |
| Malus hupehensis | miR397b | Botryosphaeria dothidea | Laccase | [98,99] | ||
| Potato | miR482d, miR397 | Potato virus A | [91,100] | |||
| Corn | miR168, miR528, miR159, miR397, miR827 | Sugarcane mosaic virus | ||||
| Rice | miR396 | Aspergillus oryzae | Flavonoids | [101,102] | ||
| Alfala | miR396 | Spodoptera litura larvae | Lignin, Flavonoids, Glucosinolates | [103] | ||
4. Plant miRNAs Mediate Plant Resistance via Cross-Kingdom Regulation
In addition to their role in regulating the expression of endogenous genes, miRNAs also function as intercellular signaling molecules, facilitating cross-species regulation between plants and other plants, microorganisms, and animals. In 2012, Zhang et al. determined that miR168a—a plant-derived miRNA—can enter the circulatory system of animals through food intake. In mice, this miRNA targets and inhibits the expression of low-density lipoprotein receptor adapter protein 1 (LDLRAP1), resulting in the cross-kingdom regulation of lipid metabolism [104]. Ultimately, this study provided a foundation for research into miRNA-mediated cross-species regulation by plants.
A study by Chen et al. found that plant miRNAs in bee diets have an inhibitory effect on the ovary and overall growth and development of worker honeybee larvae. In contrast, royal jelly does not contain plant miRNAs; therefore, young bees that consume royal jelly develop into queen bees [105]. This finding challenges the previous understanding that royal jelly contains specific beneficial substances that allow young bees to develop into queens. Moreover, plant miRNAs can fine-tune bee caste development, providing insights into the interactions and co-evolution across different biological kingdoms.
In a study by Liu et al., next-generation sequencing identified 30 plant miRNAs in a healthy Chinese population. These sequences did not align with any established sequences within the human genome; in contrast, they aligned identically with plant miRNAs commonly found in food [106].
Hence, growing evidence indicates that plant miRNAs exhibit cross-species and cross-kingdom regulation (Table 3), serving as mediators of gene silencing and as a link between animal, plant, and microbial communities.
4.1. miRNA-Mediated Regulation of Plant–Plant Gene Expression
miRNAs can play related roles throughout the whole plant [107,108]. For example, miR399 and miR156 can be transported throughout the plant, serving as signal molecules [109,110,111]. In plants overexpressing miR399 and miR156 and exposed to exogenous A. thaliana, the expression of PHO2 and SPLs is decreased, respectively. When A. thaliana plants overexpressing miR399 and WT are cultured in the same liquid medium, the expression of PHO2 in WT plants decreases, while plants overexpressing miR156 inhibit the SPL expression of nearby WT plants [112].
Cuscuta is a genus of parasitic plants with specialized physiology. Cuscuta spp. employs climbing stems to attach to other plants, forming specialized structures called haustoria at the contact sites. These haustoria can be used to extract water and nutrients from the host. As shown in Figure 2, Shahid et al. reported that, during the parasitization of A. thaliana by Cuscuta, corresponding Cuscuta miRNAs (22 nt long) target specific A. thaliana mRNAs; this increases cleavage of the mRNAs, heightens production of secondary siRNAs, and reduces mRNA content [113]. Indeed, mutations in two loci encoding the target mRNAs in A. thaliana can significantly accelerate the growth rate of Cuscuta after parasitization, suggesting that increased miRNA-mediated targeting of host mRNAs can enhance Cuscuta parasitism. Moreover, Hudzik et al. identified several miRNAs in Cuscuta that target and regulate specific host mRNAs in a cross-species manner. The targeted host mRNAs in A. thaliana include those involved in biodefense, hormone signaling, and vascular development [114].
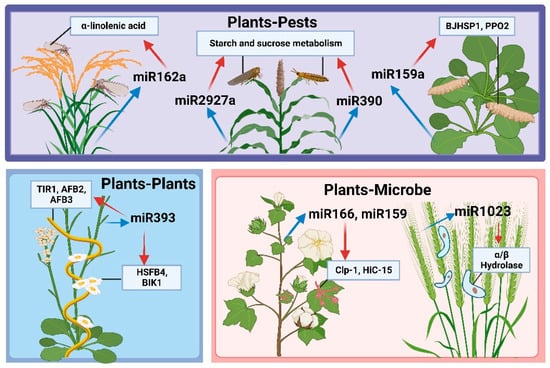
Figure 2.
Cross-kingdom miRNA regulation.
Finally, Betti et al. demonstrated that plants can secrete miRNAs. Notably, plants with a high miRNA secretion capacity influence neighboring plants with lower miRNA secretion by inducing post-transcriptional gene silencing. Ultimately, this confirms that miRNAs can function as signaling molecules, enabling plant-to-plant communication [112].
4.2. miRNA-Mediated Regulation of Plant–Microbe Gene Expression
miRNA-mediated cross-species genetic regulation is important in plant–microbe interactions. For example, Zhang et al. found that V. dahliae infection induces the expression of endogenous miRNAs miR166 and miR159 in cotton, which translocates to the pathogen. Ultimately, in V. dahlia, miR166, and miR159 inhibit the formation of micronuclei and mycelial growth via specific genetic cleavage of the pathogen virulence factors Ca2+-dependent cysteine protease (Clp-1) and isotrichodermin C-15 hydroxylase (HiC-15), respectively. This further inhibits V. dahliae colonization in cotton, enhancing cotton resistance [19].
Additionally, Meng et al. determined that miR1001 from tomatoes reduces the virulence of Botrytis cinerea in infected tomato plants by targeting ATP-dependent metallopeptidases and cysteine-type endopeptidases in B. cinerea [115]. Moreover, wheat miR1023 silences the Fusarium graminearum α/β hydrolase gene, hindering F. graminearum invasion [116]. Luo et al. identified 133 miRNAs in potatoes that are predicted targets in the genome of Phytophthora infestans. Upon further validation, miR0003, miR394, etc. can target multiple conserved P. infestans genes, providing a new approach for the control of late blight [117]. Notably, cross-kingdom regulation between plants and microbes is not limited to plants exporting specific miRNAs to silence disease-causing genes in pathogens. Pathogenic microbes can also export specific miRNAs into plants, resulting in the silencing of disease-resistant genes and accelerating infection [118,119].
4.3. miRNA-Mediated Regulation of Plant–Pest Gene Expression
Recently, plant miRNAs have been shown to protect against insect damage by regulating plant growth and development. In addition, some plant miRNAs enter the body of feeding insects and regulate insect growth and development, exerting a significant effect on natural plant–pest interactions [120]. For example, Chen et al. found that brown planthopper damage to rice induces the expression of rice miR162a. This initiates a defense response against infestation by down-regulating brown planthopper reproduction and development via transboundary silencing and inhibiting the α-linolenic acid metabolic pathways in rice, reducing brown planthopper host selectivity. Indeed, the JA-mediated defense response has created long-term evolutionary competition between the pest and host [20]. Alternatively, Zhang et al. identified plant-derived miRNAs in the hemolymph of the cruciferous cabbage moth, Cechetra minor. They reported that these miRNAs can enter the circulatory system by penetrating the midgut barrier; notably, these plant-derived miRNAs may play regulatory roles in C. minor [121]. Similarly, 13 sorghum and 3 barley miRNAs were detected in two cereal aphids, Schizaphis graminum and Sipha flava; the associated target genes were predicted to participate in aphid defense by inhibiting starch and sucrose metabolism [122]. Similar to the involvement of miRNAs in plant–microbe cross-species regulation, plant-derived miRNAs can regulate insect target genes; insect-derived miRNAs can enter the plant via the saliva, contributing to interspecies communication between the plant and insect [120,123].


Table 3.
Summary of plant cross-kingdom miRNA regulation under biological stress.
Table 3.
Summary of plant cross-kingdom miRNA regulation under biological stress.
| Plants | miRNA-Regulation | Transboundary Species | Targets | Reference(s) | |
|---|---|---|---|---|---|
| Arabidopsis thaliana | miR393 | Plants | Cuscuta campestris | Maybe TIR1, AFB2, AFB3, HSFB4, BIK1 | [113,114] |
| Terrestrial cotton | miR166, miR159 | Microbe | V. dahliae | Clp-1, HiC-15 | [19] |
| Tomato | miR1001 | Botrytis cinerea | ATP-dependent metallopeptidases and cysteine-type endopeptidases | [115] | |
| Wheat | miR1023 | Fusarium graminearum | FGSG_03101 | [116] | |
| Rice | miR162a | Pests | Brown planthopper | α-linolenic acid metabolic pathways | [20] |
| Arabidopsis thaliana | miR159a, novel-7703-5p | Moth Cechetra minor | BJHSP1, PPO2 | [121] | |
| Sorghum and Switchgrass | miR2927a | Schizaphis graminum | Starch and sucrose metabolism | [122] | |
| miR390 | Sipha flava | ||||
5. Potential Applications and Strategies for Cross-Kingdom Plant miRNAs
There is a growing body of evidence suggesting that plant miRNAs have the ability to migrate from plants to other plant species, microbes, insects, and even mammalian cells, allowing them to regulate specific genetic processes. This property allows these small molecules to serve as alternative biopesticides in modern agriculture. As previously discussed, many of the identified plant miRNAs exhibit cross-kingdom regulatory functions. Comprehensive investigations into the mechanisms behind cross-species miRNA regulation may provide important references for the design, modification, optimization, and utilization of miRNAs.
