Adar Regulates Drosophila melanogaster Spermatogenesis via Modulation of BMP Signaling
Abstract
1. Introduction
2. Results
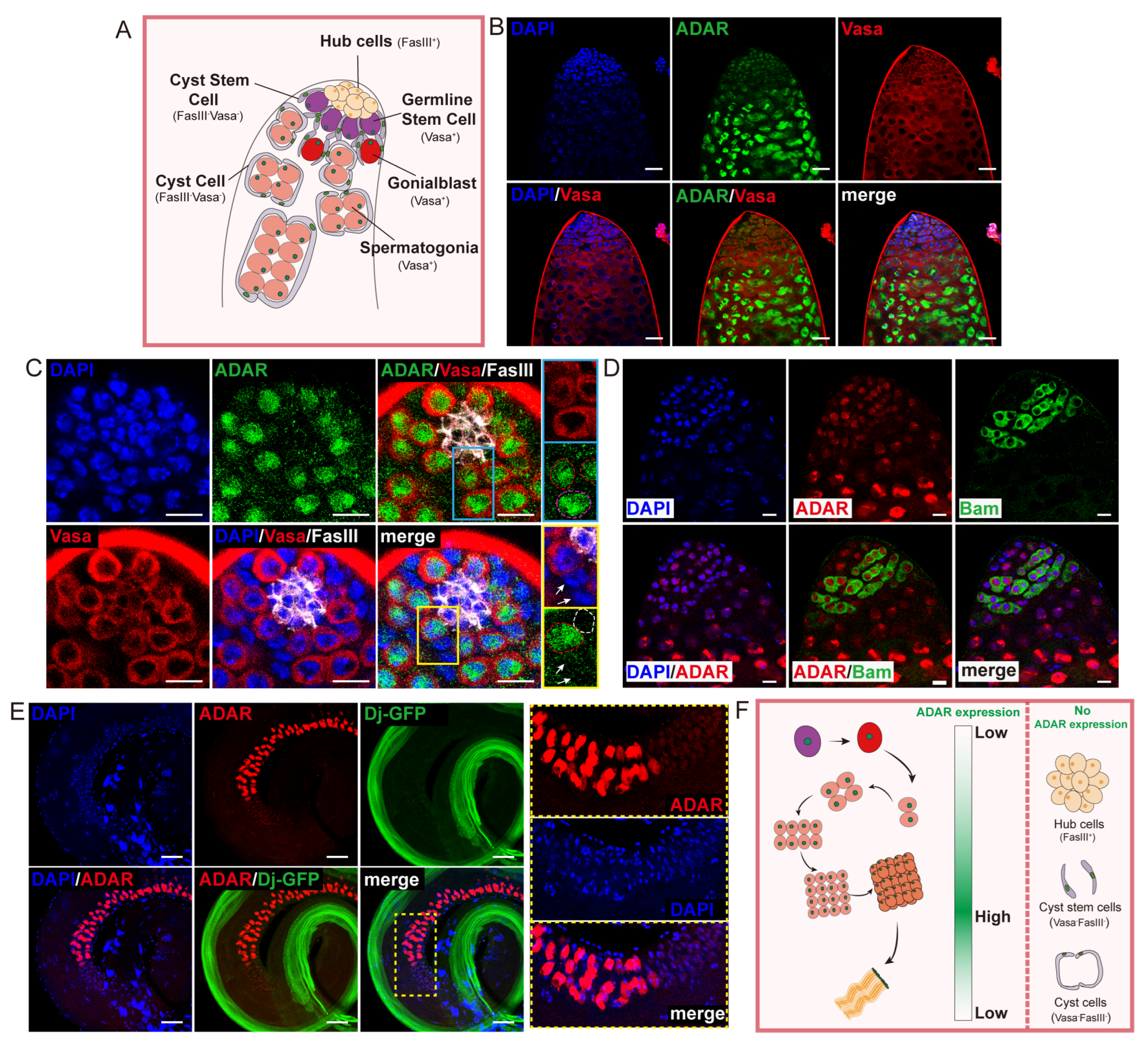
2.1. ADAR Highly Expressed in Drosophila Male Germline Cells
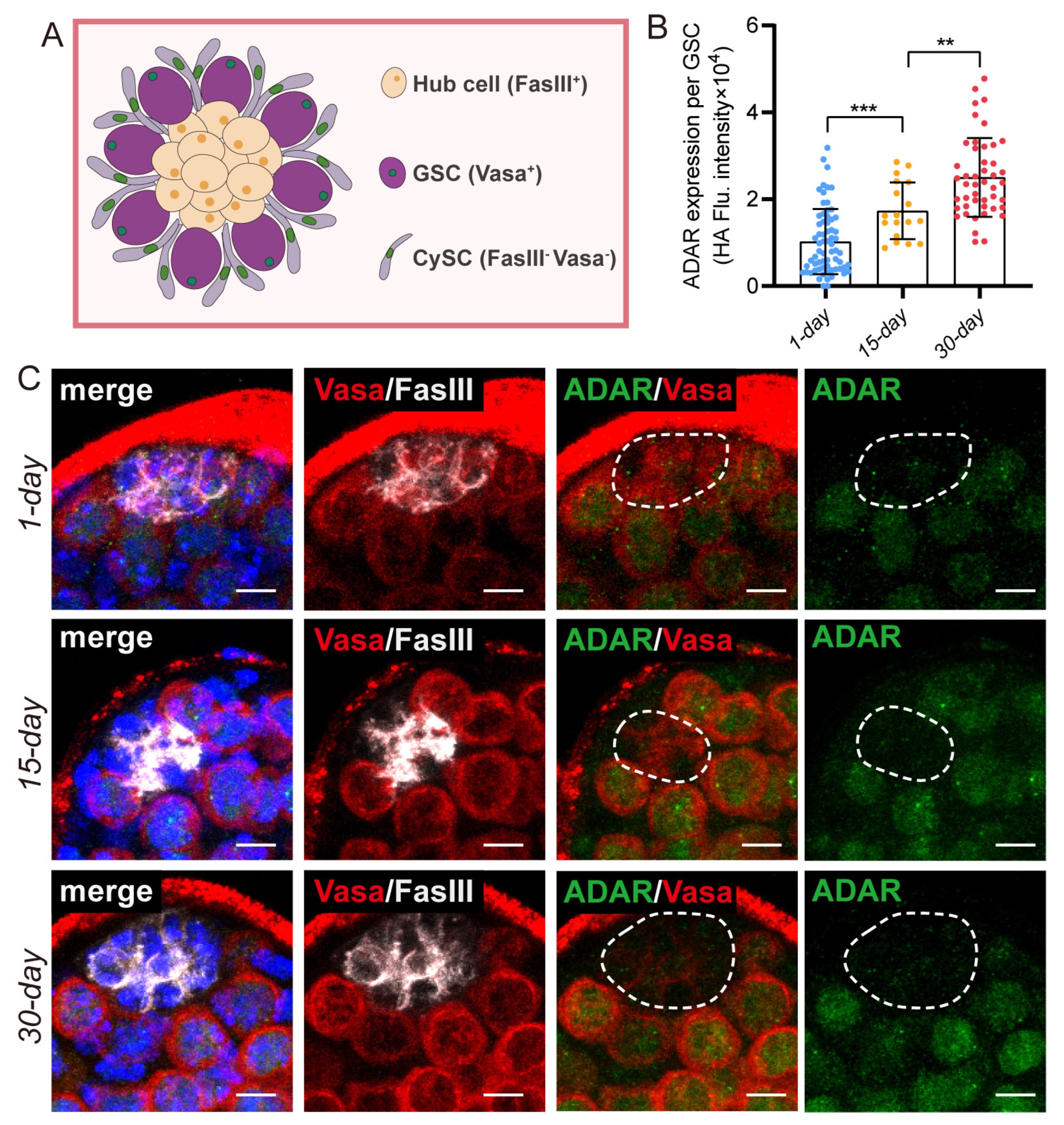
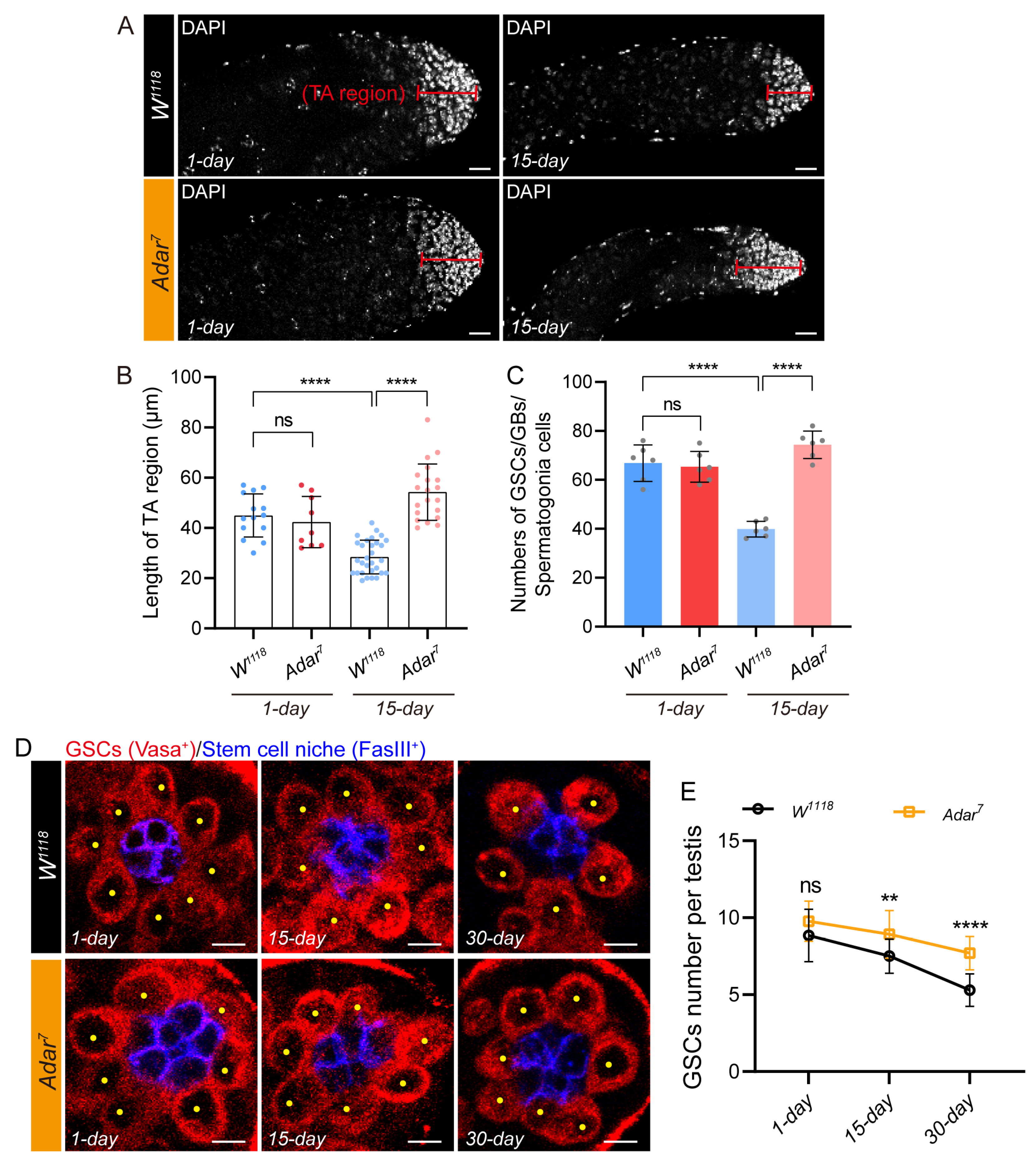
2.2. Adar Loss Suppresses Male-Germline-Cell Differentiation in Drosophila Testis upon Aging
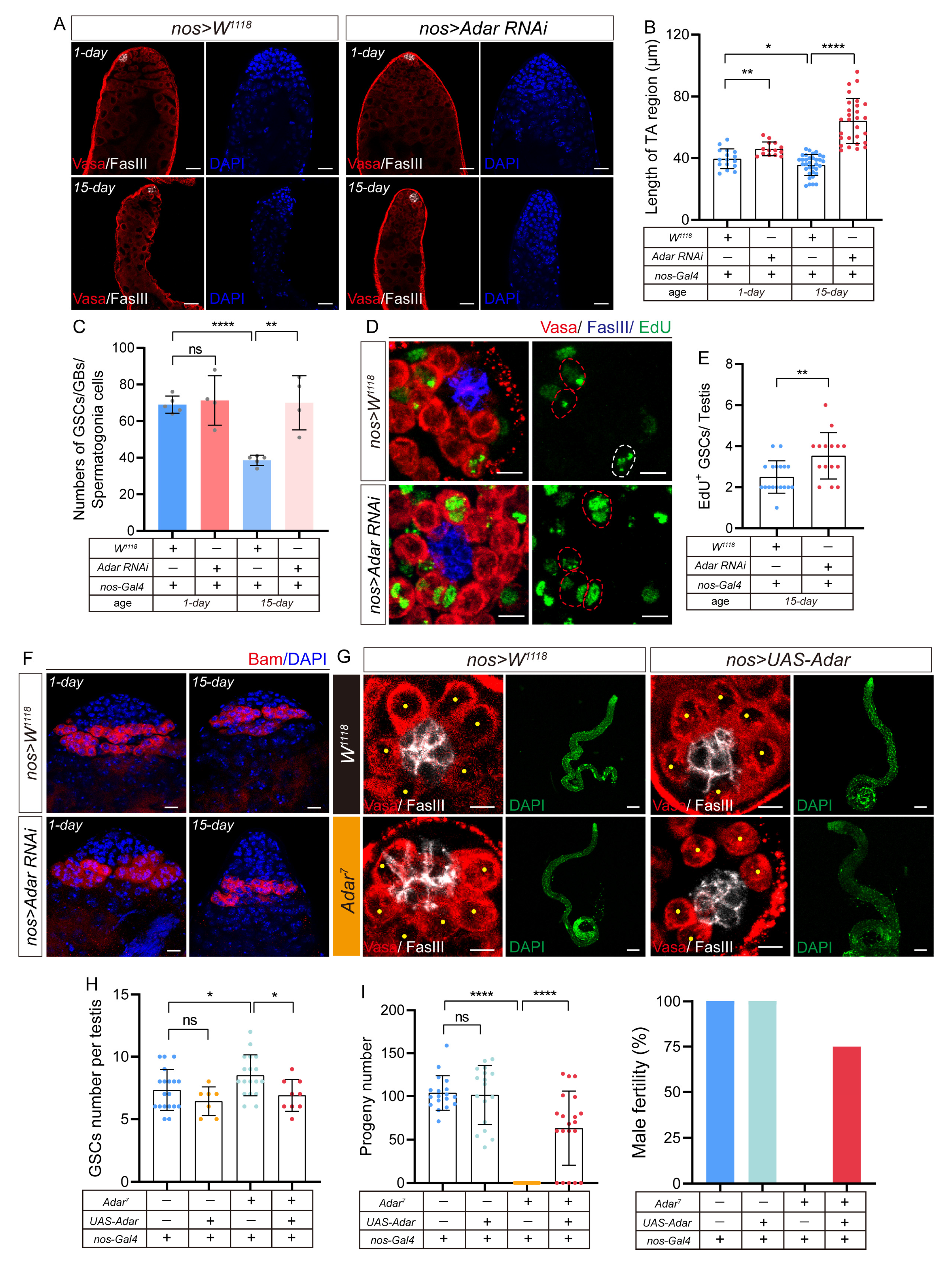
2.3. Depletion of ADAR in GSCs Leads to the Accumulation of Transit-Amplifying Germline Cells in Drosophila Testis
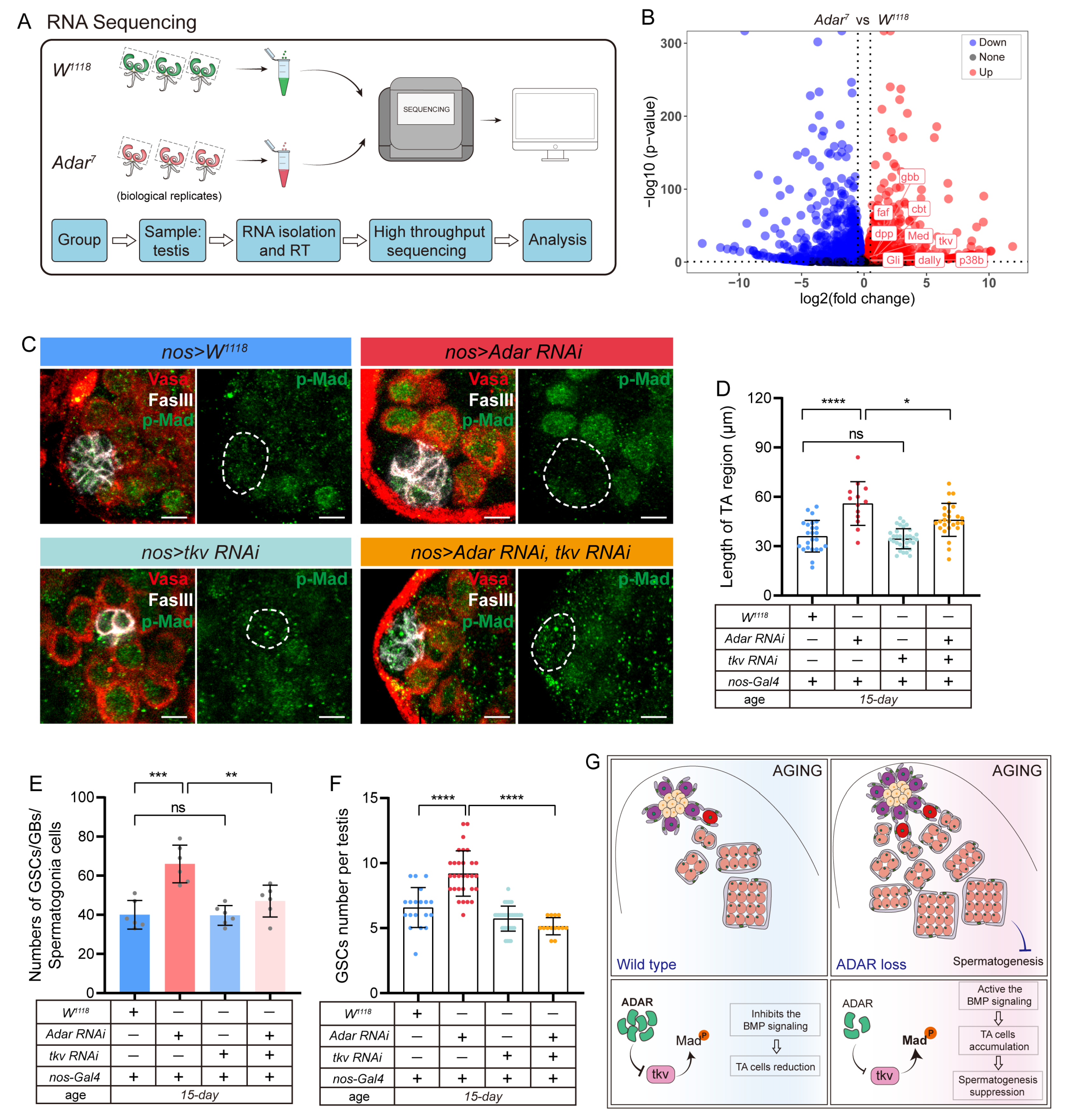
2.4. Adar Regulates Male-Germline-Stem- and Progenitor-Cell Differentiation through Modulation of BMP Signaling
3. Discussion
4. Materials and Methods
4.1. Drosophila Stocks
4.2. Generation of Knock-In, Knock-Out, and Transgenic Fly Lines
4.3. Immunofluorescence
4.4. Fluorescence-Intensity Statistics
4.5. EdU Labeling Experiments
4.6. Fertility Assays
4.7. RNA-Seq
4.8. RNA Extraction and RT-qPCR
4.9. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Neto, F.T.; Bach, P.V.; Najari, B.B.; Li, P.S.; Goldstein, M. Spermatogenesis in humans and its affecting factors. Semin. Cell Dev. Biol. 2016, 59, 10–26. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, A.; Wilkinson, M.F. Transcription and post-transcriptional regulation of spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1637–1651. [Google Scholar] [CrossRef] [PubMed]
- Dura, M.; Teissandier, A.; Armand, M.; Barau, J.; Lapoujade, C.; Fouchet, P.; Bonneville, L.; Schulz, M.; Weber, M.; Baudrin, L.G.; et al. DNMT3A-dependent DNA methylation is required for spermatogonial stem cells to commit to spermatogenesis. Nat. Genet. 2022, 54, 469–480. [Google Scholar] [CrossRef]
