Abstract
SCN1A, the gene encoding for the Nav1.1 channel, exhibits dominant interneuron-specific expression, whereby variants disrupting the channel’s function affect the initiation and propagation of action potentials and neuronal excitability causing various types of epilepsy. Dravet syndrome (DS), the first described clinical presentation of SCN1A channelopathy, is characterized by severe myoclonic epilepsy in infancy (SMEI). Variants’ characteristics and other genetic or epigenetic factors lead to extreme clinical heterogeneity, ranging from non-epileptic conditions to developmental and epileptic encephalopathy (DEE). This current study reports on findings from 343 patients referred by physicians in hospitals and tertiary care centers in Greece between 2017 and 2023. Positive family history for specific neurologic disorders was disclosed in 89 cases and the one common clinical feature was the onset of seizures, at a mean age of 17 months (range from birth to 15 years old). Most patients were specifically referred for SCN1A investigation (Sanger Sequencing and MLPA) and only five for next generation sequencing. Twenty-six SCN1A variants were detected, including nine novel causative variants (c.4567A>Τ, c.5564C>A, c.2176+2T>C, c.3646G>C, c.4331C>A, c.1130_1131delGAinsAC, c.1574_1580delCTGAGGA, c.4620A>G and c.5462A>C), and are herein presented, along with subsequent genotype–phenotype associations. The identification of novel variants complements SCN1A databases extending our expertise on genetic counseling and patient and family management including gene-based personalized interventions.
1. Introduction
Epilepsy, a complex neurological disease characterized by spontaneous and recurrent seizures, affects 70 out of 100,000 individuals worldwide (~1–2% of global population). Classified into focal onset, generalized onset and unknown onset [1], the types of seizures can help define epilepsy as focal, generalized and combined focal and generalized, while when limited information hinders the determination of epilepsy as focal or generalized, it remains unknown. On a final level of diagnosis, epilepsy syndromes refer to clusters of features that tend to present together and may include specific seizure types, EEG and imaging findings and characteristic comorbidities such as neurodevelopmental deficits [2]. Treatment with antiepileptic medications (AEDs) is the first-line and the most common treatment of choice, completely controlling seizures in almost 70% of patients albeit not curing the underlying condition. In the remaining patients pharmaco-resistant epilepsy may be managed by surgery, electrical stimulation or dietary therapies (https://www.aans.org/en/Patients/Neurosurgical-Conditions-and-Treatments/Epilepsy, last assessed in 14 May 2024). The selection of proper approaches requires characterization and understanding of pathogenic mechanisms. As a complex disease, epilepsy extends to multiple levels of pathophysiology including inherent genetic alterations in the neurotransmission machinery components or acquired lesions linked to drug or other substance toxicity, strokes or infectious diseases and traumas/injuries [3]. Channelopathies, synaptopathies and interneuronopathies, as well as epigenetic dysregulation, neurodegeneration and mitochondrial deficiencies, are some of the genetic causes of epilepsy. Of those, channelopathies were for many years considered the prominent cause of epilepsy [4,5] and, although deposed in the presence of novel findings arising from next generation sequencing (NGS) approaches, still remain a major cause of variable types of epilepsy [6,7,8].
Voltage-gated Na+ channels share a common structure containing one α and one, or more, auxiliary β subunits. Amongst the nine well-characterized α subunit proteins, Nav1.1 is enriched in the central nervous system (CNS) and more specifically to the dendrites and cell bodies of excitatory neurons. Nav1.1 comprises four homologous domains (DI–DIV) each of which contains six α transmembrane helices (S1–S6) connected by loops. The S4 helix shapes the voltage sensors and the S5 and S6 helices, with their connecting loop, shape the ion-conducting pore [9]. SCN1A, the gene encoding for the Nav1.1 channel, has 26 exons, is located at 2q24.3 and exhibits dominant interneuron-specific expression. SCN1A pathogenic or likely pathogenic variants affect the initiation and propagation of action potentials and disrupt regular neuronal excitability, leading to various types of seizures [10]. Depending on the actual impact of each variant they can be specifically categorized as loss of function (LOF), partial loss of function (pLOF), decreased excitability (DE), gain of function (GOF), increased excitability (IE) and simultaneously gain–loss of function (G-LOF). Genotype–phenotype correlations have indicated that LOF variants affecting the pore region are mainly associated with SMEI although missense variants in the same region have been recorded in patients with generalized epilepsy febrile seizure + (GEFS+) [11,12]. Focal febrile seizures are usually linked to pLOF and G-LOF variants and GEFS+ to IE, DE, pLOF or GOF variants [12]. SCN1A variants are transmitted in an autosomal dominant pattern; however, incomplete penetrance and variable expressivity have been recorded, further adding to the great variability of phenotypes, the first of which is currently known as Dravet syndrome, previously SMEI, as described by the French neurologist Charlotte Dravet in 1978 [13]. Advances in the clinical description and genetic diagnosis of many patients has allowed the classification of a broad spectrum of SCN1A-related disorders, including DS, myoclonic-atonic epilepsy (MAE), epilepsy of infancy with migrating focal seizures (EIMFS), early-infantile developmental and epileptic encephalopathy (EIDEE), GEFS+ and partial epilepsy with febrile seizure plus (PEFS+), as well as non-epileptic disorders such as familial hemiplegic migraine (FHM), autism spectrum disorders (ASD) and arthrogryposis [14,15,16,17,18]. SCN1A-related disorders are somehow considered pharmaco-resistant, and the use of sodium channel blockers, such as phenytoin, carbamazepine, oxycarbazepine and lamotrigine, is contraindicated in patients with channel haploinsufficiency, where aggravation of seizures and consequent neurodevelopmental deficits have been observed [19]. Treatment with topiramate or valproic acid [20] or enhancement of GABA receptor activity via benzodiazepines or stiripentol present a better option which combined with a ketogenic diet seems to have better results particularly in respect to cognition [21].
To date, more than 2000 SCN1A variants have been recorded (Human Gene Mutation Database Gen-Locus Specific Database https://www.hgmd.cf.ac.uk/ac/index.php last assessed in 14 May 2024, and SCN1A database http://scn1a.caae.org.cn/index.php, last assessed in 14 May 2024) and associated with variable phenotypes. Besides a rather clear correlation between truncating loss of function variants and more severe presentations, further genotype–phenotype associations remain open to question [11,20].