5.1. Artificial miRNAs Increasing the Value of Plants
Artificial miRNA (amiRNA) technology, designed according to the principles of natural miRNA generation and action, employs a small RNA molecule that targets one or more specific genes to efficiently and specifically inhibit their expression [124]. RNA interference (RNAi) is a powerful tool for studying gene function, with siRNA and short hairpin RNAs (shRNAs) the preferred choice for the transient knockdown of gene expression. In contrast, amiRNAs are used less frequently, possibly due to their more complex design and less predictable processing, potentially resulting in less efficient silencing [125]. Nonetheless, amiRNAs designed against specific targets are just as effective as shRNAs, providing not only long-term silencing but also a higher degree of safety and fewer non-specific outcomes [126,127,128]. These features make amiRNAs promising tools for genetic therapy. Artificial miRNA can significantly enhance the application value of plants, showing excellent performance in pest and microbial resistance and even playing an important role in the field of new human medicine (e.g., Table 4).
5.1.1. Artificial miRNAs in Insect Resistance
amiRNA technology holds significant potential for enhancing plant disease resistance; its application in the context of insect resistance, in particular, has been widely studied. The striped stem borer (SSB) represents a significant threat to rice (Oryza sativa) [129]. Nonetheless, the amiRNA csu-novel-260 inhibits the synthesis of ecdysteroids, one of the most important hormones regulating insect development [130]. Therefore, Hao et al. employed amiRNA technology to overexpress csu-novel-260 in rice, obtaining transgenic rice with significant SSB resistance [129]. Specifically, when SSB larvae consume csu-novel-260-containing transgenic rice, the suppression of ecdysteroid synthesis is induced, resulting in a high SSB mortality rate. In another study, Wen et al. overexpressed 13 small endogenous RNAs of SSB in rice using amiRNA technology; corresponding feeding assays revealed significantly inhibited SSB growth [131]. Specifically, pupation was delayed for 4 days when SSB larvae were continuously fed transgenic rice expressing the novel SSB miRNA candidate csu-novel-miR15 (csu-15). Gene expression analysis revealed a significant alteration (up- or down-regulation) in the expression levels of at least six individual SSB genes after oral consumption of csu-15 rice. Moreover, Liu et al. successfully engineered SSB-resistant rice (csu-53) expressing an artificial SSB endogenous miRNA (csu-novel-miR53) [132]. Subsequent feeding experiments demonstrated that consumption of csu-53 rice inhibits SSB larval growth, delays pupation time, and decreases pupal weight and emergence rate. In a 10-day feeding experiment, the miRNA mimic csu-novel-miR53 not only inhibited larval growth but also increased larval mortality.
Rice stem borers (RSB) cause massive annual economic losses. He et al. identified an insect-specific miRNA, miR-14, in RSB as a potential regulatory gene associated with molting [133]. Notably, csu-miR-14 is highly expressed at the end of each larval instar. Conversely, overexpression of csu-miR-14 on the third day of the fifth RSB instar results in high mortality and developmental abnormalities. Therefore, He et al. constructed transgenic rice with high miR-14 expression and observed high RSB resistance in the transgenic line.
The cotton bollworm Helicoverpa armigera is a major pest affecting various crops. While the expression of Bacillus thuringiensis (Bt) toxins in transgenic crops has successfully inhibited pests such as the cotton bollworm, field-evolved resistance has emerged in a wide range of pests. Therefore, new protective strategies must be sought to effectively suppress Bt-resistant insect populations. Pre-miRNA transcripts of insects can be modified into amiRNAs that target insect genes. Bally et al. expressed these modified genes in Nicotiana benthamian. The cotton bollworms that consumed transgenic Nicotiana benthamian leaves exhibit increased mortality, developmental abnormalities, and delayed growth [134]. Hence, expressing insect pre-amiRNAs in plants might represent a novel strategy for protecting plants from herbivorous insects. The ecdysone receptor (HaEcR) gene in the cotton bollworm is pivotal in regulating all developmental stages of the insect life cycle; nonetheless, this gene can be silenced by a sequence-specific amiRNA (amiRNA-HaEcR). Therefore, Yogindran et al. constructed transgenic tomatoes expressing amiRNA-319a-HaEcR; continuous feeding of the transgenic leaves to cotton bollworms reduced their overall growth rate and survival [135]. Alternatively, Agrawal et al. constructed a vector-producing amiR-24, specifically targeting the chitinase gene of the cotton bollworm [136]. When bollworm larvae consumed the leaves of transgenic tobacco plants with high amiR-24 expression, they exhibited molting anomalies and eventually died. Furthermore, Faisal et al. employed miRNA-mediated RNAi technology to develop transgenic tomato plants with resistance to aphids by silencing the peach aphid acetylcholinesterase 1 (Ace1) gene [137]. Quantitative PCR data clearly demonstrated that Ace1 was significantly downregulated in aphids raised in clamp cages on T-1 transgenic plants. Additionally, the aphid populations reared on the T-1 transgenic plants of both tomato plant cultivars exhibited a significant reduction.
5.1.2. Artificial miRNAs in Microbe Resistance
amiRNAs are widely used to investigate the resistance of plants to pathogenic microorganisms. Niu et al. constructed two amiRNA vectors by replacing the conserved miR159a backbone fragments from A. thaliana with the HC-Pro gene from the turnip mosaic virus (TuMV) and the P69 gene from the turnip yellow mosaic virus (TYMV). The results show that these two amiRNAs specifically silence target gene expression and are stably inherited in subsequent generations [138]. Additionally, genetic modification of tomato plants with an amiRNA vector targeting the silencing repressor 2b gene of the cucumber mosaic virus (CMV) induces effective resistance against CMV infection [139,140]. Moreover, genetic transformation of the precursor amiRNA backbones derived from different viral genes into the same plant can induce simultaneous resistance to multiple viruses. For example, Ai et al. introduced amiRNAs targeting the silencing suppressors HC-Pro (potato virus Y, PVY) and p25 (potato virus X) into Nicotiana tabacum. The transgenic tobacco exhibits markedly improved resistance to both diseases [141]. Similarly, Song et al. designed amiRNAs based on the PVY and tobacco etch virus (TEV) Nib and CP genes; the transgenic tobacco transfected with these amiRNAs exhibits resistance to both viruses [142].
Al-Roshdi et al. constructed amiRNAs using the miR159a hairpin structure to produce tomato plants resistant to the whitefly-transmitted tomato yellow leaf curl virus-Oman (TYLCV-OM). The transgenic tomato plants exhibit upregulated amiRNA expression, which effectively downregulates or silences the AC1/Rep transcript of TYLCV-OM, encoding enhanced tolerance against TYLCV infection [143]. Under TYLCV-OM infection conditions, viral replication is dramatically reduced in T1 transgenic tomato plants. Alternatively, Khalid et al. utilized amiRNA technology to express 14 amiRNAs targeting the conserved regions of seven TYLCLV genes and their satellite DNAs, incorporating intronic and exonic amiRNAs [144]. The resulting pAMIN14 and pAMIE14 vectors encode extensive amiRNA clusters, the silencing of which was validated by transient assays and stable transgenic N. tabacum plants. To assess the resistance of pAMIE14 and pAMIN14 transgenic plants to TYLCLV, the corresponding plasmids were transformed into the tomato cultivar A57; viral resistance was evaluated following mixed infection with TYLCLV. Overall, the pAMIN14 transgenic line exhibits a greater level of TYLCLV resistance than the pAMIE14 transgenic line, with resistance comparable to that of plants carrying the TY1 resistance gene. Additionally, zucchini resistant to the yellow mosaic virus (ZYMV) can be created via artificial miRNA-mediated methods [145]. These amiRNAs are derived from the gene encoding HC-Pro—a ZYMV protein involved in aphid transmission and cell-to-cell movement thought to be an RNAi repressor [146]. Hence, amiRNAs have considerable potential as crop antimicrobials.
Practical validation is required for amiRNA applications. Indeed, the transfer of amiRNA targeting potato spindle tuber viroids (PSTVd) into tomato plants causes developmental retardation and reproductive defects in the plants [147]. Furthermore, amiRNAs inadvertently silenced sterol glycosyltransferase 1 (SISGT1) in tomatoes, leading to impairment of the growth hormone signaling pathway and impeding the growth of amiR-PSTVd plants [148].
5.1.3. Artificial miRNAs in Human-Related Disease Resistance
Plant-derived amiRNAs based on plant gene expression have been investigated for the treatment of mammalian diseases. For instance, Kakeshpour et al. constructed plant expression vectors containing amiRNAs targeting mouse complement 3 (C3) and coagulation factor 7 (CF7) mRNAs using the rice miRNA backbone Osa-miR528 [149]. The corresponding transgenic lettuce successfully expresses primary and mature amiRNAs. Subsequently, Zhang et al. produced lettuce (Lactuca sativa L.) expressing small silencing amiRNAs that specifically inhibit the expression of the hepatitis B virus HBsAg gene at relatively low levels compared to synthetic siRNAs, effectively alleviating liver injury in p21-HBsAg transgenic mice [150]. Continuous administration of these amiRNAs to mice maintains relatively stable amiRNA levels in vivo, markedly reducing pathological damage. While overexpression via viral transduction and exogenous substances is relatively impossible without interfering with the cell’s physiological condition, amiRNAs offer a more “natural” gene therapy tool regarding their structure, biogenesis, and expression levels. This enhances the safety of plant-derived siRNAs as RNAi therapeutics.
