PA200-Mediated Proteasomal Protein Degradation and Regulation of Cellular Senescence
Abstract
1. Introduction
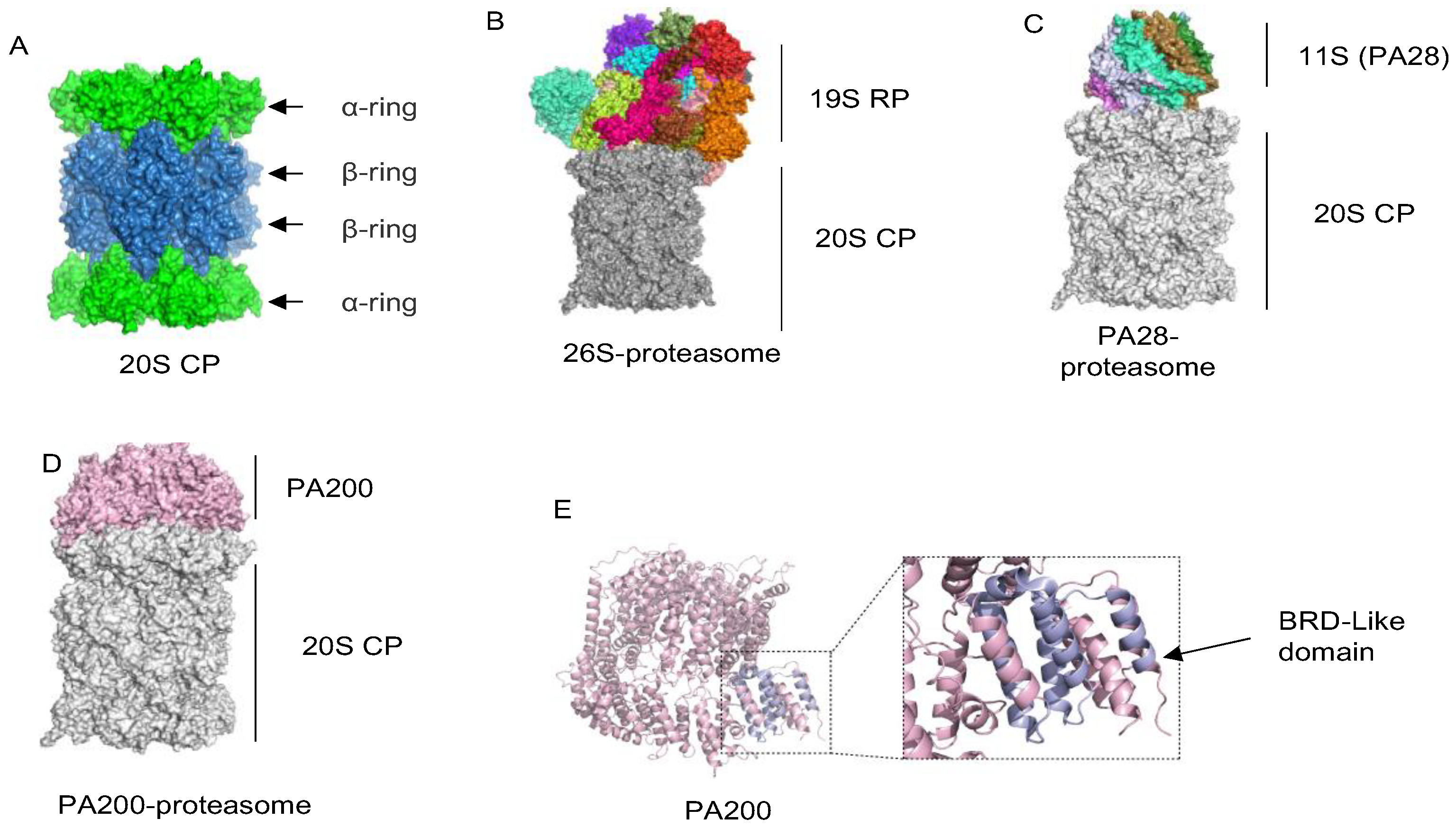
2. Overview of Proteasomes
2.1. The 20S Catalytic Particle
2.2. The 19S Regulatory Particle
2.3. Proteasome Activators PA28α, PA28β and PA28γ
2.4. The Proteasome Activator PA200/Blm10
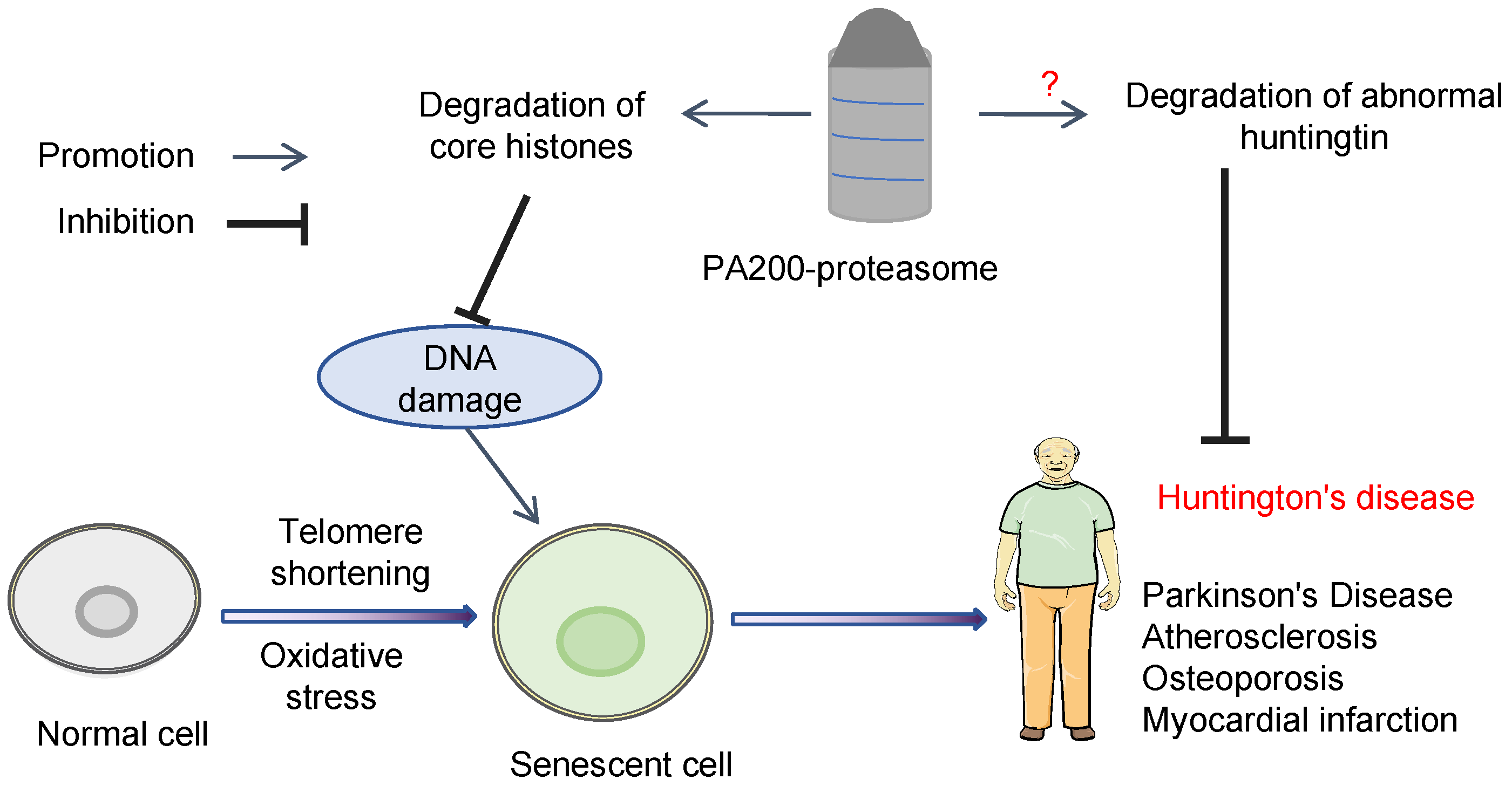
3. PA200 Plays Important Roles in Preventing Cellular Senescence
3.1. PA200-Mediated Degradation of the Core Histones during Senescence
3.2. PA200 Is Associated with Development of Certain Types of Tumors during Aging
3.3. Degradation of Exogenous N-Terminal Fragment of Huntingtin Protein by PA200/Blm10-Proteasome
3.4. PA200 Prevents Cellular Senescence in Mesenchymal Stem Cells
| Aging-Related Disease | Mechanisms | References | |
|---|---|---|---|
| Cancer | NSCLC | Reduces intracellular antigen processing and inhibits T-cell activity, leading to ICI therapy resistance. | [88] |
| HCC | Activates mTOR signaling; increases Malignant Progression of HCC | [92] | |
| Huntington’s disease | Decreases the cellular levels of the exogenous N-Htt aggregates | [110] | |
| Myocardial infarction | Depletes YAP in the nucleus; promotes the cardiac commitment of MSC. | [114] | |
4. Conclusions and Perspectives for Future Studies
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 19S RP | 19S regulatory particle |
| 20S CP | 20S core particle |
| Aha | Azidohomoalanine |
| ALS | Amyotrophic lateral sclerosis |
| Blm10 | Proteasome activator BLM10 |
| BRD | Bromodomain |
| BRDL | Brd-like domain |
| CDKN1A | Cell cycle protein-dependent kinase inhibitor 1A |
| ChIP | Chromatin immunoprecipitation |
| COVID-19 | Coronavirus Disease 2019 |
| cryo-EM | Cryo-electron microscopy |
| DNA | Deoxyribonucleic acid |
| EVs | Extracellular vesicles |
| GAHD | Genome-wide analysis of histone degradation |
| GSEA | Gene set enrichment analysis |
| HCC | Hepatocellular carcinoma |
| HCCDB | Hepatocellular carcinoma cell database |
| HD | Huntington’s disease |
| HDAC | Histone deacetylase |
| HLA | Human leukocyte antigen |
| i20S | Immunoproteasome 20S |
| IFN-γ | Interferon gamma |
| IR | Ionizing radiation |
| MAPP | Mass spectrometry analysis of proteolytic peptides |
| MSC | Mesenchymal stem cell |
| N-Htt | N-terminal fragment of huntingtin |
| NSCLC | Non-small cell lung cancer |
| OSCC | Oral squamous cell carcinoma |
| PDB | Protein data bank |
| SAGA | Spt-ada-gcn5 acetyltransferase |
| SASP | Senescence-associated secretory phenotype |
| SGF29 | SAGA complex-associated factor 29 |
| TAZ | Transcriptional coactivator with pdz-binding domain |
| TCGA | The Cancer Genome Atlas |
| TSSs | Transcription start sites |
| YAP | Yes-associated protein |
References
- Campisi, J. Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 2013, 75, 685–705. [Google Scholar] [CrossRef]
- Abdueva, D.; Wing, M.; Schaub, B.; Triche, T.; Davicioni, E. Quantitative expression profiling in formalin-fixed paraffin-embedded samples by affymetrix microarrays. J. Mol. Diagn. 2010, 12, 409–417. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef] [PubMed]
- Passos, J.F.; Nelson, G.; Wang, C.; Richter, T.; Simillion, C.; Proctor, C.J.; Miwa, S.; Olijslagers, S.; Hallinan, J.; Wipat, A.; et al. Feedback between p21 and reactive oxygen production is necessary for cell senescence. Mol. Syst. Biol. 2010, 6, 347. [Google Scholar] [CrossRef] [PubMed]