- Huang, Q.; Liu, Y.; Zhang, S.; Yap, Y.T.; Li, W.; Zhang, D.; Gardner, A.; Zhang, L.; Song, S.; Hess, R.A.; et al. Autophagy core protein ATG5 is required for elongating spermatid development, sperm individualization and normal fertility in male mice. Autophagy 2021, 17, 1753–1767. [Google Scholar] [CrossRef] [PubMed]
- Lord, T.; Nixon, B. Metabolic Changes Accompanying Spermatogonial Stem Cell Differentiation. Dev. Cell 2020, 52, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.D.; Serrano, J.B.; Martins, F.; da Cruz, E.S.O.A.B.; Rebelo, S. Nuclear envelope dynamics during mammalian spermatogenesis: New insights on male fertility. Biol. Rev. Camb. Philos. Soc. 2019, 94, 1195–1219. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Kang, Z.; Zhang, Y.; Gong, Y.; Liu, M.; Xue, Y.; He, W.; Wang, Y.; Zhang, S.; Xu, Q.; et al. HDAC3 controls male fertility through enzyme-independent transcriptional regulation at the meiotic exit of spermatogenesis. Nucleic Acids Res. 2021, 49, 5106–5123. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, M.L.; Esteves, S.C.; Lamb, D.J.; Hotaling, J.M.; Giwercman, A.; Hwang, K.; Cheng, Y.S. Male infertility. Nat. Rev. Dis. Primers 2023, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Sênos Demarco, R.; Jones, D.L. EGFR signaling promotes basal autophagy for lipid homeostasis and somatic stem cell maintenance in the Drosophila testis. Autophagy 2020, 16, 1145–1147. [Google Scholar] [CrossRef]
- Witt, E.; Benjamin, S.; Svetec, N.; Zhao, L. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. eLife 2019, 8, e47138. [Google Scholar] [CrossRef]