However, it is important to note that amiRNA technology may encounter certain challenges in agricultural applications, especially in the molecular breeding of antiviral transgenes. Moreover, all published case studies include transgenic model plants; thus, field application is not immediately available. In addition, viral genomes evolve faster than plant miRNAs, and viruses can alter the conservation of their target regions (e.g., through mutation), potentially resulting in off-target effects of the amiRNAs.


Table 4.
Artificial miRNAs increase the value of plants.
Table 4.
Artificial miRNAs increase the value of plants.
| Plant | amiRNA | Target Species | Effect | Reference(s) | |
|---|---|---|---|---|---|
| Rice | csu-novel-260 | Pests | Striped stem borer | Inhibits ecdysteroid synthesis | [129,130] |
| csu-novel-miR15 | Inhibits SSB growth | [131] | |||
| csu-novel-miR53 | Inhibits larval growth, delaying pupation time | [132] | |||
| csu-miR-14 | Rice stem borers | High mortality | [133] | ||
| Tobacco | insect pre-amiRNAs | Cotton bollworm | Increases mortality and developmental abnormalities | [134] | |
| amiRNA-319a-HaEcR | Reduces overall growth rate and survival | [135] | |||
| amiR-24 | Molting anomalies and death | [136] | |||
| A. thaliana | amiR-HC-Pro, amiR-P69 | Microbe | Turnip mosaic virus | Silence target gene expression | [138] |
| Tomato | amiR-CMV-2b | Cucumber mosaic virus | Targets CMV infection | [139,140] | |
| Tobacco | amiR-PVY-HC-Pro | Potato virus Y | Improves resistance | [141] | |
| amiR-PVX-p25 | Potato virus X | ||||
| amiR-TEV-CP | Tobacco etch virus | [142] | |||
| Tomato | amiR159a-TYLCV-OM | Tomato yellow leaf curl virus-Oman | Enhances tolerance in plants against TYLCV infection | [143] | |
| Zucchini | amiR-HC-Pro | Yellow mosaic virus | ZYMV resistance | [145] | |
| Tomato | amiR-PSTVd | Potato spindle tuber viroid | PSTVd resistance | [147] | |
| Rice | Osa-miR528-C3, Osa-miR528-CF7 | – | Mouse | Express primary and mature amiRNAs | [149] |
| Lettuce | lettuce-derived amiRNAs | Hepatitis B virus | Reduce pathological damage | [150] | |
5.2. Plant Extracellular Vesicles Can Facilitate the Cross-Species Transport of miRNAs
Extracellular vesicles (EVs) are a general term for vesicles with a membrane structure secreted by cells and represent natural nanoscale particles [151]. EVs can carry molecules such as nucleic acids, proteins, lipids, etc., and participate in specific and efficient intercellular material transfer and information exchange. EVs exist in animals, plants, and microorganisms [152]. Currently, most research on EVs focuses on mammals and those that are ~30–150 nm in size. Meanwhile, research on plants remains relatively limited. In 2009, Regente et al. discovered vesicles with a 50–200 nm diameter within the fluid collected from sunflower seeds; this represents the first known attempt to extract EVs from plants. They found that EVs can affect the normal growth and development processes of fungi, suggesting that they are involved in plant–microbial interactions [153].
To differentiate from animal EVs, plant EVs are commonly referred to as exosome-like nanovesicles (ELNVs). Although the biogenesis of ELNVs has not been fully characterized, the fusion of multi-vesicular bodies and plant-specific exocyst-positive organelles with the plasma membrane is thought to be the main source of plant ELNVs [154,155]. miRNA-rich plant ELNVs are important in plant defense against biotic stress. For example, Cai et al. reported that A. thaliana can inhibit gray mold virulence genes via the exosomal delivery of miRNAs and inhibit fungal infestation through a transboundary nucleic acid delivery mechanism [155].
The structure of plant-derived ELNVs is similar to that of animal-derived EVs. This provides insights regarding the potential biological functions of ELNVs, including information transmission in animal cells [156,157]. The ELNV structure can partially resist the external environment, preventing the degradation or inactivation of its contents [158]. In particular, the RNA in ELNVs remains active after oral administration [159,160,161]. A recent study evaluated the biological effects of miRNA159 loaded in ELNVs by isolating and employing four food-borne ELNVs as transport carriers [162]. Among them, the level of miRNA transfected into ELNVs is ≥20 times higher than that of untreated miRNA, and cell absorption is also relatively improved. Moreover, the delivery of ELNVs isolated from broccoli carrying miR167a effectively reduced pancreatic cancer cell activity by inhibiting IRS3 of the PI1K–AKT pathway [163]. Katia et al. further revealed the presence of miR156c and miR159a in ELNVs extracted from walnuts. These miRNAs can regulate the mammalian TNF-α signaling pathway in adipocytes, modulating inflammation; notably, they have also influenced human physiology by participating in molecular regulation in vivo [164].
This confirms the previously established hypothesis that utilizing the biogenesis and cross-kingdom delivery mechanisms of endogenous plant miRNAs not only provides valuable insights for the development of novel biopesticides but also holds promise for novel pharmaceutical applications.
5.3. Potential of Oral Plant miRNA in the Pharmaceutical Industry
Plant-based foods contain plant miRNAs that can enter animals and humans via oral consumption and actively participate in gene expression regulation. More specifically, dietary miRNAs can be encapsulated and released into the circulation following cellular uptake in the gastrointestinal tract; they are then transported to various tissues, similar to endogenous miRNAs [165]. These exogenous plant miRNAs subsequently participate in the regulation of gene expression, affecting physiological processes and disease development and exerting antiviral, anti-tumor, and anti-inflammatory effects.
Notably, miR2911 inhibits the replication of myriad viruses, e.g., the novel coronavirus [166], Varicella-zoster virus [167], Enterovirus 71 [168], and Influenza A viruses [166]. Zhang postulated that miR2911 is the “penicillin of virology” and suggests that it can serve as a novel therapeutic for the prevention of other viral infections [166]. In addition, miR159 is the first plant miRNA shown to inhibit tumor growth in mammals. A study conducted by Chin et al. analyzed small RNAs in the sera of 42 patients with breast cancer, revealing the presence of plant miR159 in the Western population; interestingly, higher levels of miR159 were associated with a reduced risk and progression of breast cancer [169]. Moreover, a miR156a mimic, miRNA156a, inhibits epithelial–mesenchymal transition in human nasopharyngeal cancer by targeting the 3′-UTR of junctional adhesion molecule A (JAMA) [170]. Additionally, Chen et al. demonstrated a role for plant-derived miR5338 in the treatment of benign prostatic hyperplasia in rats by inhibiting Mfn1 in the prostate [171].
SIDT1 is an essential factor in the transport of plant miRNAs. miRNA absorption is hypothesized to occur in the small intestine; however, the associated hostile environment poses a major challenge to the stability of orally administered plant miRNAs [172]. Hence, Zhang et al. reported that dietary miRNA uptake is conducted by the stomach, which is enriched in SIDT1 protein—a key transporter. Additionally, dietary miRNA uptake is markedly reduced following SIDT1 knockdown [173]. Subsequently, exogenous dietary miRNAs are transported into the cells via the intrinsic carrier protein SIDT1 in the stomach. These miRNAs are then secreted as functional factors in exosomes, protecting them from degradation in the bloodstream and facilitating their cellular uptake. This natural mammalian uptake pathway for dietary miRNAs can be readily exploited for the oral administration of therapeutic miRNAs, representing an important future direction for developing RNA-based therapeutics.
6. Discussion and Perspective
Global warming continues to expose plants to an increasing number of biological and abiotic stresses. Plant-derived miRNAs can regulate and alleviate plant stress through endogenous, cross-species, and cross-kingdom gene regulation. In this review, we have summarized the current understanding of the synthesis, self-regulation, cross-kingdom regulatory mechanisms, and potential application strategies of plant miRNAs. Overall, these plant-derived miRNAs hold potential for the research and development of novel pesticides in modern agriculture.
While a growing number of miRNAs with cross-kingdom regulatory potential have been identified, their low expression and limited utilization pose significant challenges. To promote the practical application of miRNAs in agriculture and medicine, a more rational approach to the production and modification of miRNAs is needed. In particular, this approach should take advantage of plant-specific molecular mechanisms for synthesizing and transporting miRNAs. Synthetic biology is an emerging field of biological research used to elucidate and simulate the basic laws of biosynthesis while allowing the effective artificial design and construction of novel, physiologically specific products. Synthetic biology can produce disease-specific biomarkers, acting as a powerful new tool for targeting various diseases [174]. Traditional miRNA research focuses on the genetic regulation of phenotypic traits, evaluating the regulation of mRNAs by an individual’s miRNAs. In contrast, synthetic biology is used as a bridge to combine the strengths of different species, increasing the future research potential of miRNAs.
Notably, miRNAs can be used in the cross-species regulation of plant parasites and beneficial insects [105]; this holds promise as an effective strategy for developing novel pesticides and promoting the development of modern agriculture [175]. amiRNAs can be considered species-specific insecticides, presenting a powerful alternative to traditional chemical methods [176]. These miRNAs not only support beneficial insects in eliminating diseases but can also reduce pest infestations. This can be implemented by applying pest-specific amiRNAs to the leaves and soil, disrupting key physiological pathways within pests [177]. The application of miRNAs can also be combined with existing agricultural technologies to increase crop yield and enhance crop resilience to global climate change and related agricultural challenges [178].