Findings, and subsequent genotype–phenotype associations, from a cohort of patients referred for SCN1A genetic testing are herein presented to enrich SCN1A databases, exchange acquired knowledge and enhance our expertise on patient diagnosis, prognosis and management. This is particularly significant for differential diagnosis in the extremely clinically heterogeneous SCN1A-related disorders.
2. Results
Twenty-five variants were detected, nineteen with Sanger sequencing, one with multiplex ligation-dependent probe amplification (MLPA), one after clinical exome sequencing (CES) and four after whole exome sequencing (WES). Of those, nine were novel, with no previous reports in the literature or databases available (Families 1, 3, 5, 10, 14, 15, 19, 22 and Table 1). Eleven variants were missense, six nonsense, three frameshift, two splice site, one synonymous, one partial gene deletion and one whole gene deletion (Figure 1). Subsequent segregation analysis and family studies were available in 19 families and disclosed the de novo nature in 12 cases.

Table 1.
Characteristics of the novel variants detected in the current cohort.
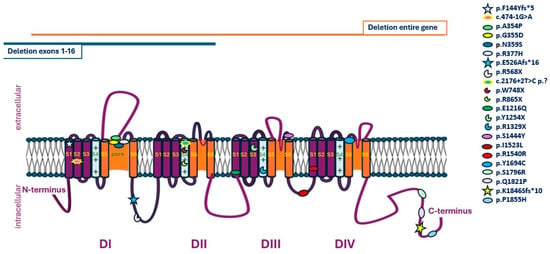
Figure 1.
Schematic representation of SCN1A variants disclosed in the current cohort.
Twenty-five patients (12 males, 13 females; mean age at referral: 4.5 years; 6 months–31 years), from 24 unrelated families, received a diagnosis of an SCN1A-related channelopathy. The earliest seizure onset was recorded in P22 on the fifth day of life and the latest in P5 and P13 at the age of 2 years (median age of onset 7.8 months). Well-recognized triggers of seizures included fever, infections and vaccinations while first symptoms varied from seizures (10), febrile seizures (5), febrile status epilepticus/complex seizures (5) and combinations of febrile and afebrile seizures (5).
Uneventful pregnancies and perinatal periods were reported for all patients while a family history of seizures or epilepsy was positive for 15 cases. The father of Patient 2 had febrile and afebrile seizures and transmitted the SCN1A:c.1064G>A variant to both his offspring, who had only febrile seizures. In Family 3, the patient experiencing her first episode of febrile seizures (FS) at the age of 6 months, while having a COVID-19 infection, inherited the SCN1A:c.2176+2T>C variant from her mother who experienced seizures between ages 6 and 15 years. The mother of Patient 5 reported epilepsy between ages 6 and 7 and has transmitted the SCN1A:c.3646G>C variant to her daughter with the onset of complex FS at age 2 during an upper respiratory infection. In Families 6 and 7, first cousins have inherited the SCN1A:c.5081A>G variant form their fathers who are brothers and both report seizures. Patient 21, a 10-month old female with status epilepticus (25 min long) during COVID-19 infection shared the same SCN1A:c.474-1G>A variant with her father (Figure 2). Table 2 summarizes data on the clinical presentation and the variants detected in all 25 cases.
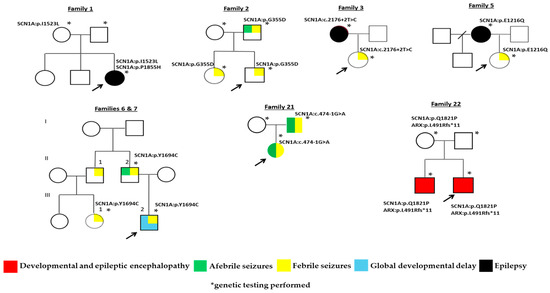
Figure 2.
Family trees of cases with transmission of pathogenic or likely pathogenic SCN1A variants.

Table 2.
Data on the clinical presentation and variants detected in cohort’s probands.
In respect to neurodevelopment, some patients presented with disorders affecting intellectual abilities including speech and/or motor skills and disorders within the autism spectrum (Patients 3, 6, 9–11, 15, 17–18, 20, 22–24).
3. Discussion
Consistent with the core phenotypic features ofDS, the first clinical presentation linked to SCN1A [13], 20/25 probands experienced their first seizure during infanthood and 16/25 had febrile seizures. In three patients, a genetic diagnosis was achieved immediately after the first episode and before the development of the complete clinical presentation. In older patients, additional types of seizures were recorded along with cognitive and learning difficulties. Triggers of a first episode included infections such as COVID-19, influenza Β or infections of the upper respiratory system (five patients), as well as vaccinations.
Cases of special interest comprise co-segregation of two variants either in SCN1A {p.(P1855H)/p.(I1523L) in Family 1} or SCN1A and ARX genes (Family 22). In the first case, p.(P1855H) is a pathogenic de novo variant considered causative, and p.(I1523L) maternally inherited and likely pathogenic. Recessive inheritance of SCN1A variants has been described before, in cases with high levels of consanguinity and variable clinical presentations, similar to those recorded in autosomal dominant transmission [23,24,25]. Variants detected were considered hypomorphic, interfering with SCN1A function and decreasing seizure threshold when combined. Studies of transgenic mouse models for the p.(R1648H) variant showed that the negative effect of the variant is somehow amplified in cases of homozygosity where mice suffer spontaneous generalized seizures and have a reduced life span [25]. In Patient 1, identification of the variants’ orientation as trans or cis was unsuccessful, hence hindering further evaluation of the possible clinical implications of a compound heterozygosity. Proline at position 1855 is very highly conserved and considered important for the protein function [26] and, although it is a novel variant, another substitution from proline to leucine p.(P1855L) has been previously recorded and reported as associated with DS [27]. SCN1A:p.I1523L is also a novel variant however a different substitution affecting the same amino acid, p.(I523T), has been recorded in a female patient with FS onset at 6 years of age, inducing developmental delay, and evolving to tonic–clonic and SE in adulthood [28].
