Establishment of Polydopamine-Modified HK-2 Cell Membrane Chromatography and Screening of Active Components from Plantago asiatica L.
Abstract
1. Introduction
2. Results and Discussion
2.1. Optimization of Combination Ratio of PDA and Silica Gel
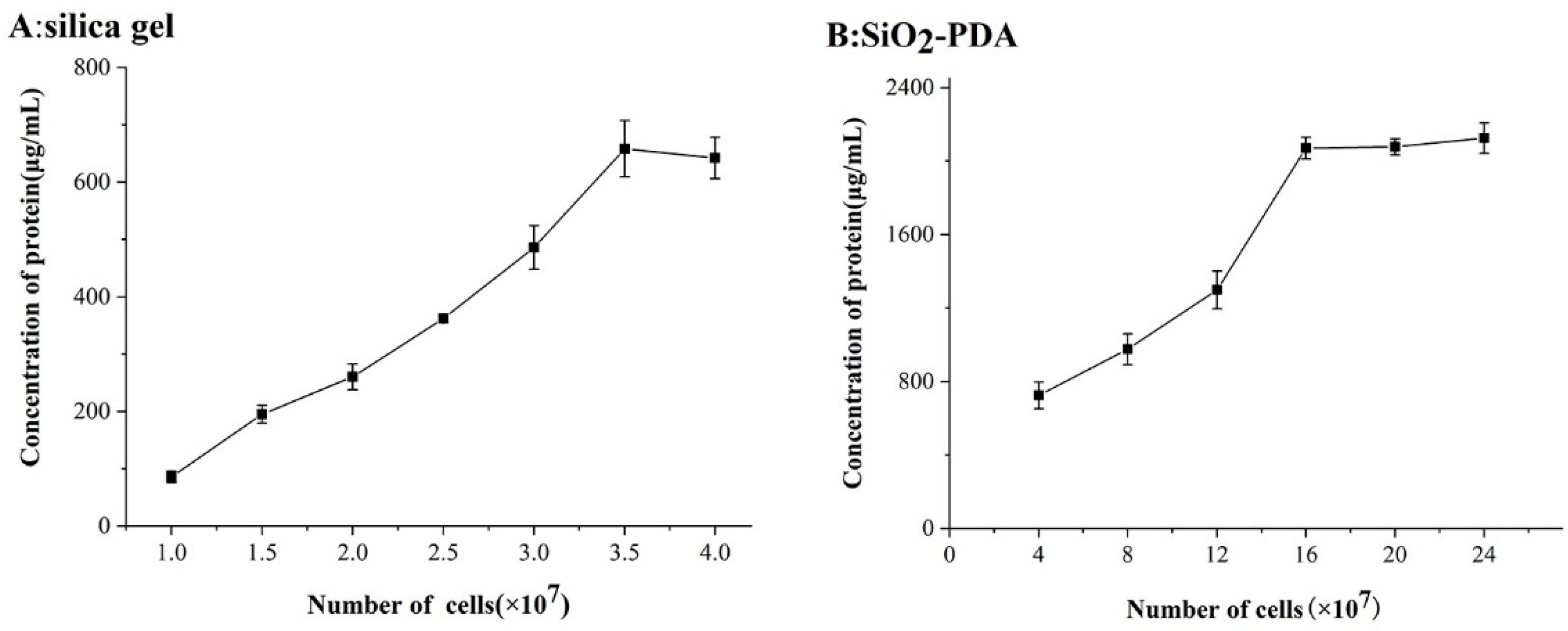
2.2. Optimization of Cell Dosage
2.3. Characterization of CMSP
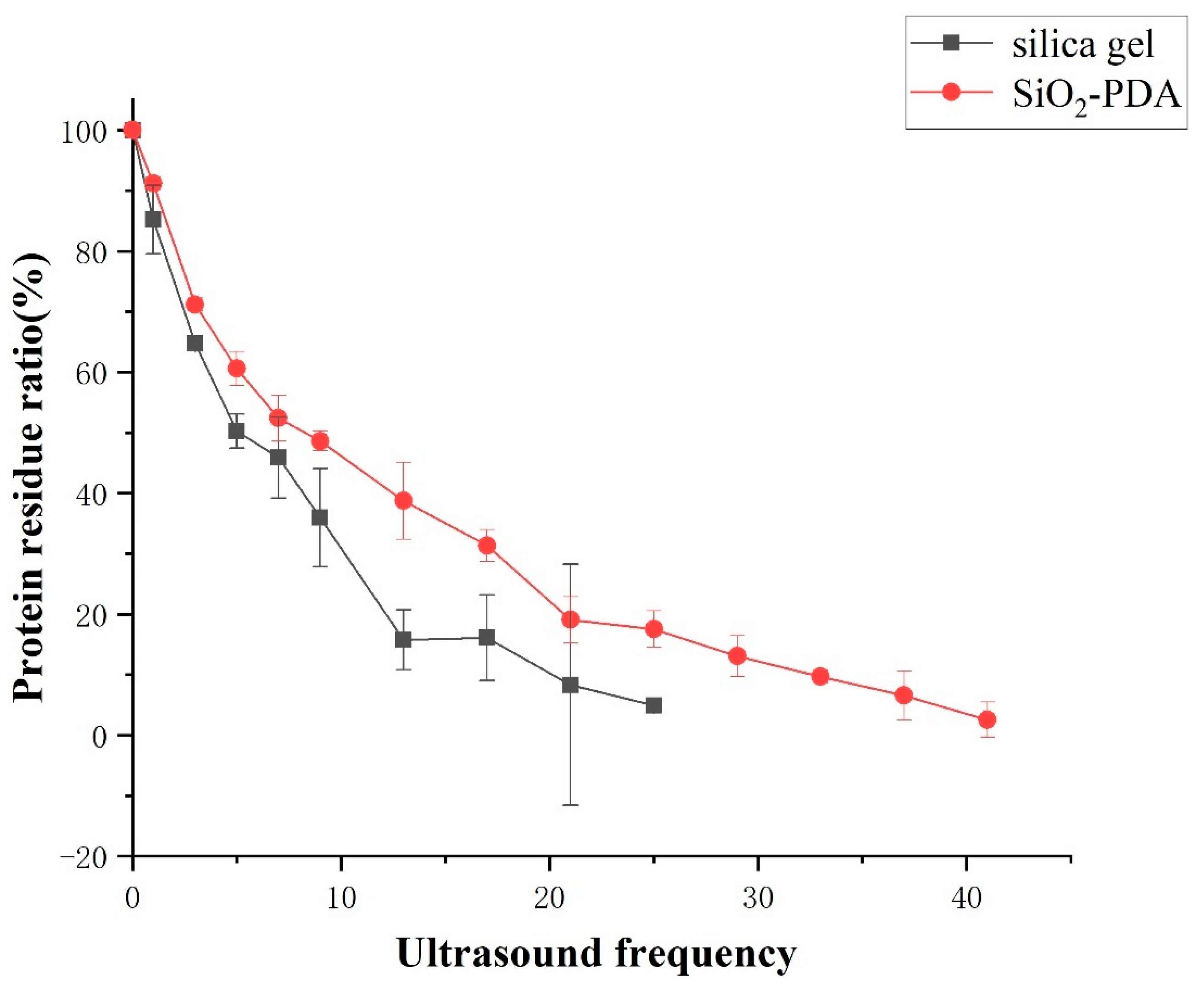
2.4. Comparison of Stability between Silica Gel and SiO2-PDA
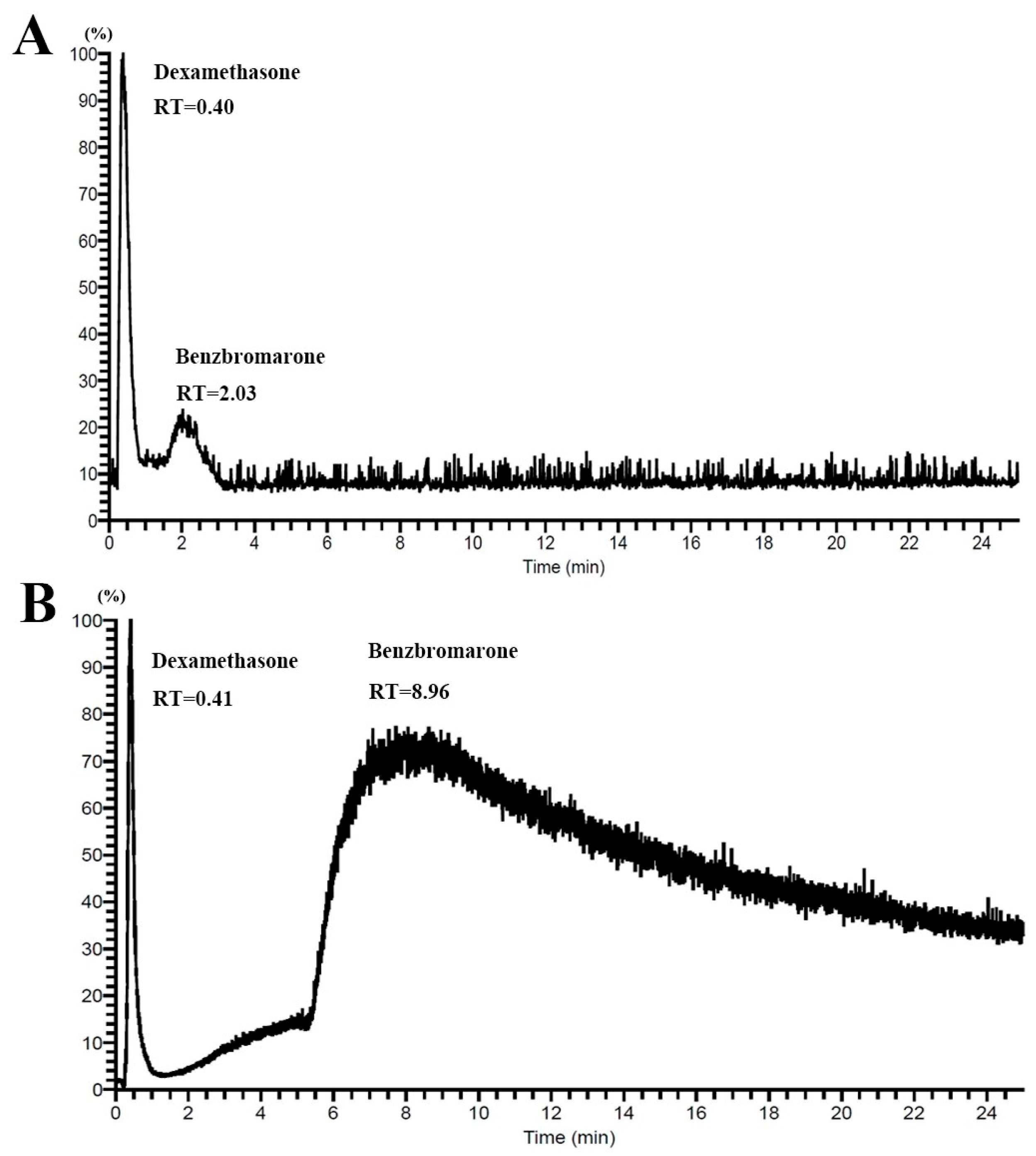
2.5. Research on the Comparative Selectivity of CMC
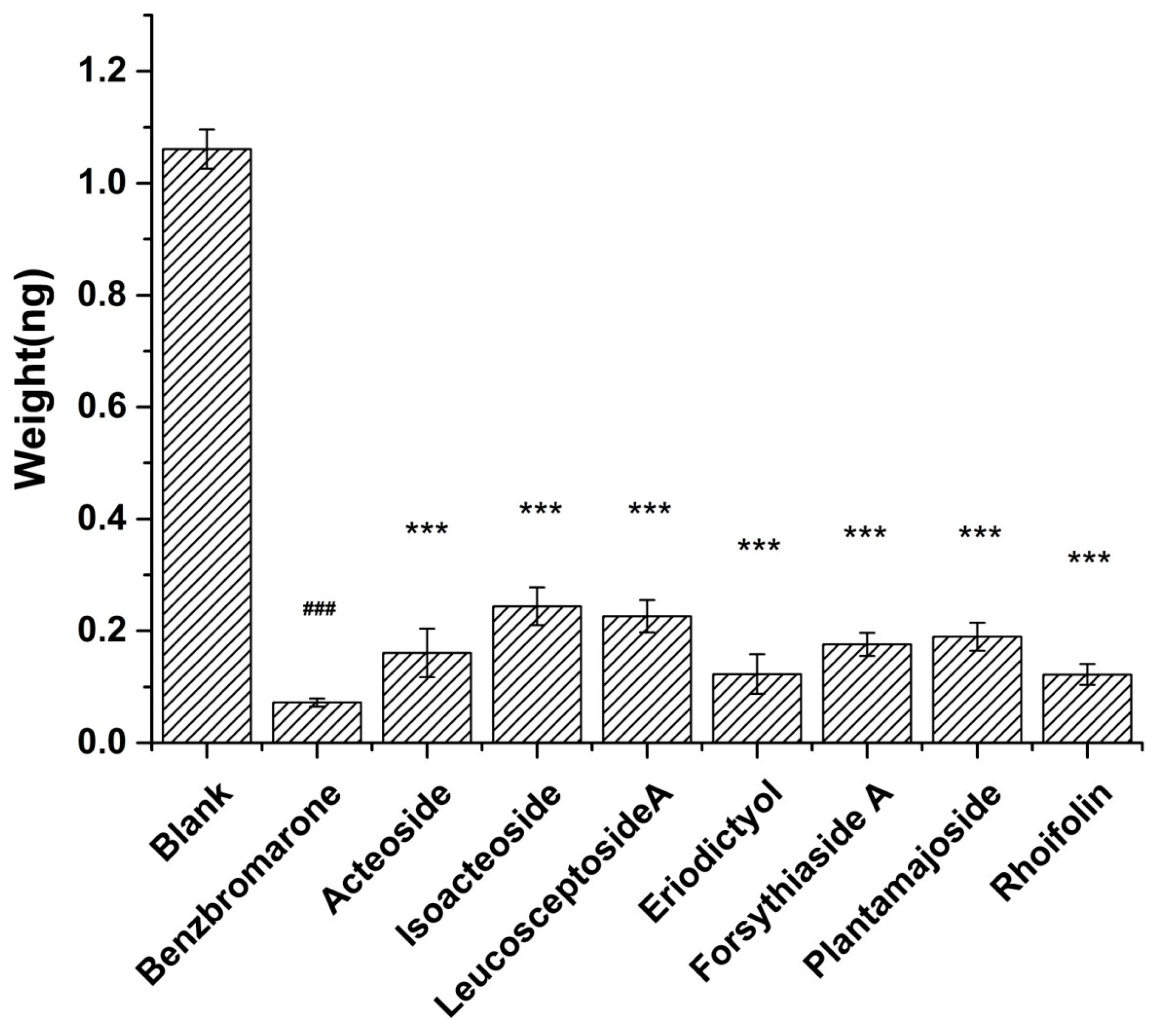
2.6. Screening Anti-Gout Compounds from the P2 Group of PAL
2.7. UA Reabsorption in HK-2 Cells
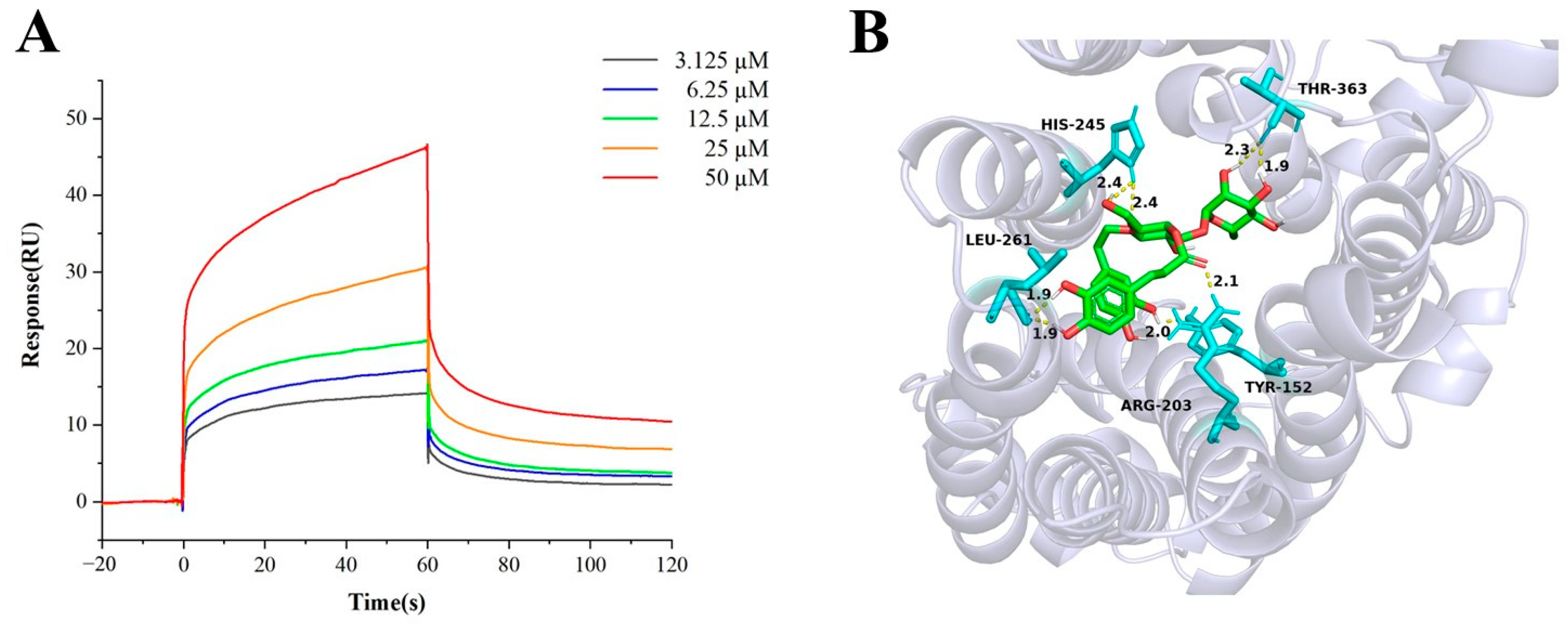
2.8. SPR Analysis and Molecular Docking
3. Materials and Methods
3.1. Materials and Reagents
3.2. Preparation and Component Identification of Samples Extract
3.3. Preparation of SiO2-PDA
3.4. Cell Culture, CMSP Preparation, and Characterization
3.5. CMC Column Preparation
3.6. CMC Analysis
3.7. UA Reabsorption of HK-2 Cells
3.8. SPR Affinity Analysis
3.9. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, L.; Yang, G.; Geng, X. Enzymatic activity and chromatographic characteristics of the cell membrane immobilized on silica surface. Chin. Sci. Bull. 1999, 44, 826–831. [Google Scholar] [CrossRef]
- He, L.; Wang, S.; Geng, X. Coating and fusing cell membranes onto a silica surface and their chromatographic characteristics. Chromatographia 2001, 54, 71–76. [Google Scholar] [CrossRef]
