Breed-Related Differential microRNA Expression and Analysis of Colostrum and Mature Milk Exosomes in Bamei and Landrace Pigs
Abstract
1. Introduction
2. Results
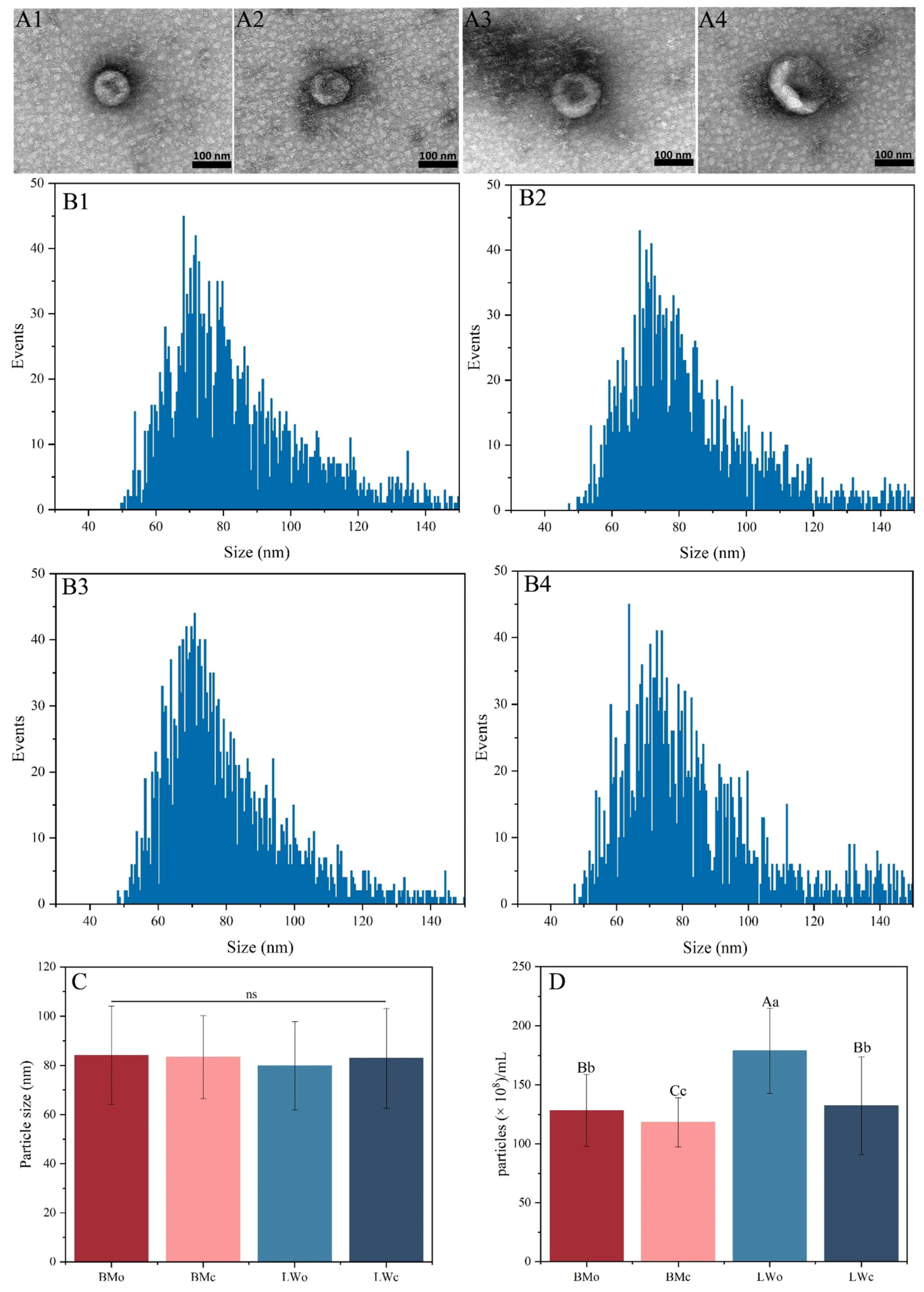
2.1. Exosome Characterization
2.2. Quality Control of Sequencing Data
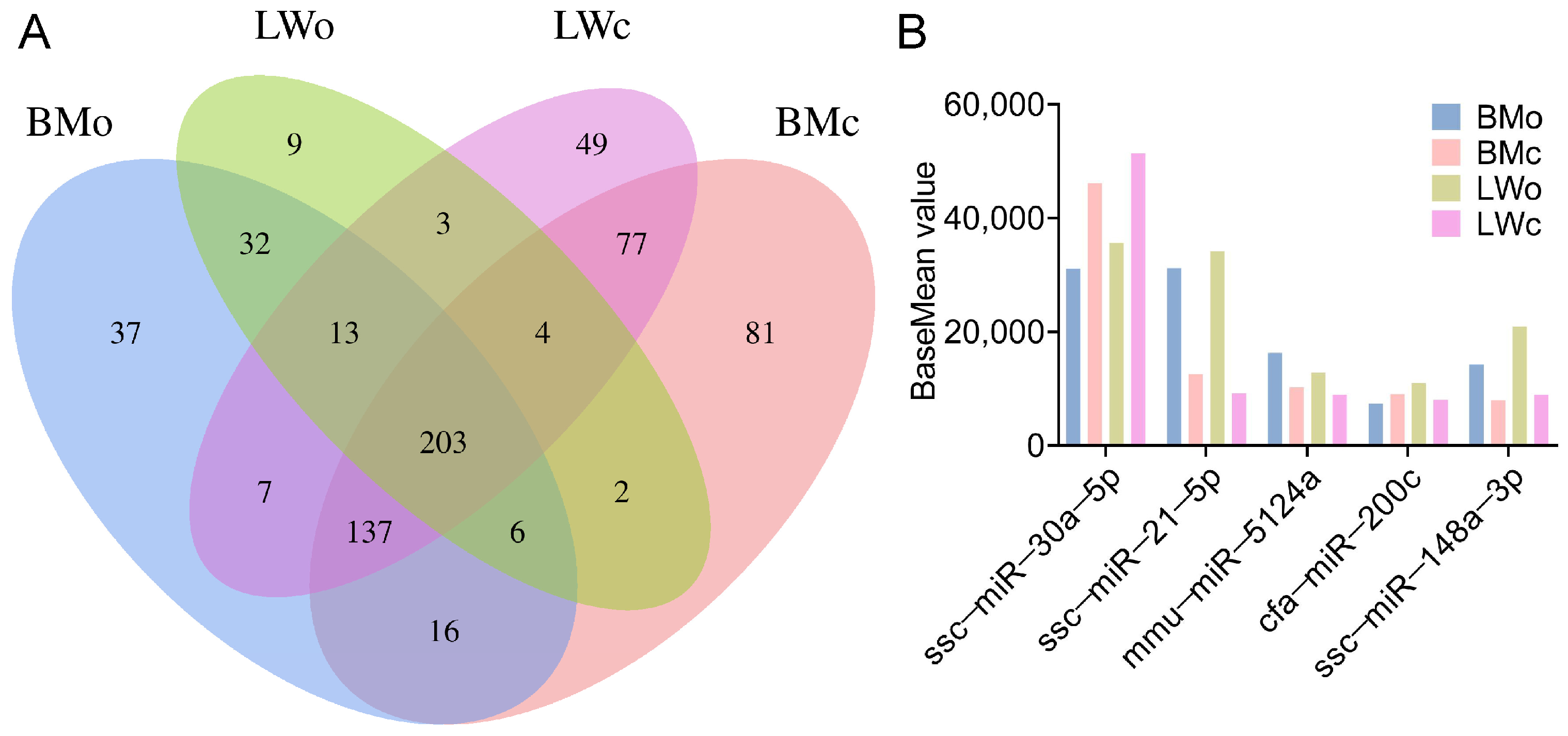
2.3. miRNA Identification and Prediction Analysis
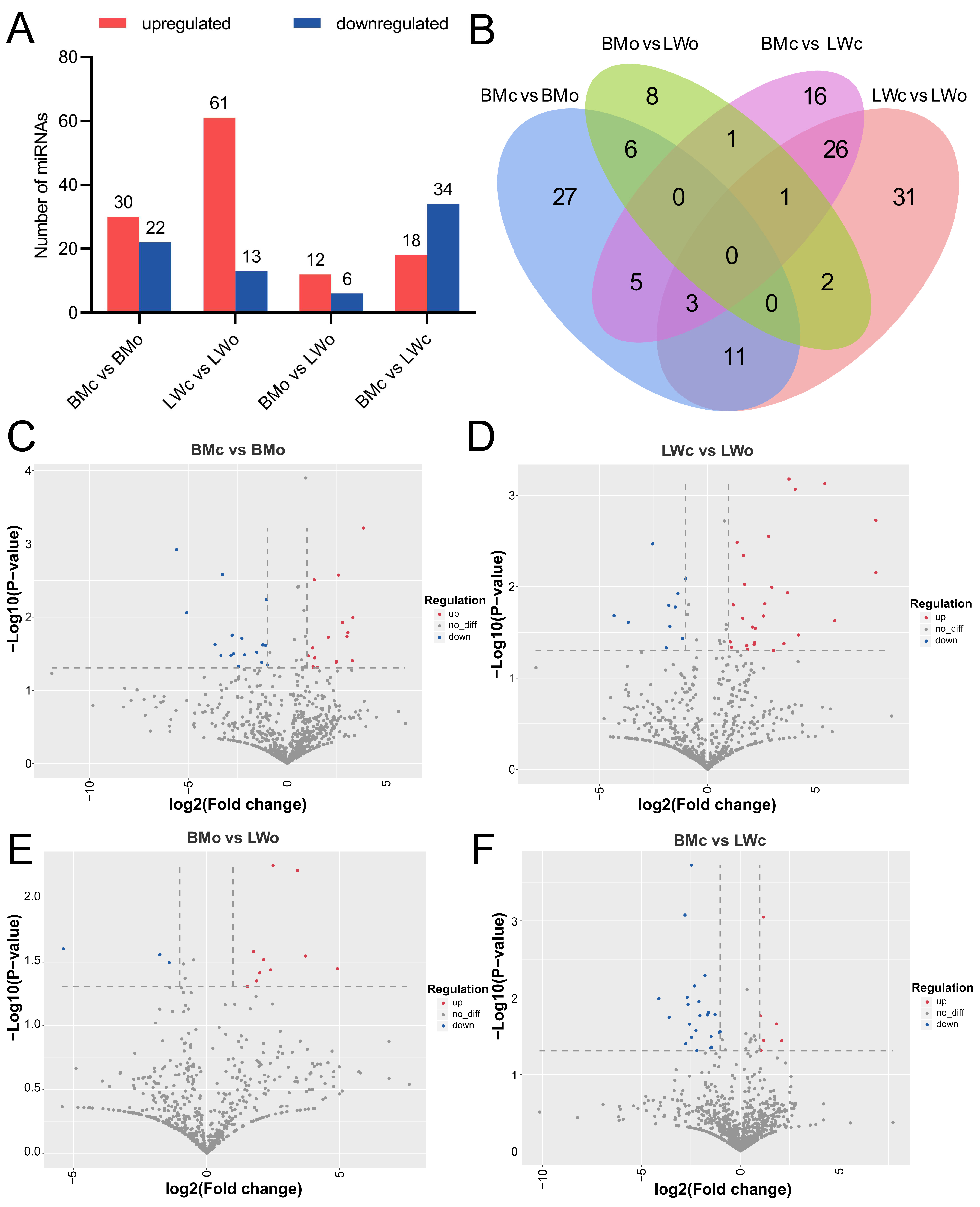
2.4. Identification of DE-miRNAs
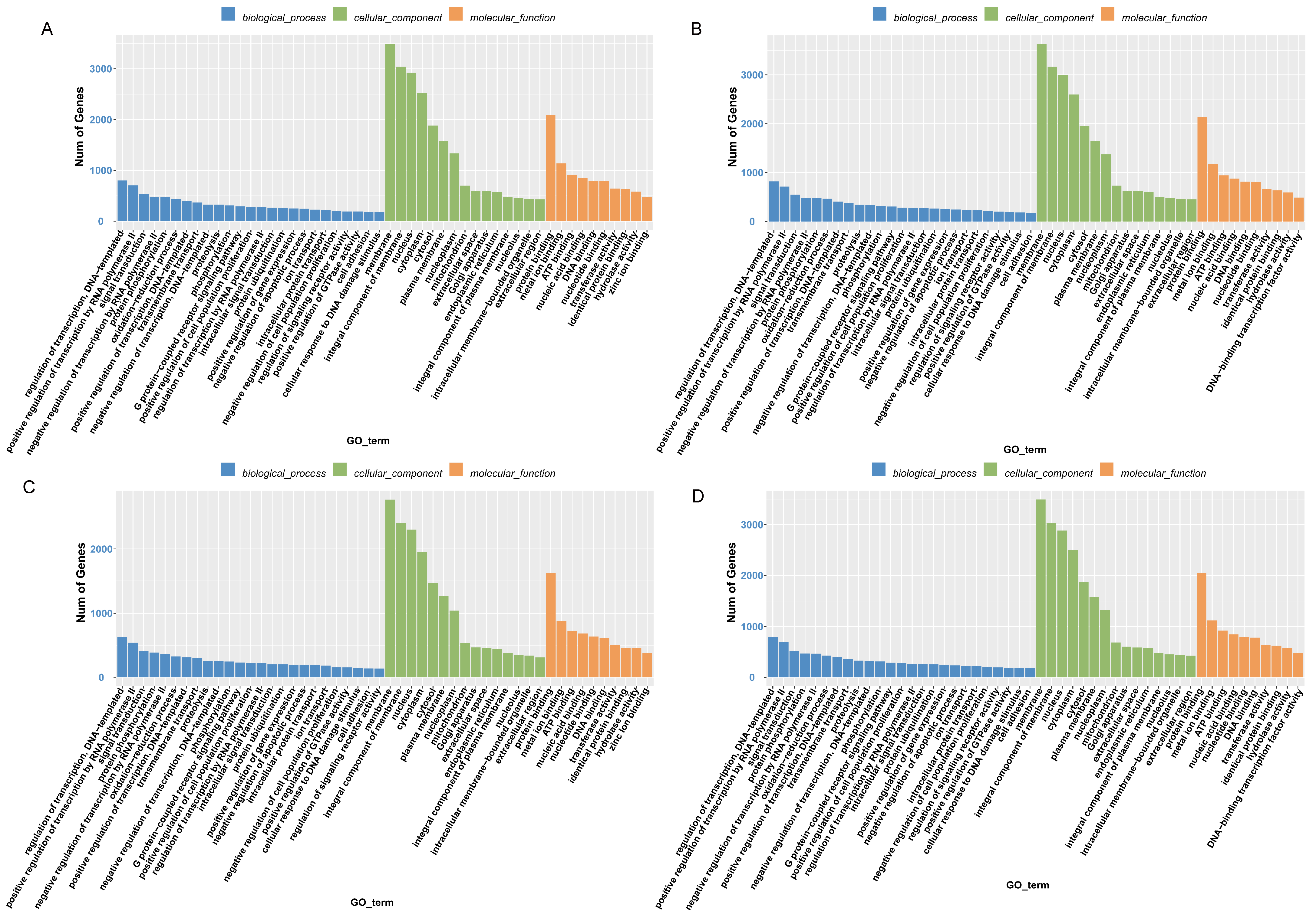
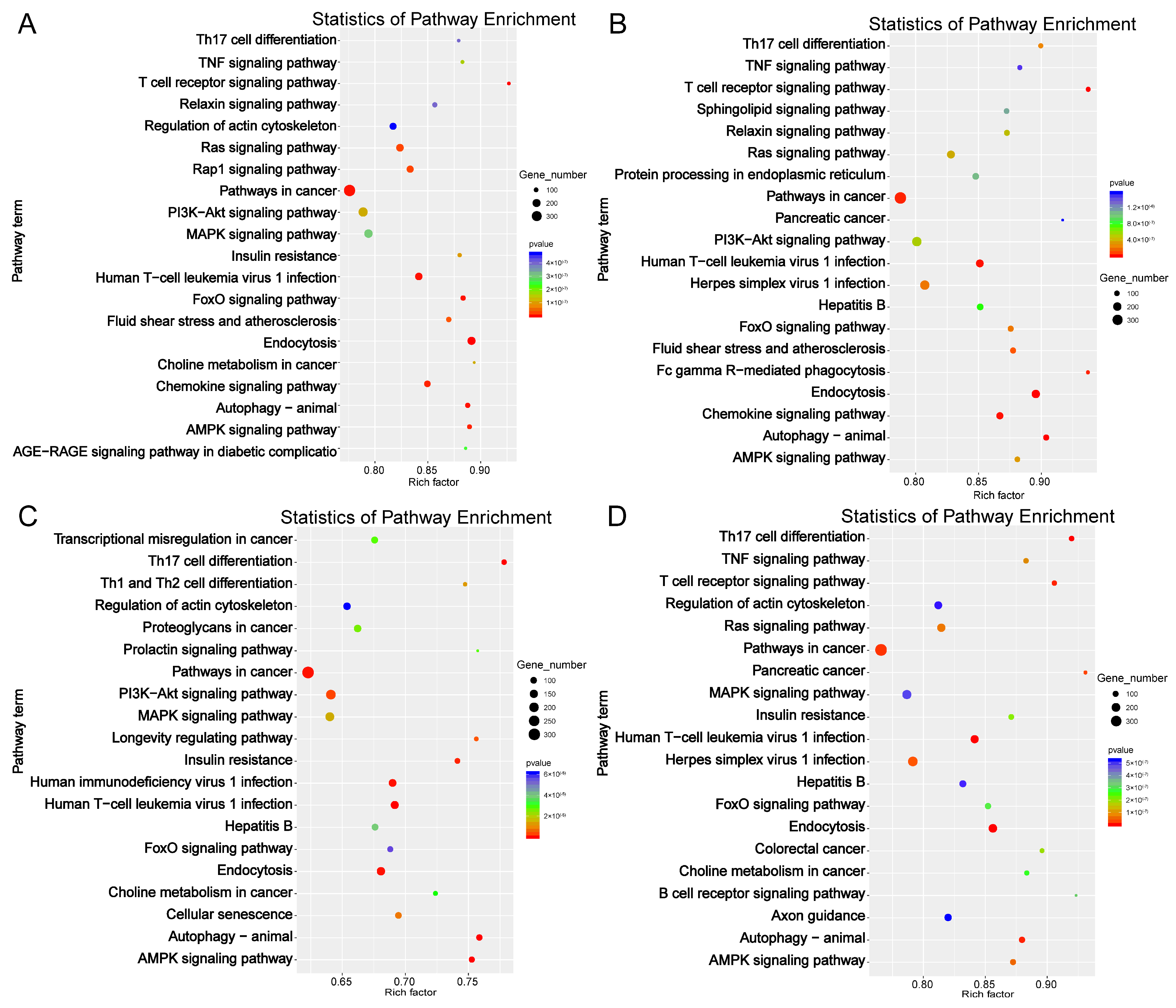
2.5. Target Gene Prediction and Functional Enrichment Analysis of DE-miRNAs
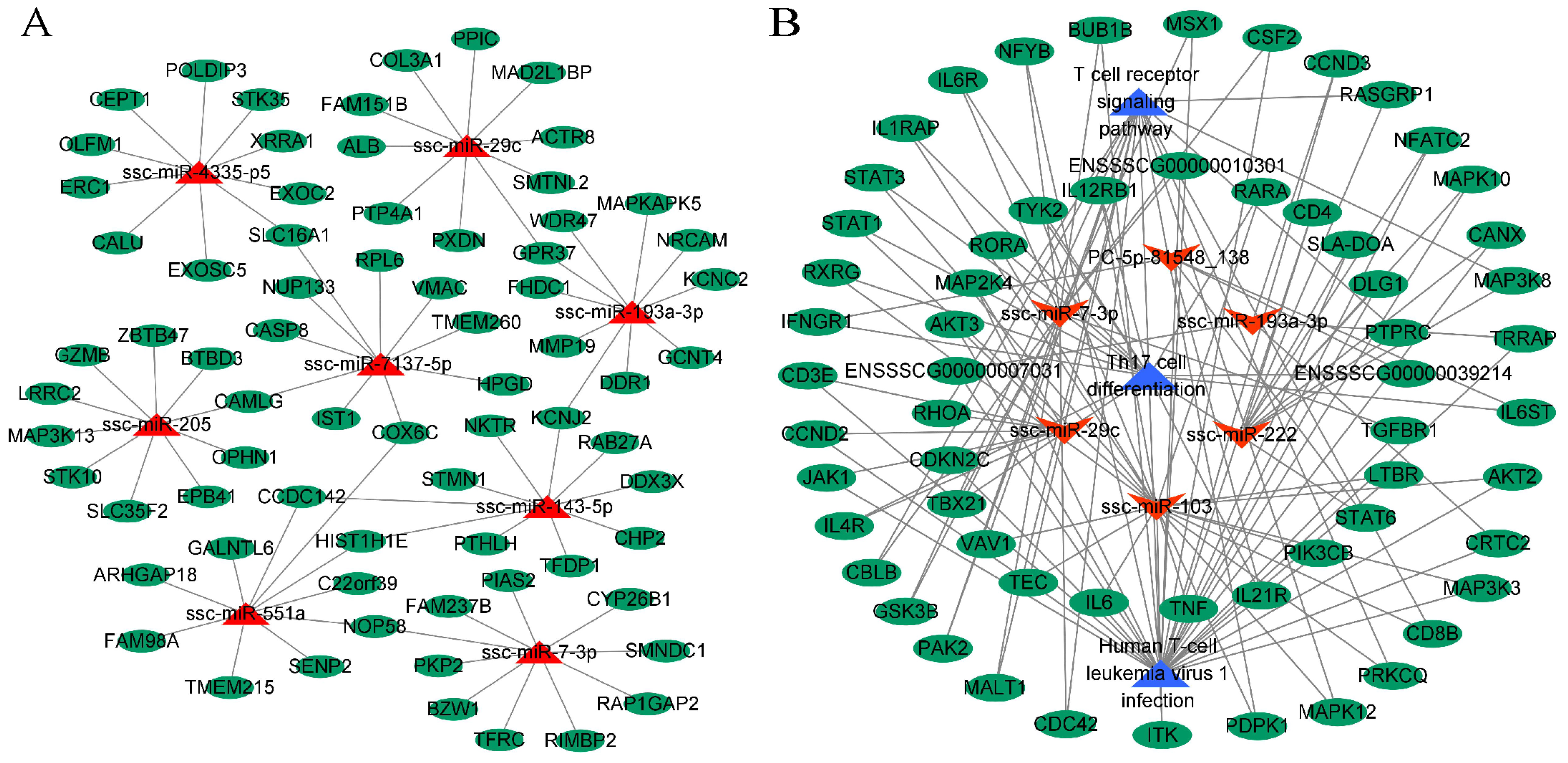
2.6. miRNA-mRNA Interaction Network Analysis
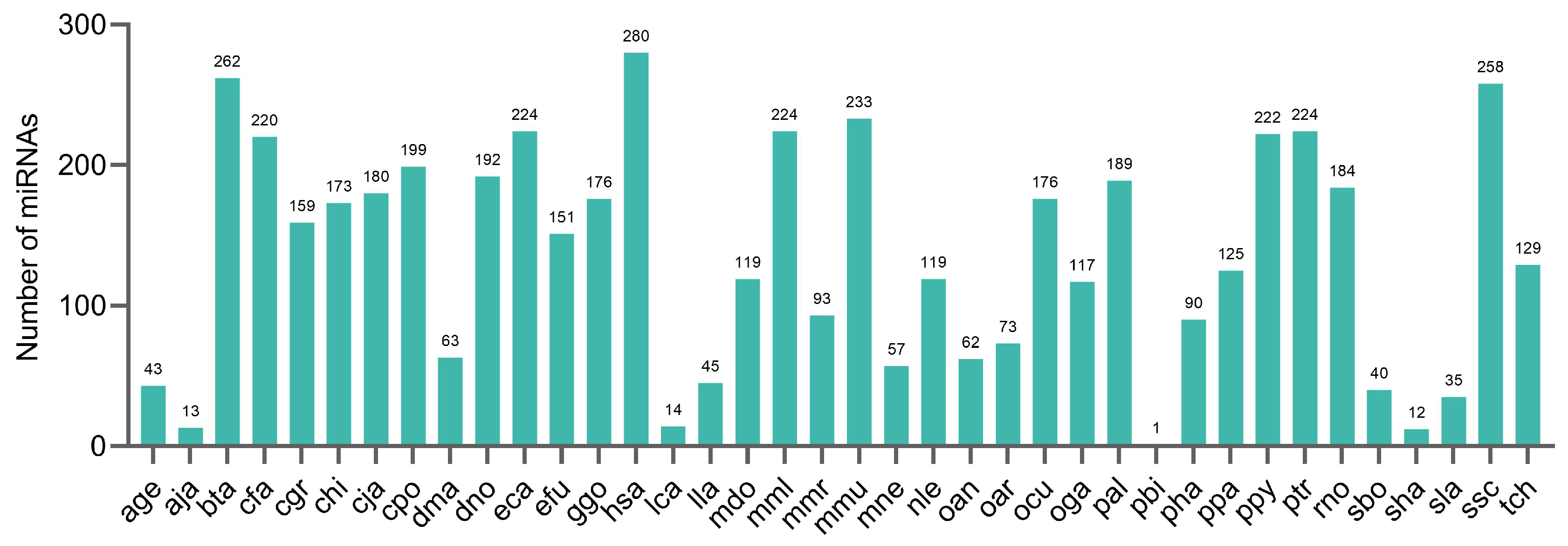
2.7. Conservation of the Identified miRNAs in Other Species
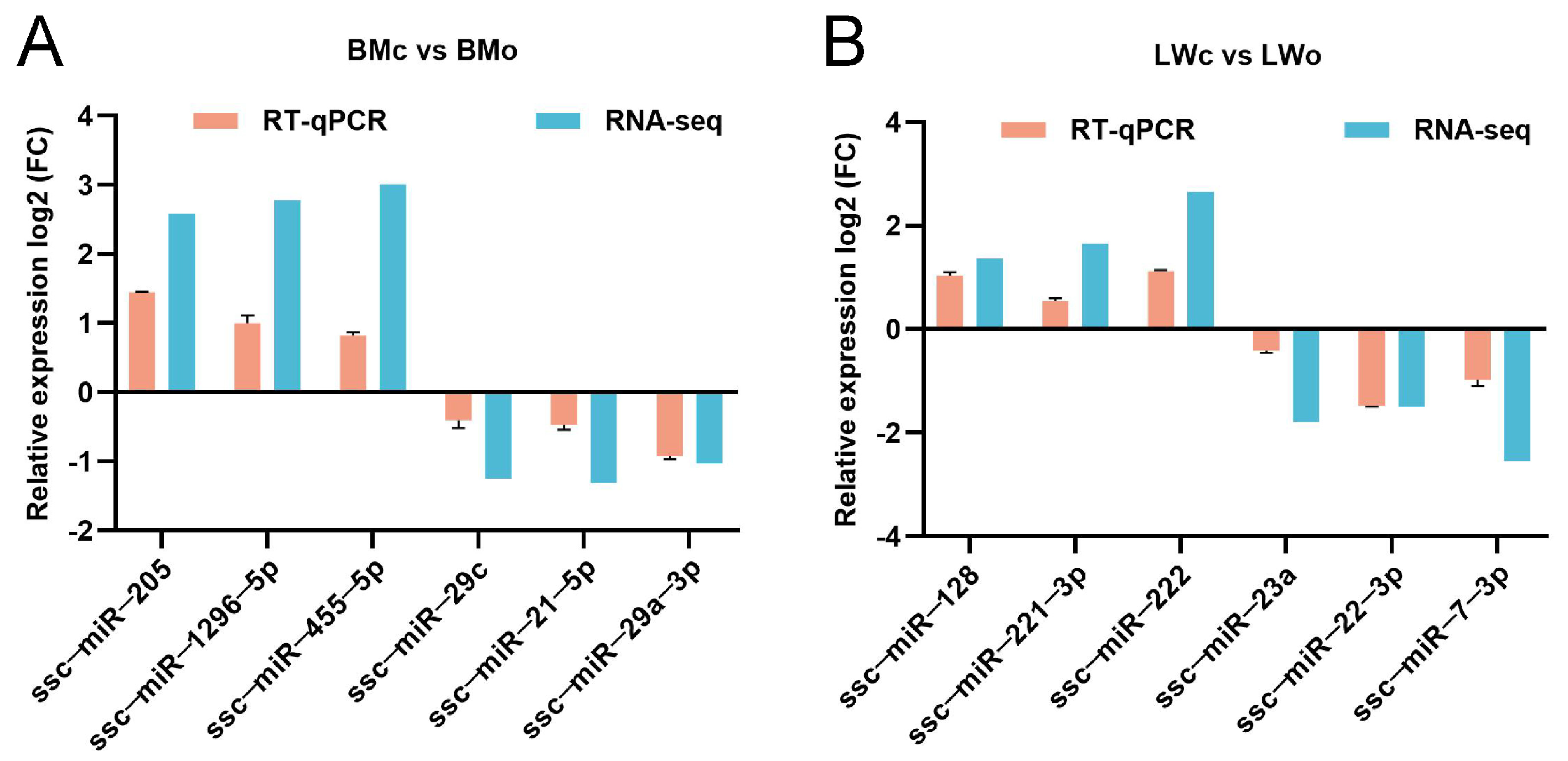
2.8. RT-qPCR Validation of DE-miRNAs
3. Discussion
4. Materials and Methods
4.1. Experimental Animal Selection
4.2. Milk Collection and Exosome Extraction
4.3. Characterization of Milk Exosomes
4.4. Library Construction and Sequencing
4.5. Raw Data Quality Control
4.6. Bioinformatics Analysis
4.7. RT-qPCR Validation of Differentially Expressed Genes
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xi, B.; Luo, J.; Gao, Y.Q.; Yang, X.L.; Guo, T.F.; Li, W.H.; Du, T.Q. Transcriptome-metabolome analysis of fatty acid of Bamei pork and Gansu Black pork in China. Bioprocess Biosyst. Eng. 2021, 44, 995–1002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, B.; Wen, X.; Sun, G. Genetic variation and relationships in the mitochondrial DNA D-loop region of Qinghai indigenous and commercial pig breeds. Cell. Mol. Biol. Lett. 2018, 23, 31. [Google Scholar] [CrossRef] [PubMed]
- Inoue, R.A.-O.X.; Tsukahara, T.A.-O. Composition and physiological functions of the porcine colostrum. Anim. Sci. J. = Nihon Chikusan Gakkaiho 2021, 92, e13618. [Google Scholar] [CrossRef] [PubMed]