In case 22, the recorded DEE was attributed to an ARX (MIM#300382) pathogenic frameshift variant in the context of the well described DEE type 1 (MIM#308350) [29]. Despite the evident association of the ARX variant with the severe phenotype of both siblings, the impact of SCN1A:p.Q1821P, a likely pathogenic maternal variant, remains elusive and may require further analysis. According to large patient series, some 3.2–7.2% may have multiple molecular diagnoses where different variants segregating independently are associated with distinct clinical entities with specific or overlapping presentations. Complex interactions between the different effects of pathogenic variants detected in more than one gene in a certain individual result in “blended phenotypes”, a term conceived to represent mixed clinical presentations [30]. Variants detected in epilepsy-related genes (DEPDC5, CHD2, SCN8A and IQSEC2) have been reported in patients with SCN1A variants and were considered to contribute to blended phenotypes comprising features of the disorder associated with the epilepsy gene and Dravet syndrome [31]. In general, the presence of possibly causative variants in phenotypically unaffected parents perplexes final classification and diagnosis. For SCN1A-related disorders, 10% of parents with (likely) pathogenic variants remain asymptomatic while offspring inheriting variants from affected parents have less than a 100% chance of developing seizures both due to incomplete penetrance and variable expressivity (https://www.ncbi.nlm.nih.gov/books/NBK1318/, SCN1A seizure disorders, Gene Reviews, last assessed in 14 May 2024). As with cases 1 and 22 the presence of variants in the apparently healthy mothers may complicate case resolution but should not be overlooked, especially in respect to prognosis, calculated risks for future pregnancies and genetic counseling.
Genotype–phenotype correlation studies assess variants in the context of type, location and impact on the channel produced including functional distribution of missense variants and truncation effects on disease onset [27]. Those affecting the crucial voltage sensor S4, or pore S5–S6 domains or their intermediate loop typically lead to severe phenotypes with multiple types of pharmaco-resistant seizures and neurodevelopmental delay [12]. In Families 11, 12 and 23, probands presenting with global developmental delay, albeit of variable severity possibly explained by their different developmental stages, all have nonsense variants affecting the S4 subunit. Variants affecting subunits S5–S6 and their connection loop were detected in Families 2, 6/7, 9, 10, 14 and 25 where, except for cases 9 and 25, a positive history for various types of seizures was reported. However, segregation analysis, available in all but Family 14, indicated transmission of variants only in Families 2 and 6/7 (Figure 2). Phenotypic variability was noted in Families 6/7, where confounding mild global developmental delay and febrile status epilepticus were the reasons for referral in the proband (III-2, Figure 2). The offspring of a male patient had a diagnosis of grand mal epilepsy and seizures initially febrile and then afebrile until the age of 18 years old. His brother (II-1, Figure 2) also reports persistent but non frequent (1–2 episodes annually) febrile seizures treated with sodium valproate and, although refusing genetic testing, seems to have transmitted the familial pathogenic variant to his daughter (III-1, Figure 2), diagnosed immediately after her first episode due to the positive family history.
Variants leading to haploinsufficiency, owing to the partial or complete loss of SCN1A, comprised deletions previously described and associated with increased severity. The 157.5kb deletion encompassing the SCN1A gene and a deletion including exons 1–16 were detected in Families 16 and 24, respectively. DEE, which is the most severe end of the SCN1A-related disease spectrum, was evident in both families, while in Family 24 additional clinical features, such as polydactyly and syndactyly (present also in the mother), were attributed to a GLI3 heterozygous pathogenic variant of maternal origin. Further supportive of the deleterious effect of abolishing gene function, frameshift or nonsense LOF variants affecting even regions outside S4, S5 and S6 helices were also detected in Families 4, 8, 15, 17, 18 and 20 where neurodevelopmental disorders such as GDD, DEE and ADHD were prominent (Table 2).
Splice site variants affecting nucleotides on ±1 or ±2 positions are widely annotated as LOF and usually considered diagnostic [32]. The SCN1A:c.474-1G>A variant, detected in Family 21, is an already known splice site variant, previously recorded de novo in a patient with classic DS [27]. Affected members of Family 21 (father and proband) present with what may seem to be clinically heterogeneous, whereby the father reports the onset of multiple seizures after a head injury at age 1 year old and through childhood while his daughter experienced the onset of afebrile focal and febrile generalized tonic–clonic seizures at 4.5 months and a status epilepticus during COVID-19 infection. WES disclosed no additional variants in epilepsy related genes. The SCN1A:c.2176+2T>C novel variant detected in Family 3 is also inherited and associated with clinical heterogeneity. The mother reports normal neurodevelopment and seizures between childhood and adolescence. Her daughter, now four years old, has motor and speech difficulties and experienced the first episode of febrile seizures at the age of 6 months during COVID-19 infection. Further supportive of variable expressivity, SCN1A:c.2176+2T>A affecting the exact same nucleotide was recorded in a patient with onset of afebrile seizure at age 5.1 months after Diphtheria-Tetanus-Pertussis (DTP) vaccination [33]. Of interest, despite the positive family history, in both cases, a genetic diagnosis was sought and achieved only after hospitalization, indicating that if not admitted they might have ended up undiagnosed.