- Moiseeva, O.; Bourdeau, V.; Roux, A.; Deschenes-Simard, X.; Ferbeyre, G. Mitochondrial dysfunction contributes to oncogene-induced senescence. Mol. Cell Biol. 2009, 29, 4495–4507. [Google Scholar] [CrossRef]
- Wiley, C.D.; Campisi, J. From Ancient Pathways to Aging Cells-Connecting Metabolism and Cellular Senescence. Cell Metab. 2016, 23, 1013–1021. [Google Scholar] [CrossRef] [PubMed]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Park, S.S.; Choi, Y.W.; Kim, J.H.; Kim, H.S.; Park, T.J. Senescent tumor cells: An overlooked adversary in the battle against cancer. Exp. Mol. Med. 2021, 53, 1834–1841. [Google Scholar] [CrossRef]
- Coppe, J.P.; Desprez, P.Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef]
- Frasca, D.; Saada, Y.B.; Garcia, D.; Friguet, B. Effects of cellular senescence on metabolic pathways in non-immune and immune cells. Mech. Ageing Dev. 2021, 194, 111428. [Google Scholar] [CrossRef]
- Yang, J.; Dungrawala, H.; Hua, H.; Manukyan, A.; Abraham, L.; Lane, W.; Mead, H.; Wright, J.; Schneider, B.L. Cell size and growth rate are major determinants of replicative lifespan. Cell Cycle 2011, 10, 144–155. [Google Scholar] [CrossRef]
- Pendergrass, W.; Angello, J.; Norwood, T.H. The relationship between cell size, the activity of DNA polymerase alpha and proliferative activity in human diploid fibroblast-like cell cultures. Exp. Gerontol. 1989, 24, 383–393. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. Hallmarks of aging: An expanding universe. Cell 2023, 186, 243–278. [Google Scholar] [CrossRef]
- Yang, J.H.; Hayano, M.; Griffin, P.T.; Amorim, J.A.; Bonkowski, M.S.; Apostolides, J.K.; Salfati, E.L.; Blanchette, M.; Munding, E.M.; Bhakta, M.; et al. Loss of epigenetic information as a cause of mammalian aging. Cell 2023, 186, 305–326.e27. [Google Scholar] [CrossRef]
- Dang, W.; Steffen, K.K.; Perry, R.; Dorsey, J.A.; Johnson, F.B.; Shilatifard, A.; Kaeberlein, M.; Kennedy, B.K.; Berger, S.L. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 2009, 459, 802–807. [Google Scholar] [CrossRef]
- Millan-Zambrano, G.; Burton, A.; Bannister, A.J.; Schneider, R. Histone post-translational modifications—Cause and consequence of genome function. Nat. Rev. Genet. 2022, 23, 563–580. [Google Scholar] [CrossRef]
- Feser, J.; Truong, D.; Das, C.; Carson, J.J.; Kieft, J.; Harkness, T.; Tyler, J.K. Elevated histone expression promotes life span extension. Mol. Cell 2010, 39, 724–735. [Google Scholar] [CrossRef]
- Ivanov, A.; Pawlikowski, J.; Manoharan, I.; van Tuyn, J.; Nelson, D.M.; Rai, T.S.; Shah, P.P.; Hewitt, G.; Korolchuk, V.I.; Passos, J.F.; et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 2013, 202, 129–143. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, K.; Xia, Z.; Chavez, M.; Pal, S.; Seol, J.H.; Chen, C.C.; Li, W.; Tyler, J.K. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev. 2014, 28, 396–408. [Google Scholar] [CrossRef]
- Kruegel, U.; Robison, B.; Dange, T.; Kahlert, G.; Delaney, J.R.; Kotireddy, S.; Tsuchiya, M.; Tsuchiyama, S.; Murakami, C.J.; Schleit, J.; et al. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011, 7, e1002253. [Google Scholar] [CrossRef]