- Boersma, E.R.; Offringa, P.J.; Muskiet, F.A.; Chase, W.M.; Simmons, I.J. Vitamin E, lipid fractions, and fatty acid composition of colostrum, transitional milk, and mature milk: An international comparative study. Am. J. Clin. Nutr. 1991, 53, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Jahan, M.; Francis, N.; Wang, B. Milk lactoferrin concentration of primiparous and multiparous sows during lactation. J. Dairy Sci. 2020, 103, 7521–7530. [Google Scholar] [CrossRef] [PubMed]
- Carr, L.E.; Bowlin, A.K.; Elolimy, A.A.; Byrum, S.D.; Washam, C.L.; Randolph, C.E.; MacLeod, S.L.; Yeruva, L. Neonatal Diet Impacts Circulatory miRNA Profile in a Porcine Model. Front. Immunol. 2020, 11, 1240. [Google Scholar] [CrossRef] [PubMed]
- Pieters, B.C.H.; Arntz, O.J.; Bennink, M.B.; Broeren, M.G.A.; van Caam, A.P.M.; Koenders, M.I.; van Lent, P.L.E.M.; van den Berg, W.B.; de Vries, M.; van der Kraan, P.M.; et al. Commercial cow milk contains physically stable extracellular vesicles expressing immunoregulatory TGF-β. PLoS ONE 2015, 10, e0121123. [Google Scholar] [CrossRef] [PubMed]
- Declerck, I.; Sarrazin, S.; Dewulf, J.; Maes, D. Sow and piglet factors determining variation of colostrum intake between and within litters. Animal 2017, 11, 1336–1343. [Google Scholar] [CrossRef]
- Del Pozo-Acebo, L.; Hazas, M.-C.L.d.L.; Tomé-Carneiro, J.; Gil-Cabrerizo, P.; San-Cristobal, R.; Busto, R.; García-Ruiz, A.; Dávalos, A. Bovine Milk-Derived Exosomes as a Drug Delivery Vehicle for miRNA-Based Therapy. Int. J. Mol. Sci. 2021, 22, 1105. [Google Scholar] [CrossRef]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.-M.; Xu, Y.-M.; Huang, L.-F.; Wang, X.-Z. Exosomes: Novel biomarkers for clinical diagnosis. ScientificWorldJournal 2015, 2015, 657086. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology function and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Stremmel, W.; Weiskirchen, R.; John, S.M.; Schmitz, G. Exosome-Derived MicroRNAs of Human Milk and Their Effects on Infant Health and Development. Biomolecules 2021, 11, 851. [Google Scholar] [CrossRef] [PubMed]
- Miao, C.; Wang, X.; Zhou, W.; Huang, J. The emerging roles of exosomes in autoimmune diseases, with special emphasis on microRNAs in exosomes. Pharmacol. Res. 2021, 169, 105680. [Google Scholar] [CrossRef] [PubMed]