Missense variants are always difficult to interpret, especially if no previous records and functional studies are available. In silico assessments may support final classification while segregation analysis may prove valuable when the variant is characterized either de novo in the proband or transmitted from an affected parent. The SCN1A:p.S1796R variant detected in Patient 13 affects a very highly conserved neutral serine on position 1796 at the C-terminus of the channel. Positively charged hydrophilic arginine presents an unfavored substitution and combined with its de novo presence seems to explain the complex febrile and afebrile seizures. SCN1A:p.E1216Q detected in proband 5 and her mother was deemed relevant to the clinical presentation of febrile and afebrile seizures. The variant is novel and according to the SCN1A database only one variant, a truncating p.(E1216X), on the same position has been recorded providing limited details on the possible effect of corresponding missense variants [34]. In respect to amino acid propensities the highly conserved glutamic acid, on position 1216, is hydrophilic with a negative charged side chain and important for the channel’s function [26]. The glutamine variant has a neutral side chain and may require functional studies towards better understanding of underlying mechanisms. Patient 10 presents with seizures and attention deficit hyperactivity disorder (ADHD) and has the de novo novel p.(S1444Y) variant. Functional data on the S1444Y variant are lacking and limited to in silico assessments. Serine 1444 is a conserved (although not highly) neutral residue, the substitution of which by tyrosine with an aromatic side chain is considered deleterious and associated with the disease [26]. The family reports seizures from the matrilineal lineage (maternal sister and niece). Targeted molecular analysis for the patient’s parents failed to disclose the variant and no further segregation was recommended. Families with a positive history of seizures and pathogenic variants detected de novo only in one proband are recorded in other cohorts as well [11]. Among genetic or non-genetic factors that could explain the unexpected absence of the specific variant in all affected family members, the well-documented phenomenon of parental mosaicism (reaching 8.6% in SCN1A [35]), as well as the possibility of diagnostic pitfalls of Sanger sequencing including human and technical errors, should be taken into account [36,37].
In Family 14, where two first cousins report febrile seizures, the parents refused to be tested, hence the de novo or inherited nature of the R377H variant could not be evaluated. Arginine 377 is a highly conserved hydrophilic variant for which a previously reported substitution by the neutral glutamine has been associated with GEFS+ or epilepsy and/or neurodevelopmental disorder (NDD) [38,39,40,41]. Histidine is also hydrophilic and might represent a better tolerated variant associated with milder presentations. The SCN1A p.R1540R synonymous variant was one of the first variants detected in this study. Characterized as appearing de novo in proband 19, it could only be assessed in silico, where Human Splicing Finder 3 [42] indicated that it possibly affects splicing through the breaking of the exonic splicing enhancer and silencer (ESEs and ESSs). Previous reports of synonymous variants with a pathogenic impact include the SCN1A:c.693A>T:p.P231P variant detected in a patient with generalized myoclonic seizures. P231P was considered pathogenic due to a predicted negative effect on normal splicing [43]. R1540R is currently re-classified as a variant of unknown significance for which further studies are needed to establish a robust association with the observed phenotype.
Clinical heterogeneity is a well acknowledged characteristic of SCN1A-related disorders whereby different presentations may be recorded even in family members with the same variant [11,44,45]. Concepts like incomplete penetrance and variable expressivity are most likely based in biology where genetic, including digenic, oligogenic and polygenic inheritance as well as double-hit contributions, and epigenetic factors are important players [30]. The impact of truncating variants on early seizure onset, and of the distribution of missense variants (depending on the functional importance of each area) may somehow explain the different clinical expression of variants. However, the phenotypic variation in identical genotypes, shown even by variable responses to antiepileptic treatment such as valproic acid, remains elusive [45]. Drug resistance in several cases indicates endogenous and somehow personalized responses possibly affected by modifying genetic and epigenetic factors. Genetic modifiers, distinct genes regulating gene expression and disease severity, include HLF, GABRA2, SCN2A, KCNQ2 and SCN8A and were identified as influencing phenotypic expression in mice models [46].
Investigation of epileptic phenotypes with NGS is expected to identify such additional genetic modifiers in cases with an SCN1A variant as well as detect a causative variant in negative cases. Whole exome sequencing was performed in 10 patients, where targeted SCN1A analysis disclosed no pathogenic variants. Pathogenic or likely pathogenic variants explaining the observed phenotypes were disclosed in five, comprising one in TGM6, one in NR2F1, one in AFF3 and two in PCDH19 [22], a gene known as a Dravet mimicker [15]. Dravet mimic genes, reported to cause phenotypes resembling DS, recorded up today include SCN2A, SCN8A, SCN9A, SCN1B, PCDH19, GABRA1, GABRG2, STXBP1, HCN1, CHD2 and KCNA2. These may also associate with distinct clinical presentations likely to shape novel clinical entities [15].
For many years now, channelopathies, mainly represented by SCN1A channelopathy, have been considered the prominent cause of epilepsy, a theory that seems to fade with advanced understanding of epileptogenesis. SCN1A-related disorders do cover a wide spectrum of clinical presentations, ranging from familial hemiplegic migraines to severe developmental epileptic encephalopathy, highlighting the variable expressivity and incomplete penetrance recorded with this gene. Targeted SCN1A Sanger sequencing, compensated by the Greek National Health System for hospitalized patients, has led to over-referrals and low diagnostic rates. Widely available NGS techniques, facilitating robust diagnosis and the additional identification of novel genes should become the first tier approach, proving to even become more cost-effective for national health systems.
Ongoing research in the field of epilepsy focuses on the identification of variants and predicted disease outcomes through in silico and functional assessments. Previous studies [47] have shown that, regardless of the reported phenotype, conventional in silico software correctly characterized benign from pathogenic variants in nearly 90% of cases, even if not able to differentiate disease severity (DS vs. GEFS+ vs. FHM), whereas patch-clamp mammalian expression systems analysis allowed evaluation of functional differences and genotype–phenotype correlations among missense variants. Braunkalus et al. [48] developed a prediction model to allow objective prognosis of disease course to DS or GEFS+ and although with expected difficulties concerning genetic and epigenetic modifiers this is expected to further assist disease management. Detection and characterization of variants is of paramount importance especially in view of emerging therapeutic approaches. Antisense oligonucleotides restoring functional SCN1A mRNA, adeno-associated virus delivered gene regulation and dCas9-based SCN1A gene activation are some of the genomics-driven therapies focusing on seizure control in SCN1A-related therapies [49,50,51].
In this context, and despite limitations concerning the small number of variants and the unavailability of extensive follow-up and functional assessments, findings from this current study are reported to enrich the SCN1A variant database and highlight related phenotypes. Acknowledgement of variants detected in patients with an SCN1A-related disorder facilitates diagnosis, prognosis and genetic counseling especially with respect to recurrence and disease risks [11,52]. Advanced understanding of SCN1A genotype–phenotype associations promotes development of gene-based interventions and better disease management crucial for the patients and their families.