- Qian, M.X.; Pang, Y.; Liu, C.H.; Haratake, K.; Du, B.Y.; Ji, D.Y.; Wang, G.F.; Zhu, Q.Q.; Song, W.; Yu, Y.; et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013, 153, 1012–1024. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Jiang, T.X.; Chen, L.B.; Zhou, W.; Liu, Y.; Gao, F.; Qiu, X.B. Proteasome subunit α4s is essential for formation of spermatoproteasomes and histone degradation during meiotic DNA repair in spermatocytes. J. Biol. Chem. 2021, 296, 100130. [Google Scholar] [CrossRef]
- Jiang, T.X.; Ma, S.; Han, X.; Luo, Z.Y.; Zhu, Q.Q.; Chiba, T.; Xie, W.; Lin, K.; Qiu, X.B. Proteasome activator PA200 maintains stability of histone marks during transcription and aging. Theranostics 2021, 11, 1458–1472. [Google Scholar] [CrossRef]
- Vernace, V.A.; Arnaud, L.; Schmidt-Glenewinkel, T.; Figueiredo-Pereira, M.E. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J. 2007, 21, 2672–2682. [Google Scholar] [CrossRef]
- Tonoki, A.; Kuranaga, E.; Tomioka, T.; Hamazaki, J.; Murata, S.; Tanaka, K.; Miura, M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell Biol. 2009, 29, 1095–1106. [Google Scholar] [CrossRef]
- Mannhaupt, G.; Schnall, R.; Karpov, V.; Vetter, I.; Feldmann, H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999, 450, 27–34. [Google Scholar] [CrossRef]
- Chen, L.B.; Ma, S.; Jiang, T.X.; Qiu, X.B. Transcriptional upregulation of proteasome activator Blm10 antagonizes cellular aging. Biochem. Biophys. Res. Commun. 2020, 532, 211–218. [Google Scholar] [CrossRef]
- Jiang, T.X.; Zhao, M.; Qiu, X.B. Substrate receptors of proteasomes. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1765–1777. [Google Scholar] [CrossRef]
- Groll, M.; Ditzel, L.; Lowe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef]
- Basler, M.; Kirk, C.J.; Groettrup, M. The immunoproteasome in antigen processing and other immunological functions. Curr. Opin. Immunol. 2013, 25, 74–80. [Google Scholar] [CrossRef]
- Lowe, J.; Stock, D.; Jap, B.; Zwickl, P.; Baumeister, W.; Huber, R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995, 268, 533–539. [Google Scholar] [CrossRef]
- Huang, X.; Luan, B.; Wu, J.; Shi, Y. An atomic structure of the human 26S proteasome. Nat. Struct. Mol. Biol. 2016, 23, 778–785. [Google Scholar] [CrossRef]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.H.; Shi, Y.; Zhang, N.; de Poot, S.A.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351, aad9421. [Google Scholar] [CrossRef]
- Deveraux, Q.; Ustrell, V.; Pickart, C.; Rechsteiner, M. A 26 S protease subunit that binds ubiquitin conjugates. J. Biol. Chem. 1994, 269, 7059–7061. [Google Scholar] [CrossRef]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef]
- Holzl, H.; Kapelari, B.; Kellermann, J.; Seemuller, E.; Sumegi, M.; Udvardy, A.; Medalia, O.; Sperling, J.; Muller, S.A.; Engel, A.; et al. The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J. Cell Biol. 2000, 150, 119–130. [Google Scholar] [CrossRef]
- Leggett, D.S.; Hanna, J.; Borodovsky, A.; Crosas, B.; Schmidt, M.; Baker, R.T.; Walz, T.; Ploegh, H.; Finley, D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 2002, 10, 495–507. [Google Scholar] [CrossRef]
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R., 3rd; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615. [Google Scholar] [CrossRef]
- Yao, T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407. [Google Scholar] [CrossRef]
- Hanna, J.; Hathaway, N.A.; Tone, Y.; Crosas, B.; Elsasser, S.; Kirkpatrick, D.S.; Leggett, D.S.; Gygi, S.P.; King, R.W.; Finley, D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 2006, 127, 99–111. [Google Scholar] [CrossRef]