- Sterzenbach, U.; Putz, U.; Low, L.-H.; Silke, J.; Tan, S.-S.; Howitt, J. Engineered Exosomes as Vehicles for Biologically Active Proteins. Mol. Ther. 2017, 25, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Izumi, H.; Sekine, K.; Ochiya, T. microRNA as a new immune-regulatory agent in breast milk. Silence 2010, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.-Y.; Chen, T.; Xi, Q.-Y.; Hou, L.-J.; Luo, J.-Y.; Zeng, B.; Li, M.; Sun, J.-J.; Zhang, Y.-L. Porcine milk exosome miRNAs protect intestinal epithelial cells against deoxynivalenol-induced damage. Biochem. Pharmacol. 2020, 175, 113898. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xie, M.-Y.; Sun, J.-J.; Ye, R.-S.; Cheng, X.; Sun, R.-P.; Wei, L.-M.; Li, M.; Lin, D.-L.; Jiang, Q.-Y.; et al. Porcine milk-derived exosomes promote proliferation of intestinal epithelial cells. Sci. Rep. 2016, 6, 33862. [Google Scholar] [CrossRef]
- Xie, M.-Y.; Hou, L.-J.; Sun, J.-J.; Zeng, B.; Xi, Q.-Y.; Luo, J.-Y.; Chen, T.; Zhang, Y.-L. Porcine Milk Exosome MiRNAs Attenuate LPS-Induced Apoptosis through Inhibiting TLR4/NF-κB and p53 Pathways in Intestinal Epithelial Cells. J. Agric. Food Chem. 2019, 67, 9477–9491. [Google Scholar] [CrossRef]
- Lin, D.; Chen, T.; Xie, M.; Li, M.; Zeng, B.; Sun, R.; Zhu, Y.; Ye, D.; Wu, J.; Sun, J.; et al. Oral Administration of Bovine and Porcine Milk Exosome Alter miRNAs Profiles in Piglet Serum. Sci. Rep. 2020, 10, 6983. [Google Scholar] [CrossRef]
- Gu, Y.; Li, M.; Wang, T.; Liang, Y.; Zhong, Z.; Wang, X.; Zhou, Q.; Chen, L.; Lang, Q.; He, Z.; et al. Lactation-related microRNA expression profiles of porcine breast milk exosomes. PLoS ONE 2012, 7, e43691. [Google Scholar] [CrossRef]