In this context, synthetic biology involves the isolation of miRNAs with multiple functions, such as miR2911, which exhibits resistance against multiple viruses. These miRNAs can be heterologously expressed at an industrial level using high-yield chassis cells, shifting from theory to practical application. Alternatively, a “backward-looking” search can identify target genes within specific pathogens or pests. RNAi can then be employed to express edible plant miRNAs that can specifically inhibit pathogens and pests. Subsequently, edible plant vesicles or animal-derived exosomes can be used to absorb and transport miRNAs throughout the body to specifically target pathogens and pests. As agriculture is vital worldwide, its full potential should be explored to determine the value of plant miRNAs in higher plants, particularly crops.
However, several limitations remain in the application of miRNAs in agriculture. For instance, it remains unclear whether genetically modified food can contaminate human genes following ingestion and whether genetically modified plants can have a genetic impact on surrounding organisms during cultivation. In this case, the concern might be whether an RNAi-active plant could adversely affect the consumer. Subsequently, upon ingestion, specific miRNAs can be encapsulated within ELNVs to mitigate a decrease in their biological activity; however, certain miRNAs remain exposed. Thus, some miRNAs demonstrate relative stability, such as miR2911, enabling them to traverse the digestive tract without degradation. Conversely, others experience functional loss after exposure to high temperatures and enzymatic hydrolysis. The reliable determination of systemic effects of orally administered plant-derived miRNAs remains elusive, and minimal absorption likely further restricts the possibility of sufficient miRNA quantities reaching the tissues or functional sites, limiting their potential biological impact [179]. Moreover, the breeding period of RNAi is lengthy, and the prerequisite for trait enhancement lies in non-interference with the original exceptional traits. To select and cultivate plants possessing resistance and high yields, a substantial number of field experiments are imperative. However, the modified plants can deviate significantly from expectations [148]; hence, the popularization and application warrant further verification.
7. Conclusions
The cross-species regulatory roles of plant miRNAs offer new prospects in agriculture, in which the understanding and application of plant miRNAs not only have the potential to enhance crop yields and resistance but also offer new insights into the development of biopesticides. Ultimately, using these miRNAs in agriculture may improve the ecological balance and reduce dependence on chemical pesticides and fertilizers. In medicine, plant miRNAs hold promise as gene therapy tools for precisely regulating specific genes. However, further research and technological development are required to address ethical, regulatory, and ecosystem impact concerns, ensuring the feasibility and sustainability of these approaches.
Author Contributions
Conceptualization, W.W., M.R. and F.L.; writing—original draft preparation, T.D.; writing—review and editing, T.D. and W.L.; funding acquisition, W.W., M.R. and F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (grant number 32301892 to W.W., U23A20182 to F.L., 31972469 to M.R.), China Postdoctoral Science Foundation (grant number 2023M733257 to W.W.), and the Innovation Program of Chinese Academy of Agricultural Sciences (F.L.). The APC was funded by National Natural Science Foundation of China (grant number 32301892 to W.W).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Vella, M.C.; Slack, F.J. C. Elegans microRNAs; WormBook: Pasadena, CA, USA, 2005; pp. 1–9. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; D’Souza, J.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1955–1986. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef]
- Reyes, R.; Alcalde, J.; Izquierdo, J.M. Depletion of T-cell intracellular antigen proteins promotes cell proliferation. Genome Biol. 2009, 10, R87. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microRNA networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Nicoloso, M.S.; Spizzo, R.; Shimizu, M.; Rossi, S.; Calin, G.A. MicroRNAs—The micro steering wheel of tumour metastases. Nat. Rev. Cancer 2009, 9, 293–302. [Google Scholar] [CrossRef]
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647. [Google Scholar] [CrossRef]
- Huang, L.; Wang, X.; Wen, C.; Yang, X.; Song, M.; Chen, J.; Wang, C.; Zhang, B.; Wang, L.; Iwamoto, A.; et al. Hsa-miR-19a is associated with lymph metastasis and mediates the TNF-α induced epithelial-to-mesenchymal transition in colorectal cancer. Sci. Rep. 2015, 5, 13350. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, Z.; Lai, D.; Sun, J.; He, C.; Chu, Z.; Ye, H.; Chen, S.; Wang, J. miR-21, miR-17 and miR-19a induced by phosphatase of regenerating liver-3 promote the proliferation and metastasis of colon cancer. Br. J. Cancer 2012, 107, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhao, Y.L.; Zhao, J.H.; Wang, S.; Jin, Y.; Chen, Z.Q.; Fang, Y.Y.; Hua, C.L.; Ding, S.W.; Guo, H.S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Q.; Yuan, L.; Shen, W.; Shi, Q.; Qi, G.; Chen, T.; Zhang, Z. Osa-miR162a Enhances the Resistance to the Brown Planthopper via α-Linolenic Acid Metabolism in Rice (Oryza sativa). J. Agric. Food Chem. 2023, 71, 11847–11859. [Google Scholar] [CrossRef]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and Their Regulatory Roles in Plant-Environment Interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Cullen, B.R. Sequence requirements for micro RNA processing and function in human cells. RNA 2003, 9, 112–123. [Google Scholar] [CrossRef]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef]
- Kurihara, Y.; Watanabe, Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. USA 2004, 101, 12753–12758. [Google Scholar] [CrossRef] [PubMed]
- Park, M.Y.; Wu, G.; Gonzalez-Sulser, A.; Vaucheret, H.; Poethig, R.S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 3691–3696. [Google Scholar] [CrossRef] [PubMed]
- Tang, G. Plant microRNAs: An insight into their gene structures and evolution. Semin. Cell Dev. Biol. 2010, 21, 782–789. [Google Scholar] [CrossRef]
- Rajagopalan, R.; Vaucheret, H.; Trejo, J.; Bartel, D.P. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006, 20, 3407–3425. [Google Scholar] [CrossRef]
- Yan, K.; Liu, P.; Wu, C.A.; Yang, G.D.; Xu, R.; Guo, Q.H.; Huang, J.G.; Zheng, C.C. Stress-induced alternative splicing provides a mechanism for the regulation of microRNA processing in Arabidopsis thaliana. Mol. Cell 2012, 48, 521–531. [Google Scholar] [CrossRef]
- Yang, G.D.; Yan, K.; Wu, B.J.; Wang, Y.H.; Gao, Y.X.; Zheng, C.C. Genomewide analysis of intronic microRNAs in rice and Arabidopsis. J. Genet. 2012, 91, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Zheng, B.; Yu, Y.; Won, S.Y.; Mo, B.; Chen, X. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 2011, 30, 814–822. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Allen, E.; Fahlgren, N.; Calamar, A.; Givan, S.A.; Carrington, J.C. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005, 138, 2145–2154. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Chen, X. Regulation of small RNA stability: Methylation and beyond. Cell Res. 2012, 22, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Kutter, C.; Svoboda, P. miRNA, siRNA, piRNA: Knowns of the unknown. RNA Biol. 2008, 5, 181–188. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.M.; Ding, Y.F.; Zhu, C. Role of miRNA in plant seed development. Yi Chuan 2015, 37, 554–560. [Google Scholar] [CrossRef]
- Dong, Q.; Hu, B.; Zhang, C. microRNAs and Their Roles in Plant Development. Front. Plant Sci. 2022, 13, 824240. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, L.; Yang, Y.; Schmid, M.; Wang, Y. miRNA Mediated Regulation and Interaction between Plants and Pathogens. Int. J. Mol. Sci. 2021, 22, 2913. [Google Scholar] [CrossRef]