- Qiu, X.B.; Ouyang, S.Y.; Li, C.J.; Miao, S.; Wang, L.; Goldberg, A.L. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006, 25, 5742–5753. [Google Scholar] [CrossRef]
- Rock, K.L.; Goldberg, A.L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu. Rev. Immunol. 1999, 17, 739–779. [Google Scholar] [CrossRef]
- Rechsteiner, M.; Realini, C.; Ustrell, V. The proteasome activator 11 S REG (PA28) and class I antigen presentation. Biochem. J. 2000, 345 Pt 1, 1–15. [Google Scholar] [CrossRef]
- Zhao, J.; Makhija, S.; Zhou, C.; Zhang, H.; Wang, Y.; Muralidharan, M.; Huang, B.; Cheng, Y. Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins. Proc. Natl. Acad. Sci. USA 2022, 119, e2207200119. [Google Scholar] [CrossRef]
- Masson, P.; Andersson, O.; Petersen, U.M.; Young, P. Identification and characterization of a Drosophila nuclear proteasome regulator. A homolog of human 11 S REGgamma (PA28gamma). J. Biol. Chem. 2001, 276, 1383–1390. [Google Scholar] [CrossRef]
- Thomas, T.A.; Smith, D.M. Proteasome activator 28gamma (PA28gamma) allosterically activates trypsin-like proteolysis by binding to the alpha-ring of the 20S proteasome. J. Biol. Chem. 2022, 298, 102140. [Google Scholar] [CrossRef]
- Realini, C.; Jensen, C.C.; Zhang, Z.; Johnston, S.C.; Knowlton, J.R.; Hill, C.P.; Rechsteiner, M. Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J. Biol. Chem. 1997, 272, 25483–25492. [Google Scholar] [CrossRef]
- Wilk, S.; Chen, W.E.; Magnusson, R.P. Properties of the nuclear proteasome activator PA28gamma (REGgamma). Arch. Biochem. Biophys. 2000, 383, 265–271. [Google Scholar] [CrossRef]
- Jonik-Nowak, B.; Menneteau, T.; Fesquet, D.; Baldin, V.; Bonne-Andrea, C.; Mechali, F.; Fabre, B.; Boisguerin, P.; de Rossi, S.; Henriquet, C.; et al. PIP30/FAM192A is a novel regulator of the nuclear proteasome activator PA28gamma. Proc. Natl. Acad. Sci. USA 2018, 115, E6477–E6486. [Google Scholar] [CrossRef]
- Frayssinhes, J.A.; Cerruti, F.; Laulin, J.; Cattaneo, A.; Bachi, A.; Apcher, S.; Coux, O.; Cascio, P. PA28gamma-20S proteasome is a proteolytic complex committed to degrade unfolded proteins. Cell Mol. Life Sci. 2021, 79, 45. [Google Scholar] [CrossRef]
- Gao, X.; Li, J.; Pratt, G.; Wilk, S.; Rechsteiner, M. Purification procedures determine the proteasome activation properties of REG gamma (PA28 gamma). Arch. Biochem. Biophys. 2004, 425, 158–164. [Google Scholar] [CrossRef]
- Li, J.; Rechsteiner, M. Molecular dissection of the 11S REG (PA28) proteasome activators. Biochimie 2001, 83, 373–383. [Google Scholar] [CrossRef]
- Groll, M.; Bajorek, M.; Kohler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef]
- Chen, D.D.; Hao, J.; Shen, C.H.; Deng, X.M.; Yun, C.H. Atomic resolution Cryo-EM structure of human proteasome activator PA28gamma. Int. J. Biol. Macromol. 2022, 219, 500–507. [Google Scholar] [CrossRef]
- Murata, S.; Kawahara, H.; Tohma, S.; Yamamoto, K.; Kasahara, M.; Nabeshima, Y.; Tanaka, K.; Chiba, T. Growth retardation in mice lacking the proteasome activator PA28gamma. J. Biol. Chem. 1999, 274, 38211–38215. [Google Scholar] [CrossRef]
- Levy-Barda, A.; Lerenthal, Y.; Davis, A.J.; Chung, Y.M.; Essers, J.; Shao, Z.; van Vliet, N.; Chen, D.J.; Hu, M.C.; Kanaar, R.; et al. Involvement of the nuclear proteasome activator PA28gamma in the cellular response to DNA double-strand breaks. Cell Cycle 2011, 10, 4300–4310. [Google Scholar] [CrossRef]