- Gubala, A.M.; Schmitz, J.F.; Kearns, M.J.; Vinh, T.T.; Bornberg-Bauer, E.; Wolfner, M.F.; Findlay, G.D. The Goddard and Saturn Genes Are Essential for Drosophila Male Fertility and May Have Arisen De Novo. Mol. Biol. Evol. 2017, 34, 1066–1082. [Google Scholar] [CrossRef]
- Zhang, Z.; Pan, C.; Zhao, Y. Hedgehog in the Drosophila testis niche: What does it do there? Protein Cell 2013, 4, 650–655. [Google Scholar] [CrossRef]
- White-Cooper, H.; Bausek, N. Evolution and spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1465–1480. [Google Scholar] [CrossRef]
- Leatherman, J.L.; Dinardo, S. Germline self-renewal requires cyst stem cells and stat regulates niche adhesion in Drosophila testes. Nat. Cell Biol. 2010, 12, 806–811. [Google Scholar] [CrossRef]
- Lehmann, R. Germline stem cells: Origin and destiny. Cell Stem Cell 2012, 10, 729–739. [Google Scholar] [CrossRef]
- Boyle, M.; Wong, C.; Rocha, M.; Jones, D.L. Decline in self-renewal factors contributes to aging of the stem cell niche in the Drosophila testis. Cell Stem Cell 2007, 1, 470–478. [Google Scholar] [CrossRef]
- Herrera, S.C.; de la Maza, D.S.; Grmai, L.; Margolis, S.; Plessel, R.; Burel, M.; O’Connor, M.; Amoyel, M.; Bach, E.A. Proliferative stem cells maintain quiescence of their niche by secreting the Activin inhibitor Follistatin. Dev. Cell 2021, 56, 2284–2294.e6. [Google Scholar] [CrossRef]
- Toledano, H.; D’Alterio, C.; Czech, B.; Levine, E.; Jones, D.L. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature 2012, 485, 605–610. [Google Scholar] [CrossRef]