- Hou, X.; Wang, S.; Zhang, T.; Ma, J.; Zhang, J.; Zhang, Y.; Lu, W.; He, H.; He, L. Recent advances in cell membrane chromatography for traditional Chinese medicines analysis. J. Pharm. Biomed. Anal. 2014, 101, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wang, C.; Liu, R.; Wang, N.; Lv, Y.; Dai, B.; He, L. Advances in cell membrane chromatography. J. Chromatogr. A 2021, 1639, 461916. [Google Scholar] [CrossRef] [PubMed]
- Qv, X.; Jiang, J.; Piao, J. Pharmacodynamic studies of Chinese medicine at levels of whole animal, cell and molecular models. Curr. Med. Chem. 2010, 17, 4521–4537. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Dai, D.; Du, H.; Sun, M.; Wang, S. Prostate cell membrane Cchromatography-LC/MS method for screening α1A-adrenoceptor antagonists from Lotus Plumule. Chromatographia 2011, 74, 375–381. [Google Scholar] [CrossRef]
- Chen, C.; Gu, Y.; Wang, R.; Chai, X.; Jiang, S.; Wang, S.; Zhu, Z.; Chen, X.; Yuan, Y. Comparative two-dimensional GPC3 overexpressing SK-Hep1 cell membrane chromatography/C18/time-of-flight mass spectrometry for screening selective GPC3 inhibitor components from Scutellariae Radix. J. Chromatogr. B 2021, 1163, 122492. [Google Scholar] [CrossRef]
- Fu, J.; Jia, Q.; Liang, P.; Wang, S.; Zhou, H.; Zhang, L.; Wang, H.; Gao, C.; Lv, Y.; Han, S.; et al. Enhanced stability designs of cell membrane chromatography for screening drug leads. J. Sep. Sci. 2022, 45, 2498–2507. [Google Scholar] [CrossRef]