- Chen, X.; Barton, L.F.; Chi, Y.; Clurman, B.E.; Roberts, J.M. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell 2007, 26, 843–852. [Google Scholar] [CrossRef]
- Li, X.; Amazit, L.; Long, W.; Lonard, D.M.; Monaco, J.J.; O’Malley, B.W. Ubiquitin- and ATP-independent proteolytic turnover of p21 by the REGgamma-proteasome pathway. Mol. Cell 2007, 26, 831–842. [Google Scholar] [CrossRef]
- Moriishi, K.; Mochizuki, R.; Moriya, K.; Miyamoto, H.; Mori, Y.; Abe, T.; Murata, S.; Tanaka, K.; Miyamura, T.; Suzuki, T.; et al. Critical role of PA28gamma in hepatitis C virus-associated steatogenesis and hepatocarcinogenesis. Proc. Natl. Acad. Sci. USA 2007, 104, 1661–1666. [Google Scholar] [CrossRef]
- Wang, X.; Tu, S.; Tan, J.; Tian, T.; Ran, L.; Rodier, J.F.; Ren, G. REG gamma: A potential marker in breast cancer and effect on cell cycle and proliferation of breast cancer cell. Med. Oncol. 2011, 28, 31–41. [Google Scholar] [CrossRef]
- Okamura, T.; Taniguchi, S.; Ohkura, T.; Yoshida, A.; Shimizu, H.; Sakai, M.; Maeta, H.; Fukui, H.; Ueta, Y.; Hisatome, I.; et al. Abnormally high expression of proteasome activator-gamma in thyroid neoplasm. J. Clin. Endocrinol. Metab. 2003, 88, 1374–1383. [Google Scholar] [CrossRef]
- Roessler, M.; Rollinger, W.; Mantovani-Endl, L.; Hagmann, M.L.; Palme, S.; Berndt, P.; Engel, A.M.; Pfeffer, M.; Karl, J.; Bodenmuller, H.; et al. Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol. Cell Proteom. 2006, 5, 2092–2101. [Google Scholar] [CrossRef]
- Seo, H.; Sonntag, K.C.; Kim, W.; Cattaneo, E.; Isacson, O. Proteasome activator enhances survival of Huntington’s disease neuronal model cells. PLoS ONE 2007, 2, e238. [Google Scholar] [CrossRef]
- Suzuki, R.; Moriishi, K.; Fukuda, K.; Shirakura, M.; Ishii, K.; Shoji, I.; Wakita, T.; Miyamura, T.; Matsuura, Y.; Suzuki, T. Proteasomal turnover of hepatitis C virus core protein is regulated by two distinct mechanisms: A ubiquitin-dependent mechanism and a ubiquitin-independent but PA28gamma-dependent mechanism. J. Virol. 2009, 83, 2389–2392. [Google Scholar] [CrossRef]
- Zhang, H.; Tu, J.; Cao, C.; Yang, T.; Gao, L. Proteasome activator PA28gamma-dependent degradation of coronavirus disease (COVID-19) nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 529, 251–256. [Google Scholar] [CrossRef]
- Fort, P.; Kajava, A.V.; Delsuc, F.; Coux, O. Evolution of proteasome regulators in eukaryotes. Genome Biol. Evol. 2015, 7, 1363–1379. [Google Scholar] [CrossRef]
- Fabre, B.; Lambour, T.; Delobel, J.; Amalric, F.; Monsarrat, B.; Burlet-Schiltz, O.; Bousquet-Dubouch, M.P. Subcellular distribution and dynamics of active proteasome complexes unraveled by a workflow combining in vivo complex cross-linking and quantitative proteomics. Mol. Cell Proteom. 2013, 12, 687–699. [Google Scholar] [CrossRef]
- Schmidt, M.; Haas, W.; Crosas, B.; Santamaria, P.G.; Gygi, S.P.; Walz, T.; Finley, D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 2005, 12, 294–303. [Google Scholar] [CrossRef]
- Guan, H.; Wang, Y.; Yu, T.; Huang, Y.; Li, M.; Saeed, A.; Perčulija, V.; Li, D.; Xiao, J.; Wang, D.; et al. Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures. PLoS Biol. 2020, 18, e3000654. [Google Scholar] [CrossRef] [PubMed]
- Dange, T.; Smith, D.; Noy, T.; Rommel, P.C.; Jurzitza, L.; Cordero, R.J.; Legendre, A.; Finley, D.; Goldberg, A.L.; Schmidt, M. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J. Biol. Chem. 2011, 286, 42830–42839. [Google Scholar] [CrossRef] [PubMed]