- Sun, Q.; Chen, X.; Yu, J.; Zen, K.; Zhang, C.-Y.; Li, L. Immune modulatory function of abundant immune-related microRNAs in microvesicles from bovine colostrum. Protein Cell 2013, 4, 197–210. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhou, H.; Zhang, W. Progress in research on genetic variations in miRNA regulatory pathway. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2015, 32, 109–112. [Google Scholar] [PubMed]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; D’Souza, J.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1955–1986. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Rajewsky, K. MicroRNA control in the immune system: Basic principles. Cell 2009, 136, 26–36. [Google Scholar] [CrossRef]
- Jia, S.; Zhai, H.; Zhao, M. MicroRNAs regulate immune system via multiple targets. Discov. Med. 2014, 18, 237–247. [Google Scholar]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Özdemir, S. Identification and comparison of exosomal microRNAs in the milk and colostrum of two different cow breeds. Gene 2020, 743, 144609. [Google Scholar] [CrossRef]
- Izumi, H.; Kosaka, N.; Shimizu, T.; Sekine, K.; Ochiya, T.; Takase, M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J. Dairy Sci. 2012, 95, 4831–4841. [Google Scholar] [CrossRef]
- Hock, A.; Miyake, H.; Li, B.; Lee, C.; Ermini, L.; Koike, Y.; Chen, Y.; Määttänen, P.; Zani, A.; Pierro, A. Breast milk-derived exosomes promote intestinal epithelial cell growth. J. Pediatr Surg. 2017, 52, 755–759. [Google Scholar] [CrossRef]
- Chen, T.; Xi, Q.Y.; Ye, R.S.; Cheng, X.; Qi, Q.E.; Wang, S.B.; Shu, G.; Wang, L.N.; Zhu, X.T.; Jiang, Q.Y.; et al. Exploration of microRNAs in porcine milk exosomes. BMC Genom. 2014, 15, 100. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Xie, Y.; Guo, J.; Su, X.; Zhao, C.; Zhang, C.; Qin, Q.; Dai, D.; Tuo, Y.; Li, Z.; et al. Comparison of porcine milk microRNA expression in milk exosomes versus whole swine milk and prediction of target genes. Arch. Anim. Breed. 2022, 65, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Luo, H.; Xiong, Z.; Wan, K.; Liao, Q.; He, H. High-throughput sequencing reveals biofluid exosomal miRNAs associated with immunity in pigs. Biosci. Biotechnol. Biochem. 2020, 84, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Liu, J.; Xu, B.; Yu, J.; Zhang, A.; Qin, L.; Liu, C.; Yang, Y. miR-21-5p inhibits inflammation injuries in LPS-treated H9c2 cells by regulating PDCD4. Am. J. Transl. Res. 2021, 13, 11450–11460. [Google Scholar] [PubMed]