- Liao, F.; He, D.; Vong, C.; Wang, L.; Chen, Z.; Zhang, T.; Luo, H.; Wang, Y. Screening of the active Ingredients in Huanglian Jiedu decoction through amide bond-Immobilized magnetic nanoparticle-assisted cell membrane chromatography. Front. Pharmacol. 2022, 13, 1087404. [Google Scholar] [CrossRef]
- Wu, C.; Wang, N.; Xu, P.; Wang, X.; Shou, D.; Zhu, Y. Preparation and application of polyvinyl alcohol-decorated cell membrane chromatography for screening anti-osteoporosis components from Liuwei Dihuang decoction-containing serum. J. Sep. Sci. 2020, 43, 2105–2114. [Google Scholar] [CrossRef]
- Hu, M.; Huang, P.; Suo, L.; Wu, F. Polydopamine-based molecularly imprinting polymers on magnetic nanoparticles for recognition and enrichment of ochratoxins prior to their determination by HPLC. Microchim. Acta 2018, 185, 300. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Niu, X.; Zhang, W.; He, Y.; Pan, J.; Yan, Y.; Qiu, F. One-pot anchoring of Pd nanoparticles on nitrogendoped carbon through dopamine self-polymerization and activity in the electrocatalytic methanol oxidation reaction. Chemsuschem 2017, 10, 976–983. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yan, L.; Zhang, Z.; Wang, J.; Luo, N. Polydopamine-coated magnetic molecularly imprinted polymer for the selective solid-phase extraction of cinnamic acid, ferulic acid and caffeic acid from radix scrophulariae sample. J. Sep. Sci. 2016, 39, 1480–1488. [Google Scholar] [CrossRef]