4. Materials and Methods
4.1. Patients
This study was approved by the institutional boards and Ethics Committees of the National & Kapodistrian University of Athens and the St. Sophia’s Children’s Hospital (approval number is 12160/23-05-16). Written informed consent was obtained from all participants or their legal guardians. The study’s cohort included 343 patients (167 female and 176 male) referred by physicians in hospitals and tertiary care centers from the Hellenic territory, between 2017 and 2023. Positive family history for specific neurologic disorders was disclosed in 89 cases and is summarized in Table 3. The one common feature of all patients was the onset of seizures, at a mean age of 17 months (range from birth to 15 years old).

Table 3.
Positive family history recorded in the cohort.
Figure 3 summarizes the types of seizures or epileptic syndromes recorded. Electroencephalographic findings were available for 51.89% (178/343) of probands and imaging for 32.06% (110/343) (Figure 4 and Figure 5 respectively). Other clinical findings included: global developmental delay (82/343, 23.90%), speech delay (22/343, 6.41%), ASD (21/343, 6.12%), microcephaly (8/343, 2.33%), disorders of intellectual development (7/343, 2.04%), hypotonia (7/343, 2.04%), coarse facial features (6/343, 1.74%) and macrocephaly (2/343, 0.74%). Poor fine motor coordination, plagiocephaly, trigonocephaly, hydrocephalus, macroglossia, hypertonia, tremor, hemiplegia, ataxia, exostoses, cryptorchidism, hypogonadism, hyponatremia and enuresis nocturna were also reported in isolated cases.
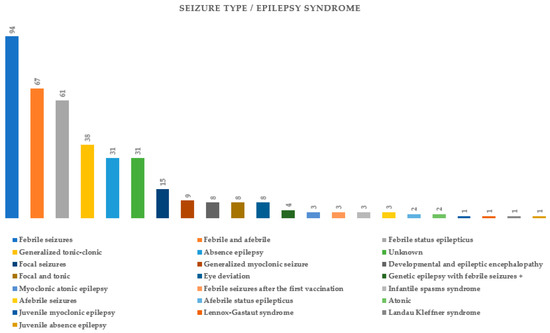
Figure 3.
Seizure types or diagnosed epileptic syndromes in probands tested for SCN1A variants.
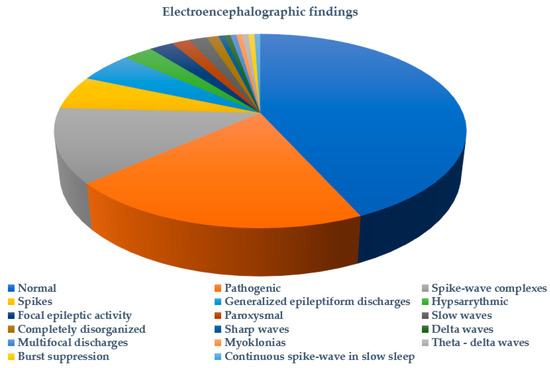
Figure 4.
Electroencephalographic findings in probands tested for SCN1A variants.
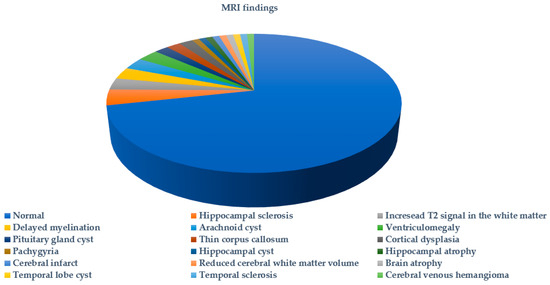
Figure 5.
MRI findings in probands tested for SCN1A variants.
4.2. Methods
Genomic DNA was extracted from peripheral blood lymphocytes with the QIAsymphony® DSP DNA technology (QIAGEN®, Hilden, Germany) according to the manufacturer’s protocols. Sanger sequencing for SCN1A was applied in 339 samples using the BigDyeTM Terminator sequencing kit (Thermo Fisher Scientific, Waltham, MA, USA) and ABI Prism 3500 Genetic Analyzer (Applied Biosystems, Woburn, MA, USA). For CNV detection, Multiplex ligation dependent probe amplification (MLPA®, kit P137, by MRC Holland, Amsterdam, The Netherlands) was performed in 41 samples (22 female and 19 male).
Whole exome sequencing (WES), performed for 4 probands, used the Human Core Exome (Twist Bioscience®, San Francisco, CA, USA) or xGenTM ExomeResearch v2 (Integrated DNA Technologies, Coralville, IA, USA) and CES was performed for 1 proband with the Clinical Exome Solution kit (by SOPHiA Genetics SA, Rolle, Switzerland) following the manufacturer’s recommendations. The resulting libraries were subjected to paired-end sequencing on an Illumina NextSeq® 500 (Illumina, San Diego, CA, USA). Library preparation and sequencing were outsourced (Genotypos-Science Labs, Athens, Greece, and NIPD Genetics, Nicosia, Cyprus). ES data quality acceptance parameters included a mean depth of coverage >50×, with >98% regions at 25×. Variant annotation was performed with the ANNOVAR algorithm and variant filtration with the VarAFT v2.16 Variant Analysis and Filtration Tool (http://varaft.eu, last assessed in 14 May 2024), or the VarSome Clinical platform (https://clinical.varsome.com/ last accessed on 14 May 2024) (Saphetor SA, Lausanne, Switzerland). Variant filtration was made for SCN1A gene (GRCh37/hg19) and CNV detection used the ExomeDepth software version 1.1.15 [53].
Variant classification followed the recommendations of the American College of Medical Genetics (ACMG) [32] and CNV classification followed those of the ACMG and Clinical Genome Resource (ClinGen) [54]. When appropriate, confirmation of biparental inheritance of short tandem repeat (STR) markers allowed the use of PS2 criterion for de novo variants.