- Epstein, Y.; Perry, N.; Volin, M.; Zohar-Fux, M.; Braun, R.; Porat-Kuperstein, L.; Toledano, H. miR-9a modulates maintenance and ageing of Drosophila germline stem cells by limiting N-cadherin expression. Nat. Commun. 2017, 8, 600. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Shiromoto, Y.; Sakurai, M.; Towers, M.; Zhang, Q.; Wu, S.; Havas, A.; Wang, L.; Berger, S.; Adams, P.D.; et al. ADAR1 downregulation by autophagy drives senescence independently of RNA editing by enhancing p16(INK4a) levels. Nat. Cell Biol. 2022, 24, 1202–1210. [Google Scholar] [CrossRef] [PubMed]
- Herzner, A.M.; Khan, Z.; Van Nostrand, E.L.; Chan, S.; Cuellar, T.; Chen, R.; Pechuan-Jorge, X.; Komuves, L.; Solon, M.; Modrusan, Z.; et al. ADAR and hnRNPC deficiency synergize in activating endogenous dsRNA-induced type I IFN responses. J. Exp. Med. 2021, 218, e20201833. [Google Scholar] [CrossRef]
- Stellos, K.; Gatsiou, A.; Stamatelopoulos, K.; Matic, L.P.; John, D.; Lunella, F.F.; Jaé, N.; Rossbach, O.; Amrhein, C.; Sigala, F.; et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat. Med. 2016, 22, 1140–1150. [Google Scholar] [CrossRef]
- Maldonado, C.; Alicea, D.; Gonzalez, M.; Bykhovskaia, M.; Marie, B. Adar is essential for optimal presynaptic function. Mol. Cell Neurosci. 2013, 52, 173–180. [Google Scholar] [CrossRef]
- Khan, A.; Paro, S.; McGurk, L.; Sambrani, N.; Hogg, M.C.; Brindle, J.; Pennetta, G.; Keegan, L.P.; O’Connell, M.A. Membrane and synaptic defects leading to neurodegeneration in Adar mutant Drosophila are rescued by increased autophagy. BMC Biol. 2020, 18, 15. [Google Scholar] [CrossRef]
- Palladino, M.J.; Keegan, L.P.; O’Connell, M.A.; Reenan, R.A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell 2000, 102, 437–449. [Google Scholar] [CrossRef]