- Yang, X.; Niu, X.H.; Mo, Z.L.; Guo, R.B.; Liu, N.J.; Zhao, P.; Liu, Z.Y. Electrochemical chiral interface based on the Michael addition/Schiff base reaction of polydopamine functionalized reduced graphene oxide. Electrochim. Acta 2019, 319, 705–715. [Google Scholar] [CrossRef]
- Ahmad, M.I.; Masood, S.; Furlanetto, D.M.; Nicolaou, S. Urate Crystals; Beyond Joints. Front. Med. 2021, 8, 649505. [Google Scholar] [CrossRef] [PubMed]
- Narang, R.K.; Dalbeth, N. Pathophysiology of Gout. Semin. Nephrol. 2020, 40, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Pascual, E.; Perdiguero, M. Gout, diuretics and the kidney. Ann. Rheum. Dis. 2006, 65, 981–982. [Google Scholar] [CrossRef]
- Ishikawa, T.; Takahashi, T.; Taniguchi, T.; Hosoya, T. Dotinurad: A novel selective urate reabsorption inhibitor for the treatment of hyperuricemia and gout. Expert Opin. Pharmacother. 2021, 22, 1397–1406. [Google Scholar] [CrossRef]
- Terkeltaub, R.; Bushinsky, D.A.; Becker, M.A. Recent developments in our understanding of the renal basis of hyperuricemia and the development of novel antihyperuricemic therapeutics. Arthritis. Res. Ther. 2006, 8, S4. [Google Scholar] [CrossRef][Green Version]
- Gao, H.; Liu, Z.; Song, F.; Xing, J.; Zheng, Z.; Liu, S. A Strategy for Identification and Structural Characterization of Compounds from Plantago asiatica L. by Liquid Chromatography-Mass Spectrometry Combined with Ion Mobility Spectrometry. Molecules 2022, 27, 4302. [Google Scholar] [CrossRef]