- Toste Rêgo, A.; da Fonseca, P.C.A. Characterization of Fully Recombinant Human 20S and 20S-PA200 Proteasome Complexes. Mol. Cell 2019, 76, 138–147.e5. [Google Scholar] [CrossRef] [PubMed]
- Millard, C.J.; Watson, P.J.; Celardo, I.; Gordiyenko, Y.; Cowley, S.M.; Robinson, C.V.; Fairall, L.; Schwabe, J.W. Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol. Cell 2013, 51, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Watson, P.J.; Millard, C.J.; Riley, A.M.; Robertson, N.S.; Wright, L.C.; Godage, H.Y.; Cowley, S.M.; Jamieson, A.G.; Potter, B.V.; Schwabe, J.W. Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nat. Commun. 2016, 7, 11262. [Google Scholar] [CrossRef]
- Hauer, M.H.; Seeber, A.; Singh, V.; Thierry, R.; Sack, R.; Amitai, A.; Kryzhanovska, M.; Eglinger, J.; Holcman, D.; Owen-Hughes, T.; et al. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat. Struct. Mol. Biol. 2017, 24, 99–107. [Google Scholar] [CrossRef]
- Mandemaker, I.K.; Geijer, M.E.; Kik, I.; Bezstarosti, K.; Rijkers, E.; Raams, A.; Janssens, R.C.; Lans, H.; Hoeijmakers, J.H.; Demmers, J.A.; et al. DNA damage-induced replication stress results in PA200-proteasome-mediated degradation of acetylated histones. EMBO Rep. 2018, 19, e45566. [Google Scholar] [CrossRef] [PubMed]
- Sadre-Bazzaz, K.; Whitby, F.G.; Robinson, H.; Formosa, T.; Hill, C.P. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell 2010, 37, 728–735. [Google Scholar] [CrossRef]
- Saul, D.; Kosinsky, R.L. Epigenetics of Aging and Aging-Associated Diseases. Int. J. Mol. Sci. 2021, 22, 401. [Google Scholar] [CrossRef]
- Borgel, J.; Guibert, S.; Li, Y.; Chiba, H.; Schubeler, D.; Sasaki, H.; Forne, T.; Weber, M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010, 42, 1093–1100. [Google Scholar] [CrossRef]
- Yan, K.; Ji, Q.; Zhao, D.; Li, M.; Sun, X.; Wang, Z.; Liu, X.; Liu, Z.; Li, H.; Ding, Y.; et al. SGF29 nuclear condensates reinforce cellular aging. Cell Discov. 2023, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Li, H.; Liu, J.; Hu, Q.; Zhang, S.; Zhang, N.; Liu, L.; Dai, Y.; Cao, D.; Li, X.; et al. Arginine hypomethylation-mediated proteasomal degradation of histone H4-an early biomarker of cellular senescence. Cell Death Differ. 2020, 27, 2697–2709. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Han, X.; Lin, K.; Jiang, T.X.; Qiu, X.B. Proteasome Activator Blm10 Regulates Transcription Especially During Aging. Curr. Genom. 2021, 22, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Loaiza, N.; Demaria, M. Cellular senescence and tumor promotion: Is aging the key? Biochim. Biophys. Acta 2016, 1865, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, A.V.; Kisseljova, N.P. DNA methylation and carcinogenesis. Biochemistry 2001, 66, 235–255. [Google Scholar] [PubMed]
- Strub, T.; Ballotti, R.; Bertolotto, C. The “ART” of Epigenetics in Melanoma: From histone “Alterations, to Resistance and Therapies”. Theranostics 2020, 10, 1777–1797. [Google Scholar] [CrossRef]
- Calcinotto, A.; Kohli, J.; Zagato, E.; Pellegrini, L.; Demaria, M.; Alimonti, A. Cellular Senescence: Aging, Cancer, and Injury. Physiol. Rev. 2019, 99, 1047–1078. [Google Scholar] [CrossRef] [PubMed]