- Luo, Y.C.; Zhou, H.; Li, Y.; Chen, J.Y.; Yang, J.H.; Chen, Y.Q.; Qu, L.H. Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett. 2006, 580, 5111–5116. [Google Scholar] [CrossRef]
- Hou, Y.; Jiang, F.; Zheng, X.; Wu, Z. Identification and analysis of oxygen responsive microRNAs in the root of wild tomato (S. habrochaites). BMC Plant Biol. 2019, 19, 100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Yu, Y.; Wang, C.Y.; Li, Z.Y.; Liu, Q.; Xu, J.; Liao, J.Y.; Wang, X.J.; Qu, L.H.; Chen, F.; et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Zhang, S.; Yu, Y.; Luo, Y.C.; Liu, Q.; Ju, C.; Zhang, Y.C.; Qu, L.H.; Lucas, W.J.; Wang, X.; et al. MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 2014, 12, 1132–1142. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Tang, S.; Ma, X.; Liu, W.; Yang, R.; Zhang, S.; Wang, N.; Song, X.; Fu, C.; Yang, R.; et al. Blocking miR528 function promotes tillering and regrowth in switchgrass. Plant Biotechnol. J. 2023. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Ding, L.; Xia, Y.; Wang, F.; Zhu, C. Emerging Roles of microRNAs in Plant Heavy Metal Tolerance and Homeostasis. J. Agric. Food Chem. 2020, 68, 1958–1965. [Google Scholar] [CrossRef]
- Benáková, M.; Ahmadi, H.; Dučaiová, Z.; Tylová, E.; Clemens, S.; Tůma, J. Effects of Cd and Zn on physiological and anatomical properties of hydroponically grown Brassica napus plants. Environ. Sci. Pollut. Res. Int. 2017, 24, 20705–20716. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Rengel, Z.; Qaswar, M.; Amir, M.; Zafar-ul-Hye, M. Arsenic and Heavy Metal (Cadmium, Lead, Mercury and Nickel) Contamination in Plant-Based Foods. In Plant and Human Health, Volume 2: Phytochemistry and Molecular Aspects; Ozturk, M., Hakeem, K.R., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 447–490. [Google Scholar] [CrossRef]
- Ali, S.; Huang, S.; Zhou, J.; Bai, Y.; Liu, Y.; Shi, L.; Liu, S.; Hu, Z.; Tang, Y. miR397-LACs mediated cadmium stress tolerance in Arabidopsis thaliana. Plant Mol. Biol. 2023, 113, 415–430. [Google Scholar] [CrossRef]
- Yue, E.; Rong, F.; Liu, Z.; Ruan, S.; Lu, T.; Qian, H. Cadmium induced a non-coding RNA microRNA535 mediates Cd accumulation in rice. J. Environ. Sci. 2023, 130, 149–162. [Google Scholar] [CrossRef]
- Yan, G.; Hua, Y.; Jin, H.; Huang, Q.; Zhou, G.; Xu, Y.; He, Y.; Zhu, Z. Sly-miR398 Participates in Cadmium Stress Acclimation by Regulating Antioxidant System and Cadmium Transport in Tomato (Solanum lycopersicum). Int. J. Mol. Sci. 2023, 24, 1953. [Google Scholar] [CrossRef]
- Riyazuddin, R.; Singh, K.; Iqbal, N.; Labhane, N.; Ramteke, P.; Singh, V.P.; Gupta, R. Unveiling the biosynthesis, mechanisms, and impacts of miRNAs in drought stress resilience in plants. Plant Physiol. Biochem. 2023, 202, 107978. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, X.; Shi, M.; Yu, J.; Guo, C. The miR159-MYB33-ABI5 module regulates seed germination in Arabidopsis. Physiol. Plant. 2022, 174, e13659. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef] [PubMed]
- Shamloo-Dashtpagerdi, R.; Shahriari, A.G.; Tahmasebi, A.; Vetukuri, R.R. Potential role of the regulatory miR1119-MYC2 module in wheat (Triticum aestivum L.) drought tolerance. Front. Plant Sci. 2023, 14, 1161245. [Google Scholar] [CrossRef]
- Briantais, J.M.; Dacosta, J.; Goulas, Y.; Ducruet, J.M.; Moya, I. Heat stress induces in leaves an increase of the minimum level of chlorophyll fluorescence, Fo: A time-resolved analysis. Photosynth. Res. 1996, 48, 189–196. [Google Scholar] [CrossRef]
- Larkindale, J.; Knight, M.R. Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 2002, 128, 682–695. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, S.N.A.; Azzeme, A.M.; Yousefi, K. Fine-Tuning Cold Stress Response Through Regulated Cellular Abundance and Mechanistic Actions of Transcription Factors. Front. Plant Sci. 2022, 13, 850216. [Google Scholar] [CrossRef] [PubMed]
- Knight, M.R.; Knight, H. Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 2012, 195, 737–751. [Google Scholar] [CrossRef]
- Matthews, C.; Arshad, M.; Hannoufa, A. Alfalfa response to heat stress is modulated by microRNA156. Physiol. Plant. 2019, 165, 830–842. [Google Scholar] [CrossRef]
- Zeng, X.; Xu, Y.; Jiang, J.; Zhang, F.; Ma, L.; Wu, D.; Wang, Y.; Sun, W. Identification of cold stress responsive microRNAs in two winter turnip rape (Brassica rapa L.) by high throughput sequencing. BMC Plant Biol. 2018, 18, 52. [Google Scholar] [CrossRef]
- Shi, X.; Jiang, F.; Wen, J.; Wu, Z. Overexpression of Solanum habrochaites microRNA319d (sha-miR319d) confers chilling and heat stress tolerance in tomato (S. lycopersicum). BMC Plant Biol. 2019, 19, 214. [Google Scholar] [CrossRef]
- Islam, W.; Tauqeer, A.; Waheed, A.; Zeng, F. MicroRNA Mediated Plant Responses to Nutrient Stress. Int. J. Mol. Sci. 2022, 23, 2562. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Fan, D.; Ding, Z.; Su, Y.; Wang, X. Cs-miR156 is involved in the nitrogen form regulation of catechins accumulation in tea plant (Camellia sinensis L.). Plant Physiol. Biochem. 2015, 97, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Q.; Davis, K.E.; Patterson, C.; Oo, S.; Liu, W.; Liu, J.; Wang, G.; Fontana, J.E.; Thornburg, T.E.; et al. Response of Root Growth and Development to Nitrogen and Potassium Deficiency as well as microRNA-Mediated Mechanism in Peanut (Arachis hypogaea L.). Front. Plant Sci. 2021, 12, 695234. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Liu, Q.; Chen, L.; Kuang, J.; Walk, T.; Wang, J.; Liao, H. Genome-wide identification of soybean microRNAs and their targets reveals their organ-specificity and responses to phosphate starvation. BMC Genom. 2013, 14, 66. [Google Scholar] [CrossRef] [PubMed]
- Thornburg, T.E.; Liu, J.; Li, Q.; Xue, H.; Wang, G.; Li, L.; Fontana, J.E.; Davis, K.E.; Liu, W.; Zhang, B.; et al. Potassium Deficiency Significantly Affected Plant Growth and Development as Well as microRNA-Mediated Mechanism in Wheat (Triticum aestivum L.). Front. Plant Sci. 2020, 11, 1219. [Google Scholar] [CrossRef]
- Safdar, H.; Amin, A.; Shafiq, Y.; Ali, A.; Yasin, R.; Sarwar, M.I. Abbas Shoukat, Maqsood Ul Hussan, Muhammad Ishtiaq Sarwar. A review: Impact of salinity on plant growth. Nat. Sci. 2019, 1, 34–40. [Google Scholar] [CrossRef]
- Goyal, V.; Jhanghel, D.; Mehrotra, S. Emerging warriors against salinity in plants: Nitric oxide and hydrogen sulphide. Physiol. Plant. 2021, 171, 896–908. [Google Scholar] [CrossRef]
- Kamran, M.; Parveen, A.; Ahmar, S.; Malik, Z.; Hussain, S.; Chattha, M.S.; Saleem, M.H.; Adil, M.; Heidari, P.; Chen, J.T. An Overview of Hazardous Impacts of Soil Salinity in Crops, Tolerance Mechanisms, and Amelioration through Selenium Supplementation. Int. J. Mol. Sci. 2019, 21, 148. [Google Scholar] [CrossRef]
- Islam, W.; Waheed, A.; Naveed, H.; Zeng, F. MicroRNAs Mediated Plant Responses to Salt Stress. Cells 2022, 11, 2806. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, L.; Yang, Z.; Wei, Y.; Dong, T. Identification and Functional Characterization of Plant MiRNA Under Salt Stress Shed Light on Salinity Resistance Improvement Through MiRNA Manipulation in Crops. Front. Plant Sci. 2021, 12, 665439. [Google Scholar] [CrossRef]
- Chen, L.; Luan, Y.; Zhai, J. Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep. 2015, 34, 2013–2025. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Li, Y.; Cai, H.; Ji, W.; Guo, D.; Zhu, Y. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta 2010, 231, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.H.; Zhou, X.Y.; Shi, S.H.; Zhang, S.; Chen, Z.H.; Ali, M.A.; Ahmed, I.M.; Wang, Y.; Wu, F. An miR156-regulated nucleobase-ascorbate transporter 2 confers cadmium tolerance via enhanced anti-oxidative capacity in barley. J. Adv. Res. 2023, 44, 23–37. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yuan, S.; Zhou, M.; Yuan, N.; Li, Z.; Hu, Q.; Bethea, F.G., Jr.; Liu, H.; Li, S.; Luo, H. Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol. J. 2019, 17, 233–251. [Google Scholar] [CrossRef]