- Ding, X.; Chen, X.; Cao, Y.; Jia, D.; Wang, D.; Zhu, Z.; Zhang, J.; Hong, Z.; Chai, Y. Quality improvements of cell membrane chromatographic column. J. Chromatogr. A 2014, 1359, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.; Cheng, J.; Si, Y.; Chen, W.; Hou, J.; Zhao, T.; Gu, Y.; Lv, L.; Hong, Z.; Zhu, Z.; et al. A stop-flow comprehensive two-dimensional HK-2 and HK-2/CIKI cell membrane chromatography comparative analysis system for screening the active ingredients from Pyrrosia calvata (Bak.) Ching against crystal-induced kidney injury. J. Pharm. Biomed. Anal. 2021, 195, 113825. [Google Scholar]
- Gupta, A.; Sharma, P.K.; Misra, A.K.; Singh, S. Lesinurad: A significant advancement or just another addition to existing therapies of gout? J. Pharmacol. Pharmacother. 2016, 7, 155–158. [Google Scholar] [CrossRef]
- Madamsetty, V.S.; Mohammadinejad, R.; Uzieliene, I.; Nabavi, N.; Dehshahri, A.; Garcia-Couce, J.; Tavakol, S.; Moghassemi, S.; Dadashzadeh, A.; Makvandi, P.; et al. Dexamethasone: Insights into pharmacologicalaspects, therapeutic mechanisms, and delivery systems. ACS Biomater. Sci. Eng. 2022, 8, 1763–1790. [Google Scholar] [CrossRef] [PubMed]
- Tin, A.; Li, Y.; Brody, J.; Nutile, T.; Chu, A.; Huffman, J.; Yang, Q.; Chen, M.; Robinson-Cohen, C.; Mace, A.; et al. Large-scale whole-exome sequencing association studies identify rare functional variants influencing serum urate levels. Nat. Commun. 2018, 9, 4228. [Google Scholar] [CrossRef]