- Robinson, J.E.; Paluch, J.; Dickman, D.K.; Joiner, W.J. ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat. Commun. 2016, 7, 10512. [Google Scholar] [CrossRef]
- Jepson, J.E.C.; Savva, Y.A.; Yokose, C.; Sugden, A.U.; Sahin, A.; Reenan, R.A. Engineered alterations in RNA editing modulate complex behavior in Drosophila: Regulatory diversity of adenosine deaminase acting on RNA (ADAR) targets. J. Biol. Chem. 2011, 286, 8325–8337. [Google Scholar] [CrossRef]
- Chawla, G.; Sokol, N.S. ADAR mediates differential expression of polycistronic microRNAs. Nucleic Acids Res. 2014, 42, 5245–5255. [Google Scholar] [CrossRef]
- Buchumenski, I.; Bartok, O.; Ashwal-Fluss, R.; Pandey, V.; Porath, H.T.; Levanon, E.Y.; Kadener, S. Dynamic hyper-editing underlies temperature adaptation in Drosophila. PLoS Genet. 2017, 13, e1006931. [Google Scholar] [CrossRef]
- Deng, P.; Khan, A.; Jacobson, D.; Sambrani, N.; McGurk, L.; Li, X.; Jayasree, A.; Hejatko, J.; Shohat-Ophir, G.; O’Connell, M.A.; et al. Adar RNA editing-dependent and -independent effects are required for brain and innate immune functions in Drosophila. Nat. Commun. 2020, 11, 1580. [Google Scholar] [CrossRef]
- Leatherman, J.L.; Dinardo, S. Zfh-1 controls somatic stem cell self-renewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell 2008, 3, 44–54. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, X.; Peng, Y.; Zhang, L.; Yu, Y.; Hua, P.; Zhu, P.; Yan, X.; Li, Y.; Zhang, L. miR-24 controls the regenerative competence of hair follicle progenitors by targeting Plk3. Cell Rep. 2021, 35, 109225. [Google Scholar] [CrossRef]