- Jodder, J.; Das, R.; Sarkar, D.; Bhattacharjee, P.; Kundu, P. Distinct transcriptional and processing regulations control miR167a level in tomato during stress. RNA Biol. 2018, 15, 130–143. [Google Scholar] [CrossRef]
- Islam, W.; Qasim, M.; Noman, A.; Adnan, M.; Tayyab, M.; Farooq, T.H.; Wei, H.; Wang, L. Plant microRNAs: Front line players against invading pathogens. Microb. Pathog. 2018, 118, 9–17. [Google Scholar] [CrossRef]
- Zhao, W.; Li, Z.; Fan, J.; Hu, C.; Yang, R.; Qi, X.; Chen, H.; Zhao, F.; Wang, S. Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. J. Exp. Bot. 2015, 66, 4653–4667. [Google Scholar] [CrossRef]
- Pinweha, N.; Asvarak, T.; Viboonjun, U.; Narangajavana, J. Involvement of miR160/miR393 and their targets in cassava responses to anthracnose disease. J. Plant Physiol. 2015, 174, 26–35. [Google Scholar] [CrossRef]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef]
- Yu, X.; Hou, Y.; Chen, W.; Wang, S.; Wang, P.; Qu, S. Malus hupehensis miR168 Targets to ARGONAUTE1 and Contributes to the Resistance against Botryosphaeria dothidea Infection by Altering Defense Responses. Plant Cell Physiol. 2017, 58, 1541–1557. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Hong, Y.H.; Liu, Y.R.; Cui, J.; Luan, Y.S. Function identification of miR394 in tomato resistance to Phytophthora infestans. Plant Cell Rep. 2021, 40, 1831–1844. [Google Scholar] [CrossRef]
- Yang, X.; Fu, T.; Yu, R.; Zhang, L.; Yang, Y.; Xiao, D.; Wang, Y.; Wang, Y.; Wang, Y. miR159a modulates poplar resistance against different fungi and bacteria. Plant Physiol. Biochem. 2023, 201, 107899. [Google Scholar] [CrossRef]
- Cheng, B.; Xu, L.; Bilal, M.S.; Huang, Q.; Niu, D.; Ma, H.; Zhou, S.; Peng, A.; Wei, G.; Chen, F.; et al. Small RNAs contribute to citrus Huanglongbing tolerance by manipulating methyl salicylate signaling and exogenous methyl salicylate primes citrus groves from emerging infection. Plant J. Cell Mol. Biol. 2023, 116, 1309–1324. [Google Scholar] [CrossRef]
- Liu, G.; Liu, F.; Zhang, D.; Zhao, T.; Yang, H.; Jiang, J.; Li, J.; Zhang, H.; Xu, X. Integrating omics reveals that miRNA-guided genetic regulation on plant hormone level and defense response pathways shape resistance to Cladosporium fulvum in the tomato Cf-10-gene-carrying line. Front. Genet. 2023, 14, 1158631. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wang, B.; Jia, P.; Wu, P.; Lu, C.; Xu, Y.; Shi, L.; Zhang, F.; Zhong, N.; Chen, A.; et al. The cotton miR530-SAP6 module activated by systemic acquired resistance mediates plant defense against Verticillium dahliae. Plant Sci. 2023, 330, 111647. [Google Scholar] [CrossRef]
- Ouyang, S.; Park, G.; Atamian, H.S.; Han, C.S.; Stajich, J.E.; Kaloshian, I.; Borkovich, K.A. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014, 10, e1004464. [Google Scholar] [CrossRef]
- Djami-Tchatchou, A.T.; Dubery, I.A. miR393 regulation of lectin receptor-like kinases associated with LPS perception in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2019, 513, 88–92. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, Y.; Zhang, Y.; Wu, C.; Wang, S.; Hao, L.; Wang, S.; Li, T. Md-miR156ab and Md-miR395 Target WRKY Transcription Factors to Influence Apple Resistance to Leaf Spot Disease. Front. Plant Sci. 2017, 8, 526. [Google Scholar] [CrossRef]
- Li, Y.; Hu, X.; Chen, J.; Wang, W.; Xiong, X.; He, C. Integrated mRNA and microRNA transcriptome analysis reveals miRNA regulation in response to PVA in potato. Sci. Rep. 2017, 7, 16925. [Google Scholar] [CrossRef]
- López-Márquez, D.; Del-Espino, Á.; Ruiz-Albert, J.; Bejarano, E.R.; Brodersen, P.; Beuzón, C.R. Regulation of plant immunity via small RNA-mediated control of NLR expression. J. Exp. Bot. 2023, 74, 6052–6068. [Google Scholar] [CrossRef]
- Sun, Y.; Zhu, Y.X.; Balint-Kurti, P.J.; Wang, G.F. Fine-Tuning Immunity: Players and Regulators for Plant NLRs. Trends Plant Sci. 2020, 25, 695–713. [Google Scholar] [CrossRef]
- Zhu, X.; Kuang, Y.; Chen, Y.; Shi, J.; Cao, Y.; Hu, J.; Yu, C.; Yang, F.; Tian, F.; Chen, H. miR2118 Negatively Regulates Bacterial Blight Resistance through Targeting Several Disease Resistance Genes in Rice. Plants 2023, 12, 3815. [Google Scholar] [CrossRef]
- Wu, J.; Yang, R.; Yang, Z.; Yao, S.; Zhao, S.; Wang, Y.; Li, P.; Song, X.; Jin, L.; Zhou, T.; et al. ROS accumulation and antiviral defence control by microRNA528 in rice. Nat. Plants 2017, 3, 16203. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Yang, Z.; Yang, R.; Huang, Y.; Guo, G.; Kong, X.; Lan, Y.; Zhou, T.; Wang, H.; Wang, W.; et al. Transcriptional Regulation of miR528 by OsSPL9 Orchestrates Antiviral Response in Rice. Mol. Plant 2019, 12, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yu, Y.; Fei, S.; Chen, Y.; Xu, Y.; Zhu, Z.; He, Y. Overexpression of Sly-miR398b Compromises Disease Resistance against Botrytis cinerea through Regulating ROS Homeostasis and JA-Related Defense Genes in Tomato. Plants 2023, 12, 2572. [Google Scholar] [CrossRef]
- Yu, X.; Gong, H.; Cao, L.; Hou, Y.; Qu, S. MicroRNA397b negatively regulates resistance of Malus hupehensis to Botryosphaeria dothidea by modulating MhLAC7 involved in lignin biosynthesis. Plant Sci. Int. J. Exp. Plant Biol. 2020, 292, 110390. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.Y.; Du, B.B.; Gao, Z.H.; Zhang, S.J.; Tu, X.T.; Chen, X.Y.; Zhang, Z.; Qu, S.C. Apple ring rot-responsive putative microRNAs revealed by high-throughput sequencing in Malus × domestica Borkh. Mol. Biol. Rep. 2014, 41, 5273–5286. [Google Scholar] [CrossRef]
- Xia, Z.; Zhao, Z.; Li, M.; Chen, L.; Jiao, Z.; Wu, Y.; Zhou, T.; Yu, W.; Fan, Z. Identification of miRNAs and their targets in maize in response to Sugarcane mosaic virus infection. Plant Physiol. Biochem. 2018, 125, 143–152. [Google Scholar] [CrossRef]
- Chandran, V.; Wang, H.; Gao, F.; Cao, X.L.; Chen, Y.P.; Li, G.B.; Zhu, Y.; Yang, X.M.; Zhang, L.L.; Zhao, Z.X.; et al. miR396-OsGRFs Module Balances Growth and Rice Blast Disease-Resistance. Front. Plant Sci. 2018, 9, 1999. [Google Scholar] [CrossRef]
- Dai, Z.; Tan, J.; Zhou, C.; Yang, X.; Yang, F.; Zhang, S.; Sun, S.; Miao, X.; Shi, Z. The OsmiR396-OsGRF8-OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa). Plant Biotechnol. J. 2019, 17, 1657–1669. [Google Scholar] [CrossRef]
- Yan, J.; Qiu, R.; Wang, K.; Liu, Y.; Zhang, W. Enhancing alfalfa resistance to Spodoptera herbivory by sequestering microRNA396 expression. Plant Cell Rep. 2023, 42, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef]
- Zhu, K.; Liu, M.; Fu, Z.; Zhou, Z.; Kong, Y.; Liang, H.; Lin, Z.; Luo, J.; Zheng, H.; Wan, P.; et al. Plant microRNAs in larval food regulate honeybee caste development. PLoS Genet. 2017, 13, e1006946. [Google Scholar] [CrossRef]
- Liu, Y.C.; Chen, W.L.; Kung, W.H.; Huang, H.D. Plant miRNAs found in human circulating system provide evidences of cross kingdom RNAi. BMC Genom. 2017, 18, 112. [Google Scholar] [CrossRef] [PubMed]
- Hyun, T.K.; Uddin, M.N.; Rim, Y.; Kim, J.Y. Cell-to-cell trafficking of RNA and RNA silencing through plasmodesmata. Protoplasma 2011, 248, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Marín-González, E.; Suárez-López, P. “And yet it moves”: Cell-to-cell and long-distance signaling by plant microRNAs. Plant Sci. Int. J. Exp. Plant Biol. 2012, 196, 18–30. [Google Scholar] [CrossRef]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.R. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. Cell Mol. Biol. 2008, 53, 731–738. [Google Scholar] [CrossRef]
- Chiang, C.P.; Li, J.L.; Chiou, T.J. Dose-dependent long-distance movement of microRNA399 duplex regulates phosphate homeostasis in Arabidopsis. New Phytol. 2023, 240, 802–814. [Google Scholar] [CrossRef]