- Toyoda, Y.; Kawamura, Y.; Nakayama, A.; Nakaoka, H.; Higashino, T.; Shimizu, S.; Ooyama, H.; Morimoto, K.; Uchida, N.; Shigesawa, R.; et al. Substantial anti-gout effect conferred by common and rare dysfunctional variants of URAT1/SLC22A12. Rheumatology 2021, 60, 5224–5232. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.O.; Ohtomo, S.; Kiyokawa, J.; Nakagawa, T.; Yamane, M.; Lee, K.J.; Kim, K.H.; Kim, B.H.; Tanaka, J.; Kawabe, Y.; et al. Stronger uricosuric effects of the novel selective URAT1 inhibitor UR-1102 lowered plasma urate in tufted capuchin monkeys to a greater extent than benzbromarone. J. Pharmacol. Exp. Ther. 2016, 357, 157–166. [Google Scholar] [CrossRef]
- Enomoto, A.; Kimura, H.; Chairoungdua, A.; Shigeta, Y.; Jutabha, P.; Cha, S.; Hosoyamada, M.; Takeda, M.; Sekine, T.; Igarashi, T.; et al. Molecular identification of a renal urate-anion exchanger that regulates blood urate levels. Nature 2002, 417, 447–452. [Google Scholar] [CrossRef]



| Compound | Calculated m/z | Observed m/z | RT on SiO2/CMC (min) | RT on SiO2-PDA/CMC (min) | Formula |
|---|---|---|---|---|---|
| acteoside | 623.1976 | 623.2022 | 0.72 | 1.23 | C29H36O15 |
| isoacteoside | 623.1976 | 623.2001 | 0.72 | 1.23 | C29H36O15 |
| forsythiaside A | 623.1976 | 623.1992 | 0.72 | 1.23 | C29H36O15 |
| plantamajoside | 639.1925 | 639.1956 | 0.76 | 1.85 | C29H36O16 |