- Cao, J.; Zhang, Y.; Mu, J.; Yang, D.; Gu, X.; Zhang, J. Exosomal miR-21-5p contributes to ovarian cancer progression by regulating CDK6. Human Cell 2021, 34, 1185–1196. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Kong, F.J.; Liang, Y.; Yang, X.M.; Yang, Y.K.; Zhong, Z.J.; Wang, Y.; Hu, Z.H.; Chen, X.H.; Gong, J.J.; et al. Screening of candidate genes related to differences in growth and development between Chinese indigenous and Western pig breeds. Physiol. Genom. 2023, 55, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Nouri, Y.; Weinkove, R.; Perret, R. T-cell intrinsic Toll-like receptor signaling: Implications for cancer immunotherapy and CAR T-cells. J. Immunother. Cancer 2021, 9, e003065. [Google Scholar] [CrossRef]
- Wu, Z.; Cai, Z.; Shi, H.; Huang, X.; Cai, M.; Yuan, K.; Huang, P.; Shi, G.; Yan, T.; Li, Z. Effective biomarkers and therapeutic targets of nerve-immunity interaction in the treatment of depression: An integrated investigation of the miRNA-mRNA regulatory networks. Aging 2022, 14, 3569–3596. [Google Scholar] [CrossRef]
- Shah, K.; Al-Haidari, A.; Sun, J.; Kazi, J.U. T cell receptor (TCR) signaling in health and disease. Signal Transduct. Target Ther. 2021, 6, 412. [Google Scholar] [CrossRef]
- Yasuda, K.; Takeuchi, Y.; Hirota, K. The pathogenicity of Th17 cells in autoimmune diseases. Semin. Immunopathol. 2019, 41, 283–297. [Google Scholar] [CrossRef]
- Wang, G.; Su, Z.; Li, H.; Xiao, L.; Li, C.; Lian, G. The role of metabolism in Th17 cell differentiation and autoimmune diseases. Int. Immunopharmacol. 2022, 103, 108450. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Zhou, M.; Chen, C.; Wu, X.; Wang, X. Role of AMPK mediated pathways in autophagy and aging. Biochimie 2022, 195, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.; Di Fiore, P.P. Endocytosis conducts the cell signaling orchestra. Cell 2006, 124, 897–900. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, M.; Piddini, E.; Fau-Vincent, J.-P.; Vincent, J.P. Endocytosis: A positive or a negative influence on Wnt signalling? Traffic 2008, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; He, P.; Huang, Y.; Li, Y.-F.; Lu, J.; Li, M.; Kurihara, H.; Luo, Z.; Meng, T.; Onishi, M.; et al. Selective autophagy of intracellular organelles: Recent research advances. Theranostics 2021, 11, 222–256. [Google Scholar] [CrossRef]
- Wu, D.J.; Adamopoulos, I.E. Autophagy and autoimmunity. Clin. Immunol. 2017, 176, 55–62. [Google Scholar] [CrossRef]
- Liao, S.-X.; Sun, P.-P.; Gu, Y.-H.; Rao, X.-M.; Zhang, L.-Y.; Ou-Yang, Y. Autophagy and pulmonary disease. Ther. Adv. Respir. Dis. 2019, 13, 1753466619890538. [Google Scholar] [CrossRef]
- Saha, S.; Panigrahi, D.P.; Patil, S.; Bhutia, S.K. Autophagy in health and disease: A comprehensive review. Biomed. Pharmacother. 2018, 104, 485–495. [Google Scholar] [CrossRef]
- van Herwijnen, M.J.C.; Driedonks, T.A.P.; Snoek, B.L.; Kroon, A.M.T.; Kleinjan, M.; Jorritsma, R.; Pieterse, C.M.J.; Hoen, E.N.M.N.-t.; Wauben, M.H.M. Abundantly Present miRNAs in Milk-Derived Extracellular Vesicles Are Conserved Between Mammals. Front. Nutr. 2018, 5, 81. [Google Scholar] [CrossRef] [PubMed]
- Zhi, L.; Yu, Y.; Li, X.; Wang, D.; Wang, D. Molecular Control of Innate Immune Response to Pseudomonas aeruginosa Infection by Intestinal let-7 in Caenorhabditis elegans. PLoS Pathog. 2017, 13, e1006152. [Google Scholar] [CrossRef] [PubMed]
- Gilles, M.-E.; Slack, F.J. Let-7 microRNA as a potential therapeutic target with implications for immunotherapy. Expert. Opin. Ther. Targets 2018, 22, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Sun, M.; Ma, K.; Liu, J. Let-7d-5p suppresses inflammatory response in neonatal rats with necrotizing enterocolitis via LGALS3-mediated TLR4/NF-κB signaling pathway. Am. J. Physiol. Cell Physiol. 2020, 319, C967–C979. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Fau-Pop, M.; Pop, M.; Fau-Salzberg, S.L.