- Bhogale, S.; Mahajan, A.S.; Natarajan, B.; Rajabhoj, M.; Thulasiram, H.V.; Banerjee, A.K. MicroRNA156: A potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014, 164, 1011–1027. [Google Scholar] [CrossRef]
- Betti, F.; Ladera-Carmona, M.J.; Weits, D.A.; Ferri, G.; Iacopino, S.; Novi, G.; Svezia, B.; Kunkowska, A.B.; Santaniello, A.; Piaggesi, A.; et al. Exogenous miRNAs induce post-transcriptional gene silencing in plants. Nat. Plants 2021, 7, 1379–1388. [Google Scholar] [CrossRef]
- Shahid, S.; Kim, G.; Johnson, N.R.; Wafula, E.; Wang, F.; Coruh, C.; Bernal-Galeano, V.; Phifer, T.; dePamphilis, C.W.; Westwood, J.H.; et al. MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 2018, 553, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Hudzik, C.; Maguire, S.; Guan, S.; Held, J.; Axtell, M.J. Trans-species microRNA loci in the parasitic plant Cuscuta campestris have a U6-like snRNA promoter. Plant Cell 2023, 35, 1834–1847. [Google Scholar] [CrossRef]
- Meng, X.; Jin, W.; Wu, F. Novel tomato miRNA miR1001 initiates cross-species regulation to suppress the conidiospore germination and infection virulence of Botrytis cinerea in vitro. Gene 2020, 759, 145002. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Peng, D. Wheat microRNA1023 suppresses invasion of Fusarium graminearum via targeting and silencing FGSG_03101. J. Plant Interact. 2018, 13, 514–521. [Google Scholar] [CrossRef]
- Luo, M.; Sun, X.; Xu, M.; Tian, Z. Identification of miRNAs Involving Potato-Phytophthora infestans Interaction. Plants 2023, 12, 461. [Google Scholar] [CrossRef] [PubMed]
- Mathur, M.; Nair, A.; Kadoo, N. Plant-pathogen interactions: MicroRNA-mediated trans-kingdom gene regulation in fungi and their host plants. Genomics 2020, 112, 3021–3035. [Google Scholar] [CrossRef]
- Sharma, S.; Sett, S.; Das, T.; Prasad, A.; Prasad, M. Recent perspective of non-coding RNAs at the nexus of plant-pathogen interaction. Plant Physiol. Biochem. 2023, 201, 107852. [Google Scholar] [CrossRef]
- Chi, X.; Wang, Z.; Wang, Y.; Liu, Z.; Wang, H.; Xu, B. Cross-Kingdom Regulation of Plant-Derived miRNAs in Modulating Insect Development. Int. J. Mol. Sci. 2023, 24, 7978. [Google Scholar] [CrossRef]
- Zhang, L.L.; Jing, X.D.; Chen, W.; Wang, Y.; Lin, J.H.; Zheng, L.; Dong, Y.H.; Zhou, L.; Li, F.F.; Yang, F.Y.; et al. Host Plant-Derived miRNAs Potentially Modulate the Development of a Cosmopolitan Insect Pest, Plutella xylostella. Biomolecules 2019, 9, 602. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, C.; Dou, Y.; Yu, B.; Liu, Y.; Heng-Moss, T.M.; Lu, G.; Wachholtz, M.; Bradshaw, J.D.; Twigg, P.; et al. Insect and plant-derived miRNAs in greenbug (Schizaphis graminum) and yellow sugarcane aphid (Sipha flava) revealed by deep sequencing. Gene 2017, 599, 68–77. [Google Scholar] [CrossRef]
- Beran, F.; Petschenka, G. Sequestration of Plant Defense Compounds by Insects: From Mechanisms to Insect-Plant Coevolution. Annu. Rev. Entomol. 2022, 67, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Kotowska-Zimmer, A.; Pewinska, M.; Olejniczak, M. Artificial miRNAs as therapeutic tools: Challenges and opportunities. Wiley Interdiscip. Rev. RNA 2021, 12, e1640. [Google Scholar] [CrossRef]
- Moore, C.B.; Guthrie, E.H.; Huang, M.T.; Taxman, D.J. Short hairpin RNA (shRNA): Design, delivery, and assessment of gene knockdown. Methods Mol. Biol. 2010, 629, 141–158. [Google Scholar] [CrossRef]
- Sliva, K.; Schnierle, B.S. Selective gene silencing by viral delivery of short hairpin RNA. Virol. J. 2010, 7, 248. [Google Scholar] [CrossRef]
- Boudreau, R.L.; Martins, I.; Davidson, B.L. Artificial microRNAs as siRNA shuttles: Improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. J. Am. Soc. Gene Ther. 2009, 17, 169–175. [Google Scholar] [CrossRef] [PubMed]
- McBride, J.L.; Boudreau, R.L.; Harper, S.Q.; Staber, P.D.; Monteys, A.M.; Martins, I.; Gilmore, B.L.; Burstein, H.; Peluso, R.W.; Polisky, B.; et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: Implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. USA 2008, 105, 5868–5873. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Weng, Z.; Li, H.; Kong, Z.; Zhou, Z.; Li, F.; Ma, W.; Lin, Y.; Chen, H. Transgenic rice overexpressing insect endogenous microRNA csu-novel-260 is resistant to striped stem borer under field conditions. Plant Biotechnol. J. 2021, 19, 421–423. [Google Scholar] [CrossRef]
- He, K.; Sun, Y.; Xiao, H.; Ge, C.; Li, F.; Han, Z. Multiple miRNAs jointly regulate the biosynthesis of ecdysteroid in the holometabolous insects, Chilo suppressalis. RNA 2017, 23, 1817–1833. [Google Scholar] [CrossRef]
- Jiang, S.; Wu, H.; Liu, H.; Zheng, J.; Lin, Y.; Chen, H. The overexpression of insect endogenous small RNAs in transgenic rice inhibits growth and delays pupation of striped stem borer (Chilo suppressalis). Pest Manag. Sci. 2017, 73, 1453–1461. [Google Scholar] [CrossRef]
- Liu, H.; Shen, E.; Wu, H.; Ma, W.; Chen, H.; Lin, Y. Trans-kingdom expression of an insect endogenous microRNA in rice enhances resistance to striped stem borer Chilo suppressalis. Pest Manag. Sci. 2022, 78, 770–777. [Google Scholar] [CrossRef]
- He, K.; Xiao, H.; Sun, Y.; Ding, S.; Situ, G.; Li, F. Transgenic microRNA-14 rice shows high resistance to rice stem borer. Plant Biotechnol. J. 2019, 17, 461–471. [Google Scholar] [CrossRef]
- Bally, J.; Fishilevich, E.; Doran, R.L.; Lee, K.; de Campos, S.B.; German, M.A.; Narva, K.E.; Waterhouse, P.M. Plin-amiR, a pre-microRNA-based technology for controlling herbivorous insect pests. Plant Biotechnol. J. 2020, 18, 1925–1932. [Google Scholar] [CrossRef]
- Yogindran, S.; Rajam, M.V. Host-derived artificial miRNA-mediated silencing of ecdysone receptor gene provides enhanced resistance to Helicoverpa armigera in tomato. Genomics 2021, 113, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, A.; Rajamani, V.; Reddy, V.S.; Mukherjee, S.K.; Bhatnagar, R.K. Transgenic plants over-expressing insect-specific microRNA acquire insecticidal activity against Helicoverpa armigera: An alternative to Bt-toxin technology. Transgenic Res. 2015, 24, 791–801. [Google Scholar] [CrossRef]
- Faisal, M.; Abdel-Salam, E.M.; Alatar, A.A. Artificial microRNA-Based RNA Interference and Specific Gene Silencing for Developing Insect Resistance in Solanum lycopersicum. Agronomy 2021, 11, 136. [Google Scholar] [CrossRef]
- Niu, Q.W.; Lin, S.S.; Reyes, J.L.; Chen, K.C.; Wu, H.W.; Yeh, S.D.; Chua, N.H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428. [Google Scholar] [CrossRef]
- Miao, S.; Liang, C.; Li, J.; Baker, B.; Luo, L. Polycistronic Artificial microRNA-Mediated Resistance to Cucumber Green Mottle Mosaic Virus in Cucumber. Int. J. Mol. Sci. 2021, 22, 12237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, H.; Zhang, J.; Zhang, C.; Gong, P.; Ziaf, K.; Xiao, F.; Ye, Z. Expression of artificial microRNAs in tomato confers efficient and stable virus resistance in a cell-autonomous manner. Transgenic Res. 2011, 20, 569–581. [Google Scholar] [CrossRef]
- Ai, T.; Zhang, L.; Gao, Z.; Zhu, C.X.; Guo, X. Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. 2011, 13, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.Z.; Han, Q.J.; Jiang, F.; Sun, R.Z.; Fan, Z.H.; Zhu, C.X.; Wen, F.J. Effects of the sequence characteristics of miRNAs on multi-viral resistance mediated by single amiRNAs in transgenic tobacco. Plant Physiol. Biochem. 2014, 77, 90–98. [Google Scholar] [CrossRef]
- Al-Roshdi, M.R.; Ammara, U.; Khan, J.; Al-Sadi, A.M.; Shahid, M.S. Artificial microRNA-mediated resistance against Oman strain of tomato yellow leaf curl virus. Front. Plant Sci. 2023, 14, 1164921. [Google Scholar] [CrossRef]
- Khalid, A.; Zhang, X.; Ji, H.; Yasir, M.; Farooq, T.; Dai, X.; Li, F. Large Artificial microRNA Cluster Genes Confer Effective Resistance against Multiple Tomato Yellow Leaf Curl Viruses in Transgenic Tomato. Plants 2023, 12, 2179. [Google Scholar] [CrossRef] [PubMed]