- Javitt, A.; Shmueli, M.D.; Kramer, M.P.; Kolodziejczyk, A.A.; Cohen, I.J.; Radomir, L.; Sheban, D.; Kamer, I.; Litchfield, K.; Bab-Dinitz, E.; et al. The proteasome regulator PSME4 modulates proteasome activity and antigen diversity to abrogate antitumor immunity in NSCLC. Nat. Cancer 2023, 4, 629–647. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Chan, K.W.; Bao, J.Y.; Wong, K.Y.; Chen, J.N.; Kwan, P.S.; Tang, K.H.; Fu, L.; Qin, Y.R.; Lok, S.; et al. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res. 2012, 72, 6024–6035. [Google Scholar] [CrossRef]
- Jagannathan, S.; Vad, N.; Vallabhapurapu, S.; Vallabhapurapu, S.; Anderson, K.C.; Driscoll, J.J. MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib. Leukemia 2015, 29, 727–738. [Google Scholar] [CrossRef]
- Pisamai, S.; Roytrakul, S.; Phaonakrop, N.; Jaresitthikunchai, J.; Suriyaphol, G. Proteomic analysis of canine oral tumor tissues using MALDI-TOF mass spectrometry and in-gel digestion coupled with mass spectrometry (GeLC MS/MS) approaches. PLoS ONE 2018, 13, e0200619. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Huang, H.; Huang, W.; Ji, R.; Chen, J.; Wu, S.; Wang, L.; Huang, T.; Sheng, Y.; Yan, H.; et al. PSME4 Activates mTOR Signaling and Promotes the Malignant Progression of Hepatocellular Carcinoma. Int. J. Gen. Med. 2022, 15, 885–895. [Google Scholar] [CrossRef]
- Yu, Z.; Wei, X.; Liu, L.; Sun, H.; Fang, T.; Wang, L.; Li, Y.; Sui, W.; Wang, K.; He, Y.; et al. Indirubin-3’-monoxime acts as proteasome inhibitor: Therapeutic application in multiple myeloma. EBioMedicine 2022, 78, 103950. [Google Scholar] [CrossRef]
- Guo, Y.; Dong, X.; Jin, J.; He, Y. The Expression Patterns and Prognostic Value of the Proteasome Activator Subunit Gene Family in Gastric Cancer Based on Integrated Analysis. Front. Cell Dev. Biol. 2021, 9, 663001. [Google Scholar] [CrossRef] [PubMed]
- Maag, J.L.V.; Fisher, O.M.; Levert-Mignon, A.; Kaczorowski, D.C.; Thomas, M.L.; Hussey, D.J.; Watson, D.I.; Wettstein, A.; Bobryshev, Y.V.; Edwards, M.; et al. Novel Aberrations Uncovered in Barrett’s Esophagus and Esophageal Adenocarcinoma Using Whole Transcriptome Sequencing. Mol. Cancer Res. 2017, 15, 1558–1569. [Google Scholar] [CrossRef]
- Zhu, H.; Sun, J.; Zhang, C.; Li, P.; Tan, C.; Yang, M.; Zhao, G. Cellular Senescence in Non-Small Cell Lung Cancer. Front. Biosci. 2023, 28, 357. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Oncology meets immunology: The cancer-immunity cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef]
- Johnson, D.B.; Nebhan, C.A.; Moslehi, J.J.; Balko, J.M. Immune-checkpoint inhibitors: Long-term implications of toxicity. Nat. Rev. Clin. Oncol. 2022, 19, 254–267. [Google Scholar] [CrossRef]
- Rousseau, A.; Bertolotti, A. Regulation of proteasome assembly and activity in health and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 697–712. [Google Scholar] [CrossRef]
- Rock, K.L.; Reits, E.; Neefjes, J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol. 2016, 37, 724–737. [Google Scholar] [CrossRef]
- Gaczynska, M.; Rock, K.L.; Goldberg, A.L. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature 1993, 365, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.C.; Peters, H.L.; Taguchi, A.; Katayama, H.; Wang, H.; Momin, A.; Jolly, M.K.; Celiktas, M.; Rodriguez-Canales, J.; Liu, H.; et al. Immunoproteasome deficiency is a feature of non-small cell lung cancer with a mesenchymal phenotype and is associated with a poor outcome. Proc. Natl. Acad. Sci. USA 2016, 113, E1555–E1564. [Google Scholar] [CrossRef] [PubMed]