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Love, M.; Fau-Huber, W.; Huber, W.; Fau-Anders, S.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Nam, J.-W.; Rissland, O.S.; Koppstein, D.; Abreu-Goodger, C.; Jan, C.H.; Agarwal, V.; Yildirim, M.A.; Rodriguez, A.; Bartel, D.P. Global analyses of the effect of different cellular contexts on microRNA targeting. Mol. Cell 2014, 53, 1031–1043. [Google Scholar] [CrossRef]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
| Sample | Total Reads | Total Bases | Valid Reads | Valid Ratio% | Q20% | Q30% | GC% |
|---|---|---|---|---|---|---|---|
| BMo-1 | 38,893,587 | 1,983,572,937 | 9,441,552 | 24.28 | 98.06 | 94.19 | 57.34 |
| BMo-2 | 35,837,285 | 1,827,701,535 | 7,261,431 | 20.26 | 97.98 | 94.08 | 56.19 |
| BMo-3 | 33,766,916 | 1,722,112,716 | 8,741,118 | 25.89 | 97.93 | 94.08 | 57.40 |
| BMc-1 | 40,678,884 | 2,074,623,084 | 11,340,306 | 27.88 | 98.08 | 94.34 | 54.35 |
| BMc-2 | 40,108,481 | 2,045,532,531 | 11,316,963 | 28.22 | 98.05 | 94.30 | 56.13 |
| BMc-3 | 31,262,225 | 1,594,373,475 | 9,570,925 | 30.61 | 97.70 | 93.30 | 55.17 |
| LWo-1 | 36,276,785 | 1,850,116,035 | 6,949,261 | 19.16 | 97.51 | 93.03 | 57.01 |
| LWo-2 | 38,368,116 | 1,956,773,916 | 4,574,542 | 11.92 | 98.07 | 94.33 | 53.28 |
| LWo-3 | 33,638,761 | 1,715,576,811 | 2,902,916 | 8.63 | 97.73 | 93.55 | 57.57 |
| LWc-1 | 33,048,336 | 1,685,465,136 | 11,280,944 | 34.13 | 98.11 | 94.34 | 54.90 |
| LWc-2 | 39,577,542 | 2,018,454,642 | 10,166,522 | 25.69 | 98.15 | 94.5 | 55.78 |
| LWc-3 | 32,290,639 | 1,646,822,589 | 10,117,959 | 31.33 | 98.11 | 94.44 | 56.05 |
| miRNA Name | miRNA Sequence (5′-3′) | Groups | Up/Down |
|---|---|---|---|
| ssc-miR-340 | TTATAAAGCAATGAGACTGATT | BMc vs. BMo | up |
| ssc-miR-30e-5p | TGTAAACATCCTTGACTGGAAGCT | BMc vs. BMo | up |
| ssc-miR-205 | TCCTTCATTCCACCGGAGTCTGT | LWc vs. LWo | up |
| ssc-miR-221-3p | AGCTACATTGTCTGCTGGGTTT | LWc vs. LWo | up |
| ssc-miR-29a-3p | TAGCACCATCTGAAATCGGTTA | BMc vs. LWc | up |
| ssc-miR-103 | AGCAGCATTGTACAGGGCTATGA | BMc vs. LWc | up |