| leucoseptoside A | 637.2132 | 637.2151 | 0.79 | 2.50 | C30H38O15 |
| eriodictyol | 287.0556 | 287.0560 | 1.05 | 2.75 | C15H12O6 |
| rhoifolin | 577.1557 | 577.1572 | 1.35 | 2.95 | C27H30O14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.; Liu, Z.; Song, F.; Xing, J.; Zheng, Z.; Hou, Z.; Liu, S. Establishment of Polydopamine-Modified HK-2 Cell Membrane Chromatography and Screening of Active Components from Plantago asiatica L. Int. J. Mol. Sci. 2024, 25, 1153. https://doi.org/10.3390/ijms25021153
Gao H, Liu Z, Song F, Xing J, Zheng Z, Hou Z, Liu S. Establishment of Polydopamine-Modified HK-2 Cell Membrane Chromatography and Screening of Active Components from Plantago asiatica L. International Journal of Molecular Sciences. 2024; 25(2):1153. https://doi.org/10.3390/ijms25021153
Chicago/Turabian StyleGao, Hongxue, Zhiqiang Liu, Fengrui Song, Junpeng Xing, Zhong Zheng, Zong Hou, and Shu Liu. 2024. "Establishment of Polydopamine-Modified HK-2 Cell Membrane Chromatography and Screening of Active Components from Plantago asiatica L." International Journal of Molecular Sciences 25, no. 2: 1153. https://doi.org/10.3390/ijms25021153
APA StyleGao, H., Liu, Z., Song, F., Xing, J., Zheng, Z., Hou, Z., & Liu, S. (2024). Establishment of Polydopamine-Modified HK-2 Cell Membrane Chromatography and Screening of Active Components from Plantago asiatica L. International Journal of Molecular Sciences, 25(2), 1153. https://doi.org/10.3390/ijms25021153