Author Contributions
D.V. performed Sanger sequencing, MLPA, exome sequencing data analysis, variant interpretation, segregation analysis and drafted the article. V.T., M.K., P.V., G.N., T.T., R.P., K.K. (Konstantina Kosma), A.K., I.T. and P.M. examined patients and provided clinical information. K.K. (Kyriaki Kekou) performed part of the data analysis. J.T.-S. supervised the study and reviewed the article. C.S. performed data analysis, supervised the study and critically reviewed the article. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was approved by the institutional boards and Ethics Committees of the National & Kapodistrian University of Athens and the St. Sophia’s Children’s Hospital (approval number 12160/23-05-16).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors would like to thank all families participating in this study and Efthalia Daniel for her technical assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Fisher, R.S.; Cross, J.H.; D’Souza, C.; French, J.A.; Haut, S.R.; Higurashi, N.; Hirsch, E.; Jansen, F.E.; Lagae, L.; Moshé, S.L.; et al. Instruction manual for the ILAE 2017 operational classification of seizure types. Epilepsia 2017, 58, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Scheffer, I.E.; Berkovic, S.; Capovilla, G.; Connolly, M.B.; French, J.; Guilhoto, L.; Hirsch, E.; Jain, S.; Mathern, G.W.; Moshé, S.L.; et al. ILAE classification of the epilepsies: Position paper of the ILAE Commission for Classification and Terminology. Epilepsia 2017, 58, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Zheng, T.; Marra, V.; Yang, D.; Liu, J.K. Multi-scale modelling of the epileptic brain: Advantages of computational therapy exploration. J. Neural Eng. 2024, 21, 021002. [Google Scholar] [CrossRef] [PubMed]
- Gokben, S.; Onay, H.; Yilmaz, S.; Atik, T.; Serdaroglu, G.; Tekin, H.; Ozkinay, F. Targeted next generation sequencing: The diagnostic value in early-onset epileptic encephalopathy. Acta Neurol. Belg. 2017, 117, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Mei, D.; Parrini, E.; Marini, C.; Guerrini, R. The Impact of Next-Generation Sequencing on the Diagnosis and Treatment of Epilepsy in Paediatric Patients. Mol. Diagn. Ther. 2017, 21, 357–373. [Google Scholar] [CrossRef]
- Staley, K. Molecular mechanisms of epilepsy. Nat. Neurosci. 2015, 18, 367–372. [Google Scholar] [CrossRef]
- Stafstrom, C.E. Epilepsy: A review of selected clinical syndromes and advances in basic science. J. Cereb. Blood Flow Metab. 2006, 26, 983–1004. [Google Scholar] [CrossRef]
- Perucca, E.; French, J.; Bialer, M. Development of new antiepileptic drugs: Challenges, incentives, and recent advances. Lancet Neurol. 2007, 6, 793–804. [Google Scholar] [CrossRef]
- Brunklaus, A.; Lal, D. Sodium channel epilepsies and neurodevelopmental disorders: From disease mechanisms to clinical application. Dev. Med. Child Neurol. 2020, 62, 784–792. [Google Scholar] [CrossRef]
- Stafstrom, C.E. Severe epilepsy syndromes of early childhood: The link between genetics and pathophysiology with a focus on SCN1A mutations. J. Child Neurol. 2009, 24, 15s–23s. [Google Scholar] [CrossRef]
- Chen, C.; Fang, F.; Wang, X.; Lv, J.; Wang, X.; Jin, H. Phenotypic and Genotypic Characteristics of SCN1A Associated Seizure Diseases. Front. Mol. Neurosci. 2022, 15, 821012. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Xu, H.Q.; Yu, L.; Lin, G.W.; He, N.; Su, T.; Shi, Y.W.; Li, B.; Wang, J.; Liu, X.R.; et al. The SCN1A mutation database: Updating information and analysis of the relationships among genotype, functional alteration, and phenotype. Hum. Mutat. 2015, 36, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Dravet, C. The core Dravet syndrome phenotype. Epilepsia 2011, 52 (Suppl. 2), 3–9. [Google Scholar] [CrossRef]
- Scheffer, I.E.; Nabbout, R. SCN1A-related phenotypes: Epilepsy and beyond. Epilepsia 2019, 60 (Suppl. 3), S17–S24. [Google Scholar] [CrossRef] [PubMed]
- Steel, D.; Symonds, J.D.; Zuberi, S.M.; Brunklaus, A. Dravet syndrome and its mimics: Beyond SCN1A. Epilepsia 2017, 58, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Mei, D.; Cetica, V.; Marini, C.; Guerrini, R. Dravet syndrome as part of the clinical and genetic spectrum of sodium channel epilepsies and encephalopathies. Epilepsia 2019, 60 (Suppl. 3), S2–S7. [Google Scholar] [CrossRef] [PubMed]
- Goldberg-Stern, H.; Aharoni, S.; Afawi, Z.; Bennett, O.; Appenzeller, S.; Pendziwiat, M.; Kuhlenbäumer, G.; Basel-Vanagaite, L.; Shuper, A.; Korczyn, A.D.; et al. Broad phenotypic heterogeneity due to a novel SCN1A mutation in a family with genetic epilepsy with febrile seizures plus. J. Child Neurol. 2014, 29, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Jaber, D.; Gitiaux, C.; Blesson, S.; Marguet, F.; Buard, D.; Varela Salgado, M.; Kaminska, A.; Saada, J.; Fallet-Bianco, C.; Martinovic, J.; et al. De novo mutations of SCN1A are responsible for arthrogryposis broadening the SCN1A-related phenotypes. J. Med. Genet. 2021, 58, 737–742. [Google Scholar] [CrossRef]