- Kawase, E.; Wong, M.D.; Ding, B.C.; Xie, T. Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 2004, 131, 1365–1375. [Google Scholar] [CrossRef]
- Schulz, C.; Kiger, A.A.; Tazuke, S.I.; Yamashita, Y.M.; Pantalena-Filho, L.C.; Jones, D.L.; Wood, C.G.; Fuller, M.T. A misexpression screen reveals effects of bag-of-marbles and TGF beta class signaling on the Drosophila male germ-line stem cell lineage. Genetics 2004, 167, 707–723. [Google Scholar] [CrossRef]
- Inaba, M.; Buszczak, M.; Yamashita, Y.M. Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature 2015, 523, 329–332. [Google Scholar] [CrossRef]
- Zheng, Q.; Wang, Y.; Vargas, E.; DiNardo, S. magu is required for germline stem cell self-renewal through BMP signaling in the Drosophila testis. Dev. Biol. 2011, 357, 202–210. [Google Scholar] [CrossRef]
- Alfano, M.; Tascini, A.S.; Pederzoli, F.; Locatelli, I.; Nebuloni, M.; Giannese, F.; Garcia-Manteiga, J.M.; Tonon, G.; Amodio, G.; Gregori, S.; et al. Aging, inflammation and DNA damage in the somatic testicular niche with idiopathic germ cell aplasia. Nat. Commun. 2021, 12, 5205. [Google Scholar] [CrossRef]
- Shi, G.; Bai, Y.; Zhang, X.; Su, J.; Pang, J.; He, Q.; Zeng, P.; Ding, J.; Xiong, Y.; Zhang, J.; et al. Bend family proteins mark chromatin boundaries and synergistically promote early germ cell differentiation. Protein Cell 2022, 13, 721–741. [Google Scholar] [CrossRef]
- Artoni, F.; Kreipke, R.E.; Palmeira, O.; Dixon, C.; Goldberg, Z.; Ruohola-Baker, H. Loss of foxo rescues stem cell aging in Drosophila germ line. eLife 2017, 6, e27842. [Google Scholar] [CrossRef]