- Berbati, M.; Kaldis, A.; Voloudakis, A. Efficient artificial microRNA-mediated resistance against zucchini yellow mosaic virus in zucchini via agroinfiltration. J. Virol. Methods 2023, 321, 114805. [Google Scholar] [CrossRef] [PubMed]
- Valli, A.A.; Gallo, A.; Rodamilans, B.; López-Moya, J.J.; García, J.A. The HCPro from the Potyviridae family: An enviable multitasking Helper Component that every virus would like to have. Mol. Plant Pathol. 2018, 19, 744–763. [Google Scholar] [CrossRef]
- Cisneros, A.E.; Lisón, P.; Campos, L.; López-Tubau, J.M.; Altabella, T.; Ferrer, A.; Daròs, J.A.; Carbonell, A. Down-regulation of tomato STEROL GLYCOSYLTRANSFERASE 1 perturbs plant development and facilitates viroid infection. J. Exp. Bot. 2023, 74, 1564–1578. [Google Scholar] [CrossRef]
- Chávez, A.; Castillo, N.; López-Tubau, J.M.; Atanasov, K.E.; Fernández-Crespo, E.; Camañes, G.; Altabella, T.; Ferrer, A. Tomato STEROL GLYCOSYLTRANSFERASE 1 silencing unveils a major role of steryl glycosides in plant and fruit development. Environ. Exp. Bot. 2023, 206, 105181. [Google Scholar] [CrossRef]
- Kakeshpour, T.; Tamang, T.M.; Park, W.D.; Manohar, M.; Yang, J.; Hirschi, K.D.; Park, S. Expression of mouse small interfering RNAs in lettuce using artificial microRNA technology. Biotechniques 2020, 68, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sang, X.; Hou, D.; Chen, J.; Gu, H.; Zhang, Y.; Li, J.; Yang, D.; Zhu, H.; Yang, X.; et al. Plant-derived RNAi therapeutics: A strategic inhibitor of HBsAg. Biomaterials 2019, 210, 83–93. [Google Scholar] [CrossRef] [PubMed]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Regente, M.; Corti-Monzón, G.; Maldonado, A.M.; Pinedo, M.; Jorrín, J.; de la Canal, L. Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett. 2009, 583, 3363–3366. [Google Scholar] [CrossRef]
- Robinson, D.G.; Ding, Y.; Jiang, L. Unconventional protein secretion in plants: A critical assessment. Protoplasma 2016, 253, 31–43. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Lian, M.Q.; Chng, W.H.; Liang, J.; Yeo, H.Q.; Lee, C.K.; Belaid, M.; Tollemeto, M.; Wacker, M.G.; Czarny, B.; Pastorin, G. Plant-derived extracellular vesicles: Recent advancements and current challenges on their use for biomedical applications. J. Extracell. Vesicles 2022, 11, e12283. [Google Scholar] [CrossRef]
- An, Q.; van Bel, A.J.; Hückelhoven, R. Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal. Behav. 2007, 2, 4–7. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Yan, H.; Han, X.; Weng, L.; Wei, Q.; Sun, X.; Lu, W.; Wei, Q.; Ye, J.; Cai, X.; et al. Ginseng-derived nanoparticles alter macrophage polarization to inhibit melanoma growth. J. Immunother. Cancer 2019, 7, 326. [Google Scholar] [CrossRef] [PubMed]
- Pomatto, M.A.C.; Gai, C.; Negro, F.; Massari, L.; Deregibus, M.C.; Grange, C.; De Rosa, F.G.; Camussi, G. Plant-Derived Extracellular Vesicles as a Delivery Platform for RNA-Based Vaccine: Feasibility Study of an Oral and Intranasal SARS-CoV-2 Vaccine. Pharmaceutics 2023, 15, 974. [Google Scholar] [CrossRef]
- Del Pozo-Acebo, L.; López de Las Hazas, M.C.; Tomé-Carneiro, J.; Del Saz-Lara, A.; Gil-Zamorano, J.; Balaguer, L.; Chapado, L.A.; Busto, R.; Visioli, F.; Dávalos, A. Therapeutic potential of broccoli-derived extracellular vesicles as nanocarriers of exogenous miRNAs. Pharmacol. Res. 2022, 185, 106472. [Google Scholar] [CrossRef]
- Pomatto, M.A.C.; Gai, C.; Negro, F.; Massari, L.; Deregibus, M.C.; De Rosa, F.G.; Camussi, G. Oral Delivery of mRNA Vaccine by Plant-Derived Extracellular Vesicle Carriers. Cells 2023, 12, 1826. [Google Scholar] [CrossRef]
- López de Las Hazas, M.C.; Tomé-Carneiro, J.; Del Pozo-Acebo, L.; Del Saz-Lara, A.; Chapado, L.A.; Balaguer, L.; Rojo, E.; Espín, J.C.; Crespo, C.; Moreno, D.A.; et al. Therapeutic potential of plant-derived extracellular vesicles as nanocarriers for exogenous miRNAs. Pharmacol. Res. 2023, 198, 106999. [Google Scholar] [CrossRef]
- Wang, X.; Wu, B.; Sun, G.; He, W.; Gao, J.; Huang, T.; Liu, J.; Zhou, Q.; He, X.; Zhang, S.; et al. Selenium Biofortification Enhanced miR167a Expression in Broccoli Extracellular Vesicles Inducing Apoptosis in Human Pancreatic Cancer Cells by Targeting IRS1. Int. J. Nanomed. 2023, 18, 2431–2446. [Google Scholar] [CrossRef]
- Aquilano, K.; Ceci, V.; Gismondi, A.; De Stefano, S.; Iacovelli, F.; Faraonio, R.; Di Marco, G.; Poerio, N.; Minutolo, A.; Minopoli, G.; et al. Adipocyte metabolism is improved by TNF receptor-targeting small RNAs identified from dried nuts. Commun. Biol. 2019, 2, 317. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, D.; Chen, X.; Li, J.; Li, L.; Bian, Z.; Sun, F.; Lu, J.; Yin, Y.; Cai, X.; et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 2010, 39, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Li, X.; Liu, J.; Dong, L.; Chen, Q.; Liu, J.; Kong, H.; Zhang, Q.; Qi, X.; Hou, D.; et al. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015, 25, 39–49. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, H.; Sun, X.; Ding, M.; Tao, G.; Li, X. Honeysuckle-derived microRNA2911 directly inhibits varicella-zoster virus replication by targeting IE62 gene. J. Neurovirol. 2019, 25, 457–463. [Google Scholar] [CrossRef]
- Li, X.; Huang, Y.; Sun, M.; Ji, H.; Dou, H.; Hu, J.; Yan, Y.; Wang, X.; Chen, L. Honeysuckle-encoded microRNA2911 inhibits Enterovirus 71 replication via targeting VP1 gene. Antivir. Res. 2018, 152, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Chin, A.R.; Fong, M.Y.; Somlo, G.; Wu, J.; Swiderski, P.; Wu, X.; Wang, S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016, 26, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Cai, L.; Tian, Y.; Tu, Y.; Qiu, H.; Xie, G.; Huang, D.; Zheng, R.; Zhang, W. miR156a Mimic Represses the Epithelial-Mesenchymal Transition of Human Nasopharyngeal Cancer Cells by Targeting Junctional Adhesion Molecule A. PLoS ONE 2016, 11, e0157686. [Google Scholar] [CrossRef]
- Chen, X.; Wu, R.Z.; Zhu, Y.Q.; Ren, Z.M.; Tong, Y.L.; Yang, F.; Dai, G.H. Study on the inhibition of Mfn1 by plant-derived miR5338 mediating the treatment of BPH with rape bee pollen. BMC Complement. Altern. Med. 2018, 18, 38. [Google Scholar] [CrossRef]
- O’Neill, M.J.; Bourre, L.; Melgar, S.; O’Driscoll, C.M. Intestinal delivery of non-viral gene therapeutics: Physiological barriers and preclinical models. Drug Discov. Today 2011, 16, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, F.; Dong, L.; Wu, H.; Xu, J.; Li, H.; Wang, J.; Zhou, Z.; Liu, C.; Wang, Y.; et al. SIDT1-dependent absorption in the stomach mediates host uptake of dietary and orally administered microRNAs. Cell Res. 2021, 31, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Cubillos-Ruiz, A.; Guo, T.; Sokolovska, A.; Miller, P.F.; Collins, J.J.; Lu, T.K.; Lora, J.M. Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov. 2021, 20, 941–960. [Google Scholar] [CrossRef] [PubMed]
- Joga, M.R.; Zotti, M.J.; Smagghe, G.; Christiaens, O. RNAi Efficiency, Systemic Properties, and Novel Delivery Methods for Pest Insect Control: What We Know So Far. Front. Physiol. 2016, 7, 553. [Google Scholar] [CrossRef] [PubMed]
- Yogindran, S.; Rajam, M.V. Artificial miRNA-mediated silencing of ecdysone receptor (EcR) affects larval development and oogenesis in Helicoverpa armigera. Insect Biochem. Mol. Biol. 2016, 77, 21–30. [Google Scholar] [CrossRef]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.W.; Broeck, J.V. RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 2018, 9, 1912. [Google Scholar] [CrossRef]
- Chen, J.; Teotia, S.; Lan, T.; Tang, G. MicroRNA Techniques: Valuable Tools for Agronomic Trait Analyses and Breeding in Rice. Front. Plant Sci. 2021, 12, 744357. [Google Scholar] [CrossRef]
- Dávalos, A.; Henriques, R.; Latasa, M.J.; Laparra, M.; Coca, M. Literature review of baseline information on non-coding RNA (ncRNA) to support the risk assessment of ncRNA-based genetically modified plants for food and feed. EFSA Support. Publ. 2019, 16, 1688E. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).