| ssc-miR-141 | TAACACTGTCTGGTAAAGATGGC | BMo vs. LWo | down |
| ssc-miR-22-3p | AAGCTGCCAGTTGAAGAACTGT | BMo vs. LWo | down |
| miRNA Name | Primer Sequence (5′-3′) |
|---|---|
| ssc-miR-205 | TCCTTCATTCCACCGGAGTCT |
| ssc-miR-1296-5p | TTAGGGCCCTGGCTCCATCT |
| ssc-miR-455-5p | TGTGCCTTTGGACTACATCG |
| ssc-miR-29c | TAGCACCATTTGAAATCGG |
| ssc-miR-21-5p | GCTTATCAGACTGATGTTG |
| ssc-miR-29a-3p | TAGCACCATCTGAAATCGGT |
| ssc-miR-128 | TCACAGTGAACCGGTCTCT |
| ssc-miR-221-3p | AGCTACATTGTCTGCTGGGT |
| ssc-miR-222 | CTACATCTGGCTACTGGGT |
| ssc-miR-23a | TCACATTGCCAGGGATTTCC |
| ssc-miR-22-3p | AGCTGCCAGTTGAAGAACTGT |
| ssc-miR-7-3p | CAACAAATCACAGTCTGCC |
| U6-F | GGAACGATACAGAGAAGATTAGC |
| U6-R | TGGAACGCTTCACGAATTTGCG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Shang, X.; Zhang, S.; Yang, Q.; Yan, Z.; Wang, P.; Gao, X.; Gun, S.; Huang, X. Breed-Related Differential microRNA Expression and Analysis of Colostrum and Mature Milk Exosomes in Bamei and Landrace Pigs. Int. J. Mol. Sci. 2024, 25, 667. https://doi.org/10.3390/ijms25010667
Li J, Shang X, Zhang S, Yang Q, Yan Z, Wang P, Gao X, Gun S, Huang X. Breed-Related Differential microRNA Expression and Analysis of Colostrum and Mature Milk Exosomes in Bamei and Landrace Pigs. International Journal of Molecular Sciences. 2024; 25(1):667. https://doi.org/10.3390/ijms25010667
Chicago/Turabian StyleLi, Jie, Xuefeng Shang, Sen Zhang, Qiaoli Yang, Zunqiang Yan, Pengfei Wang, Xiaoli Gao, Shuangbao Gun, and Xiaoyu Huang. 2024. "Breed-Related Differential microRNA Expression and Analysis of Colostrum and Mature Milk Exosomes in Bamei and Landrace Pigs" International Journal of Molecular Sciences 25, no. 1: 667. https://doi.org/10.3390/ijms25010667
APA StyleLi, J., Shang, X., Zhang, S., Yang, Q., Yan, Z., Wang, P., Gao, X., Gun, S., & Huang, X. (2024). Breed-Related Differential microRNA Expression and Analysis of Colostrum and Mature Milk Exosomes in Bamei and Landrace Pigs. International Journal of Molecular Sciences, 25(1), 667. https://doi.org/10.3390/ijms25010667