- Herati, A.S.; Zhelyazkova, B.H.; Butler, P.R.; Lamb, D.J. Age-related alterations in the genetics and genomics of the male germ line. Fertil. Steril. 2017, 107, 319–323. [Google Scholar] [CrossRef]
- Brunet, A.; Goodell, M.A.; Rando, T.A. Ageing and rejuvenation of tissue stem cells and their niches. Nat. Rev. Mol. Cell Biol. 2023, 24, 45–62. [Google Scholar] [CrossRef]
- Jasper, H. Intestinal Stem Cell Aging: Origins and Interventions. Annu. Rev. Physiol. 2020, 82, 203–226. [Google Scholar] [CrossRef]
- Hong, X.; Campanario, S.; Ramírez-Pardo, I.; Grima-Terrén, M.; Isern, J.; Muñoz-Cánoves, P. Stem cell aging in the skeletal muscle: The importance of communication. Ageing Res. Rev. 2022, 73, 101528. [Google Scholar] [CrossRef]
- Sênos Demarco, R.; Uyemura, B.S.; D’Alterio, C.; Jones, D.L. Mitochondrial fusion regulates lipid homeostasis and stem cell maintenance in the Drosophila testis. Nat. Cell Biol. 2019, 21, 710–720. [Google Scholar] [CrossRef]
- Chandrasekhara, C.; Ranjan, R.; Urban, J.A.; Davis, B.E.M.; Ku, W.L.; Snedeker, J.; Zhao, K.; Chen, X. A single N-terminal amino acid determines the distinct roles of histones H3 and H3.3 in the Drosophila male germline stem cell lineage. PLoS Biol. 2023, 21, e3002098. [Google Scholar]
- Dolezal, D.; Liu, Z.; Zhou, Q.; Pignoni, F. Fly LMBR1/LIMR-type protein Lilipod promotes germ-line stem cell self-renewal by enhancing BMP signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 13928–13933. [Google Scholar] [CrossRef]
- Michel, M.; Raabe, I.; Kupinski, A.P.; Pérez-Palencia, R.; Bökel, C. Local BMP receptor activation at adherens junctions in the Drosophila germline stem cell niche. Nat. Commun. 2011, 2, 415. [Google Scholar] [CrossRef]