- Scheffer, I.E.; Liao, J. Deciphering the concepts behind “Epileptic encephalopathy” and “Developmental and epileptic encephalopathy”. Eur. J. Paediatr. Neurol. EJPN Off. J. Eur. Paediatr. Neurol. Soc. 2020, 24, 11–14. [Google Scholar] [CrossRef]
- Zhou, P.; He, N.; Zhang, J.W.; Lin, Z.J.; Wang, J.; Yan, L.M.; Meng, H.; Tang, B.; Li, B.M.; Liu, X.R.; et al. Novel mutations and phenotypes of epilepsy-associated genes in epileptic encephalopathies. Genes Brain Behav. 2018, 17, e12456. [Google Scholar] [CrossRef]
- Wheless, J.W.; Fulton, S.P.; Mudigoudar, B.D. Dravet Syndrome: A Review of Current Management. Pediatr. Neurol. 2020, 107, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Veltra, D.; Tilemis, F.N.; Marinakis, N.M.; Svingou, M.; Mitrakos, A.; Kosma, K.; Tsoutsou, I.; Makrythanasis, P.; Theodorou, V.; Katsalouli, M.; et al. Combined exome analysis and exome depth assessment achieve a high diagnostic yield in an epilepsy case series, revealing significant genomic heterogeneity and novel mechanisms. Expert Rev. Mol. Diagn. 2023, 23, 85–103. [Google Scholar] [CrossRef]
- Brunklaus, A.; Ellis, R.; Stewart, H.; Aylett, S.; Reavey, E.; Jefferson, R.; Jain, R.; Chakraborty, S.; Jayawant, S.; Zuberi, S.M. Homozygous mutations in the SCN1A gene associated with genetic epilepsy with febrile seizures plus and Dravet syndrome in 2 families. Eur. J. Paediatr. Neurol. EJPN Off. J. Eur. Paediatr. Neurol. Soc. 2015, 19, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Marco Hernández, A.V.; Tomás Vila, M.; Caro Llopis, A.; Monfort, S.; Martinez, F. Case Report: Novel Homozygous Likely Pathogenic SCN1A Variant with Autosomal Recessive Inheritance and Review of the Literature. Front. Neurol. 2021, 12, 784892. [Google Scholar] [CrossRef]
- Martin, M.S.; Dutt, K.; Papale, L.A.; Dubé, C.M.; Dutton, S.B.; de Haan, G.; Shankar, A.; Tufik, S.; Meisler, M.H.; Baram, T.Z.; et al. Altered function of the SCN1A voltage-gated sodium channel leads to gamma-aminobutyric acid-ergic (GABAergic) interneuron abnormalities. J. Biol. Chem. 2010, 285, 9823–9834. [Google Scholar] [CrossRef]
- Stephenson, J.D.; Laskowski, R.A.; Nightingale, A.; Hurles, M.E.; Thornton, J.M. VarMap: A web tool for mapping genomic coordinates to protein sequence and structure and retrieving protein structural annotations. Bioinformatics 2019, 35, 4854–4856. [Google Scholar] [CrossRef] [PubMed]
- Zuberi, S.M.; Brunklaus, A.; Birch, R.; Reavey, E.; Duncan, J.; Forbes, G.H. Genotype-phenotype associations in SCN1A-related epilepsies. Neurology 2011, 76, 594–600. [Google Scholar] [CrossRef]
- Catarino, C.B.; Liu, J.Y.; Liagkouras, I.; Gibbons, V.S.; Labrum, R.W.; Ellis, R.; Woodward, C.; Davis, M.B.; Smith, S.J.; Cross, J.H.; et al. Dravet syndrome as epileptic encephalopathy: Evidence from long-term course and neuropathology. Brain J. Neurol. 2011, 134, 2982–3010. [Google Scholar] [CrossRef] [PubMed]
- Kato, M.; Dobyns, W.B. X-linked lissencephaly with abnormal genitalia as a tangential migration disorder causing intractable epilepsy: Proposal for a new term, “interneuronopathy”. J. Child Neurol. 2005, 20, 392–397. [Google Scholar] [CrossRef]
- Posey, J.E.; Harel, T.; Liu, P.; Rosenfeld, J.A.; James, R.A.; Coban Akdemir, Z.H.; Walkiewicz, M.; Bi, W.; Xiao, R.; Ding, Y.; et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N. Engl. J. Med. 2017, 376, 21–31. [Google Scholar] [CrossRef]
- Martins Custodio, H.; Clayton, L.M.; Bellampalli, R.; Pagni, S.; Silvennoinen, K.; Caswell, R.; Brunklaus, A.; Guerrini, R.; Koeleman, B.P.C.; Lemke, J.R.; et al. Widespread genomic influences on phenotype in Dravet syndrome, a ‘monogenic’ condition. Brain J. Neurol. 2023, 146, 3885–3897. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Verbeek, N.E.; van der Maas, N.A.; Jansen, F.E.; van Kempen, M.J.; Lindhout, D.; Brilstra, E.H. Prevalence of SCN1A-related dravet syndrome among children reported with seizures following vaccination: A population-based ten-year cohort study. PLoS ONE 2013, 8, e65758. [Google Scholar] [CrossRef]
- Yang, X.; Liu, A.; Xu, X.; Yang, X.; Zeng, Q.; Ye, A.Y.; Yu, Z.; Wang, S.; Huang, A.Y.; Wu, X.; et al. Genomic mosaicism in paternal sperm and multiple parental tissues in a Dravet syndrome cohort. Sci. Rep. 2017, 7, 15677. [Google Scholar] [CrossRef]
- Xu, X.; Yang, X.; Wu, Q.; Liu, A.; Yang, X.; Ye, A.Y.; Huang, A.Y.; Li, J.; Wang, M.; Yu, Z.; et al. Amplicon Resequencing Identified Parental Mosaicism for Approximately 10% of “de novo” SCN1A Mutations in Children with Dravet Syndrome. Hum. Mutat. 2015, 36, 861–872. [Google Scholar] [CrossRef]
- Djémié, T.; Weckhuysen, S.; von Spiczak, S.; Carvill, G.L.; Jaehn, J.; Anttonen, A.K.; Brilstra, E.; Caglayan, H.S.; de Kovel, C.G.; Depienne, C.; et al. Pitfalls in genetic testing: The story of missed SCN1A mutations. Mol. Genet. Genom. Med. 2016, 4, 457–464. [Google Scholar] [CrossRef]