- Kalaora, S.; Lee, J.S.; Barnea, E.; Levy, R.; Greenberg, P.; Alon, M.; Yagel, G.; Bar Eli, G.; Oren, R.; Peri, A.; et al. Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma. Nat. Commun. 2020, 11, 896. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Guo, J.; Jin, W.; Chang, C.; Guo, X.; Xu, C. A combined proteomic and metabolomic analyses of the priming phase during rat liver regeneration. Arch. Biochem. Biophys. 2020, 693, 108567. [Google Scholar] [CrossRef] [PubMed]
- Newsholme, P.; Procopio, J.; Lima, M.M.; Pithon-Curi, T.C.; Curi, R. Glutamine and glutamate—Their central role in cell metabolism and function. Cell Biochem. Funct. 2003, 21, 1–9. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef] [PubMed]
- Blickwedehl, J.; Olejniczak, S.; Cummings, R.; Sarvaiya, N.; Mantilla, A.; Chanan-Khan, A.; Pandita, T.K.; Schmidt, M.; Thompson, C.B.; Bangia, N. The proteasome activator PA200 regulates tumor cell responsiveness to glutamine and resistance to ionizing radiation. Mol. Cancer Res. 2012, 10, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Walker, F.O. Huntington’s disease. Lancet 2007, 369, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Finkbeiner, S. Huntington’s Disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a007476. [Google Scholar] [CrossRef]
- Aladdin, A.; Yao, Y.; Yang, C.; Kahlert, G.; Ghani, M.; Király, N.; Boratkó, A.; Uray, K.; Dittmar, G.; Tar, K. The Proteasome Activators Blm10/PA200 Enhance the Proteasomal Degradation of N-Terminal Huntingtin. Biomolecules 2020, 10, 1581. [Google Scholar] [CrossRef]
- Lee, H.J.; Alirzayeva, H.; Koyuncu, S.; Rueber, A.; Noormohammadi, A.; Vilchez, D. Cold temperature extends longevity and prevents disease-related protein aggregation through PA28gamma-induced proteasomes. Nat. Aging 2023, 3, 546–566. [Google Scholar] [CrossRef] [PubMed]
- Driskill, J.H.; Pan, D. Control of stem cell renewal and fate by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2023, 24, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Mura-Meszaros, A.; Tollot, M.; Krishnan, M.S.; Grundl, M.; Neubert, L.; Groth, M.; Rodriguez-Fraticelli, A.; Svendsen, A.F.; Campaner, S.; et al. Taz protects hematopoietic stem cells from an aging-dependent decrease in PU.1 activity. Nat. Commun. 2022, 13, 5187. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, Y.S.; Ahn, Y.; Eom, G.H.; Yoon, S. PSME4 determines mesenchymal stem cell fate towards cardiac commitment through YAP1 degradation. Korean J. Physiol. Pharmacol. 2023, 27, 407–416. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, P.; Sun, Y.; Jiang, T.-X.; Qiu, X.-B. PA200-Mediated Proteasomal Protein Degradation and Regulation of Cellular Senescence. Int. J. Mol. Sci. 2024, 25, 5637. https://doi.org/10.3390/ijms25115637
Wen P, Sun Y, Jiang T-X, Qiu X-B. PA200-Mediated Proteasomal Protein Degradation and Regulation of Cellular Senescence. International Journal of Molecular Sciences. 2024; 25(11):5637. https://doi.org/10.3390/ijms25115637
Chicago/Turabian StyleWen, Pei, Yan Sun, Tian-Xia Jiang, and Xiao-Bo Qiu. 2024. "PA200-Mediated Proteasomal Protein Degradation and Regulation of Cellular Senescence" International Journal of Molecular Sciences 25, no. 11: 5637. https://doi.org/10.3390/ijms25115637
APA StyleWen, P., Sun, Y., Jiang, T.-X., & Qiu, X.-B. (2024). PA200-Mediated Proteasomal Protein Degradation and Regulation of Cellular Senescence. International Journal of Molecular Sciences, 25(11), 5637. https://doi.org/10.3390/ijms25115637