- Rahman, M.S.; Akhtar, N.; Jamil, H.M.; Banik, R.S.; Asaduzzaman, S.M. TGF-β/BMP signaling and other molecular events: Regulation of osteoblastogenesis and bone formation. Bone Res. 2015, 3, 15005. [Google Scholar] [CrossRef]
- Akiyama, T.; Gibson, M.C. Decapentaplegic and growth control in the developing Drosophila wing. Nature 2015, 527, 375–378. [Google Scholar] [CrossRef]
- Du, G.; Xiong, L.; Li, X.; Zhuo, Z.; Zhuang, X.; Yu, Z.; Wu, L.; Xiao, D.; Liu, Z.; Jie, M.; et al. Peroxisome Elevation Induces Stem Cell Differentiation and Intestinal Epithelial Repair. Dev. Cell 2020, 53, 169–184.e11. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Fan, X.; Fu, F.; Zhu, Y.; Luo, G.; Chen, H. Adar Regulates Drosophila melanogaster Spermatogenesis via Modulation of BMP Signaling. Int. J. Mol. Sci. 2024, 25, 5643. https://doi.org/10.3390/ijms25115643
Zhang Q, Fan X, Fu F, Zhu Y, Luo G, Chen H. Adar Regulates Drosophila melanogaster Spermatogenesis via Modulation of BMP Signaling. International Journal of Molecular Sciences. 2024; 25(11):5643. https://doi.org/10.3390/ijms25115643
Chicago/Turabian StyleZhang, Qian, Xinxin Fan, Fang Fu, Yuedan Zhu, Guanzheng Luo, and Haiyang Chen. 2024. "Adar Regulates Drosophila melanogaster Spermatogenesis via Modulation of BMP Signaling" International Journal of Molecular Sciences 25, no. 11: 5643. https://doi.org/10.3390/ijms25115643
APA StyleZhang, Q., Fan, X., Fu, F., Zhu, Y., Luo, G., & Chen, H. (2024). Adar Regulates Drosophila melanogaster Spermatogenesis via Modulation of BMP Signaling. International Journal of Molecular Sciences, 25(11), 5643. https://doi.org/10.3390/ijms25115643