- de Lange, I.M.; Koudijs, M.J.; van ‘t Slot, R.; Sonsma, A.C.M.; Mulder, F.; Carbo, E.C.; van Kempen, M.J.A.; Nijman, I.J.; Ernst, R.F.; Savelberg, S.M.C.; et al. Assessment of parental mosaicism in SCN1A-related epilepsy by single-molecule molecular inversion probes and next-generation sequencing. J. Med. Genet. 2019, 56, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Zucca, C.; Redaelli, F.; Epifanio, R.; Zanotta, N.; Romeo, A.; Lodi, M.; Veggiotti, P.; Airoldi, G.; Panzeri, C.; Romaniello, R.; et al. Cryptogenic epileptic syndromes related to SCN1A: Twelve novel mutations identified. Arch. Neurol. 2008, 65, 489–494. [Google Scholar] [CrossRef]
- Cetica, V.; Chiari, S.; Mei, D.; Parrini, E.; Grisotto, L.; Marini, C.; Pucatti, D.; Ferrari, A.; Sicca, F.; Specchio, N.; et al. Clinical and genetic factors predicting Dravet syndrome in infants with SCN1A mutations. Neurology 2017, 88, 1037–1044. [Google Scholar] [CrossRef]
- Lindy, A.S.; Stosser, M.B.; Butler, E.; Downtain-Pickersgill, C.; Shanmugham, A.; Retterer, K.; Brandt, T.; Richard, G.; McKnight, D.A. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia 2018, 59, 1062–1071. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, Y.; Sun, H.; Liu, X.; Yang, X.; Xiong, H.; Jiang, Y.; Bao, X.; Wang, S.; Yang, Z.; et al. Early clinical features and diagnosis of Dravet syndrome in 138 Chinese patients with SCN1A mutations. Brain Dev. 2014, 36, 676–681. [Google Scholar] [CrossRef] [PubMed]
- Desmet, F.O.; Hamroun, D.; Lalande, M.; Collod-Beroud, G.; Claustres, M.; Beroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef] [PubMed]
- Lemke, J.R.; Riesch, E.; Scheurenbrand, T.; Schubach, M.; Wilhelm, C.; Steiner, I.; Hansen, J.; Courage, C.; Gallati, S.; Bürki, S.; et al. Targeted next generation sequencing as a diagnostic tool in epileptic disorders. Epilepsia 2012, 53, 1387–1398. [Google Scholar] [CrossRef]
- Brunklaus, A.; Leu, C.; Gramm, M.; Pérez-Palma, E.; Iqbal, S.; Lal, D. Time to move beyond genetics towards biomedical data-driven translational genomic research in severe paediatric epilepsies. Eur. J. Paediatr. Neurol. EJPN Off. J. Eur. Paediatr. Neurol. Soc. 2020, 24, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Schuster, J.; Lu, X.; Dang, Y.; Klar, J.; Wenz, A.; Dahl, N.; Chen, X. Epigenetic insights into GABAergic development in Dravet Syndrome iPSC and therapeutic implications. bioRxiv 2023. [Google Scholar] [CrossRef]
- Kearney, J.A. Genetic modifiers of neurological disease. Curr. Opin. Genet. Dev. 2011, 21, 349–353. [Google Scholar] [CrossRef]
- Brunklaus, A.; Schorge, S.; Smith, A.D.; Ghanty, I.; Stewart, K.; Gardiner, S.; Du, J.; Pérez-Palma, E.; Symonds, J.D.; Collier, A.C.; et al. SCN1A variants from bench to bedside-improved clinical prediction from functional characterization. Hum. Mutat. 2020, 41, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Brunklaus, A.; Pérez-Palma, E.; Ghanty, I.; Xinge, J.; Brilstra, E.; Ceulemans, B.; Chemaly, N.; de Lange, I.; Depienne, C.; Guerrini, R.; et al. Development and Validation of a Prediction Model for Early Diagnosis of SCN1A-Related Epilepsies. Neurology 2022, 98, e1163–e1174. [Google Scholar] [CrossRef]
- Yamagata, T.; Raveau, M.; Kobayashi, K.; Miyamoto, H.; Tatsukawa, T.; Ogiwara, I.; Itohara, S.; Hensch, T.K.; Yamakawa, K. CRISPR/dCas9-based Scn1a gene activation in inhibitory neurons ameliorates epileptic and behavioral phenotypes of Dravet syndrome model mice. Neurobiol. Dis. 2020, 141, 104954. [Google Scholar] [CrossRef]
- Colasante, G.; Lignani, G.; Brusco, S.; Di Berardino, C.; Carpenter, J.; Giannelli, S.; Valassina, N.; Bido, S.; Ricci, R.; Castoldi, V.; et al. dCas9-Based Scn1a Gene Activation Restores Inhibitory Interneuron Excitability and Attenuates Seizures in Dravet Syndrome Mice. Mol. Ther. 2020, 28, 235–253. [Google Scholar] [CrossRef]
- Tanenhaus, A.; Stowe, T.; Young, A.; McLaughlin, J.; Aeran, R.; Lin, I.W.; Li, J.; Hosur, R.; Chen, M.; Leedy, J.; et al. Cell-Selective Adeno-Associated Virus-Mediated SCN1A Gene Regulation Therapy Rescues Mortality and Seizure Phenotypes in a Dravet Syndrome Mouse Model and Is Well Tolerated in Nonhuman Primates. Hum. Gene Ther. 2022, 33, 579–597. [Google Scholar] [CrossRef] [PubMed]
- Zeng, B.; Zhang, H.; Lu, Q.; Fu, Q.; Yan, Y.; Lu, W.; Ma, P.; Feng, C.; Qin, J.; Luo, L.; et al. Identification of five novel SCN1A variants. Front. Behav. Neurosci. 2023, 17, 1272748. [Google Scholar] [CrossRef] [PubMed]
- Tilemis, F.N.; Marinakis, N.M.; Veltra, D.; Svingou, M.; Kekou, K.; Mitrakos, A.; Tzetis, M.; Kosma, K.; Makrythanasis, P.; Traeger-Synodinos, J.; et al. Germline CNV Detection through Whole-Exome Sequencing (WES) Data Analysis Enhances Resolution of Rare Genetic Diseases. Genes 2023, 14, 1490. [Google Scholar] [CrossRef]
- Riggs, E.R.; Andersen, E.F.; Cherry, A.M.; Kantarci, S.; Kearney, H.; Patel, A.; Raca, G.; Ritter, D.I.; South, S.T.; Thorland, E.C.; et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet. Med. Off. J. Am. Coll. Med. Genet. 2020, 22, 245–257. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).