Different Transcriptome Features of Peripheral Blood Mononuclear Cells in Non-Emphysematous Chronic Obstructive Pulmonary Disease
Abstract
1. Introduction
2. Results
2.1. Differential Gene Expression and Pathway Analysis in PBMCs between Patients with COPD and Healthy Controls
2.2. Differential Gene Expression and Pathway Analysis in PBMCs between Non-Emphysematous and Emphysematous COPD
3. Discussion
3.1. COPD vs. HCs
Hematopoietic Cell Lineage
3.2. Emphysematous vs. Non-Emphysematous COPD
3.2.1. GO Terms Related to T Lymphocytes
3.2.2. Keratan Sulfate Biosynthesis and Metabolic Process
3.2.3. ECM-Receptor Interaction
3.2.4. Focal Adhesion
3.2.5. Hedgehog Signaling Pathway
3.2.6. Angiogenesis Regulation
3.2.7. Interim Summary Regarding the Genetic Profile of Non-Emphysematous COPD
3.3. Study Limitations
4. Materials and Methods
4.1. Participants
4.2. Isolation of PBMCs
4.3. Total RNA Extraction, mRNA Library Preparation, and 3’ RNA Sequencing
4.4. 3’ RNA Sequencing Data Analysis
4.5. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Halpin, D.M.G.; Criner, G.J.; Papi, A.; Singh, D.; Anzueto, A.; Martinez, F.J.; Agusti, A.A.; Vogelmeier, C.F. Global Initiative for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease. The 2020 GOLD Science Committee Report on COVID-19 and Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2021, 203, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Lokke, A.; Lange, P.; Scharling, H.; Fabricius, P.; Vestbo, J. Developing COPD: A 25 year follow up study of the general population. Thorax 2006, 61, 935–939. [Google Scholar] [CrossRef] [PubMed]
- Myronenko, O.; Foris, V.; Crnkovic, S.; Olschewski, A.; Rocha, S.; Nicolls, M.R.; Olschewski, H. Endotyping COPD: Hypoxia-inducible factor-2 as a molecular “switch” between the vascular and airway phenotypes? Eur. Respir. Rev. Off. J. Eur. Respir. Soc. 2023, 32, 220173. [Google Scholar] [CrossRef] [PubMed]
- Kapellos, T.S.; Bonaguro, L.; Gemund, I.; Reusch, N.; Saglam, A.; Hinkley, E.R.; Schultze, J.L. Human Monocyte Subsets and Phenotypes in Major Chronic Inflammatory Diseases. Front. Immunol. 2019, 10, 2035. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, S.; Li, Q.; Li, D.; Li, Y.; Xie, X.; Yuan, L.; Lin, Z.; Lin, F.; Wei, X.; et al. Gene Expression Trajectories from Normal Nonsmokers to COPD Smokers and Disease Progression Discriminant Modeling in Response to Cigarette Smoking. Dis. Markers 2022, 2022, 9354286. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.A.; Tsai, M.J.; Jian, S.F.; Sheu, C.C.; Kuo, P.L. Systematic analysis of transcriptomic profiles of COPD airway epithelium using next-generation sequencing and bioinformatics. Int. J. Chronic Obstr. Pulm. Dis. 2018, 13, 2387–2398. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; An, S.; Dai, L.; Xu, S.; Liu, D.; Wang, L.; Zhang, R.; Wang, F.; Wang, Z. Identification of Potential Differentially-Methylated/Expressed Genes in Chronic Obstructive Pulmonary Disease. COPD 2023, 20, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Poliska, S.; Csanky, E.; Szanto, A.; Szatmari, I.; Mesko, B.; Szeles, L.; Dezso, B.; Scholtz, B.; Podani, J.; Kilty, I.; et al. Chronic obstructive pulmonary disease-specific gene expression signatures of alveolar macrophages as well as peripheral blood monocytes overlap and correlate with lung function. Respiration 2011, 81, 499–510. [Google Scholar] [CrossRef]
- Bahr, T.M.; Hughes, G.J.; Armstrong, M.; Reisdorph, R.; Coldren, C.D.; Edwards, M.G.; Schnell, C.; Kedl, R.; LaFlamme, D.J.; Reisdorph, N.; et al. Peripheral blood mononuclear cell gene expression in chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2013, 49, 316–323. [Google Scholar] [CrossRef]
- Roffel, M.P.; Bracke, K.R.; Heijink, I.H.; Maes, T. miR-223: A Key Regulator in the Innate Immune Response in Asthma and COPD. Front. Med. 2020, 7, 196. [Google Scholar] [CrossRef]
- Tan, Y.; Qiao, Y.; Chen, Z.; Liu, J.; Guo, Y.; Tran, T.; Tan, K.S.; Wang, D.Y.; Yan, Y. FGF2, an Immunomodulatory Factor in Asthma and Chronic Obstructive Pulmonary Disease (COPD). Front. Cell Dev. Biol. 2020, 8, 223. [Google Scholar] [CrossRef] [PubMed]
- Salter, B.M.; Sehmi, R. Hematopoietic Processes in Eosinophilic Asthma. Chest 2017, 152, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Mahor, D.; Kumari, V.; Vashisht, K.; Galgalekar, R.; Samarth, R.M.; Mishra, P.K.; Banerjee, N.; Dixit, R.; Saluja, R.; De, S.; et al. Elevated serum matrix metalloprotease (MMP-2) as a candidate biomarker for stable COPD. BMC Pulm. Med. 2020, 20, 302. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, J.T.; Plosa, E.J.; Sucre, J.M.; van der Meer, R.; Dave, S.; Gutor, S.; Nichols, D.S.; Gulleman, P.M.; Jetter, C.S.; Han, W.; et al. Neutrophilic inflammation during lung development disrupts elastin assembly and predisposes adult mice to COPD. J. Clin. Investig. 2021, 131, e139481. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Takahashi, K.; Shiozaki, K.; Yamaguchi, K.; Moriya, S.; Hosono, M.; Shima, H.; Miyagi, T. Potentiation of epidermal growth factor-mediated oncogenic transformation by sialidase NEU3 leading to Src activation. PLoS ONE 2015, 10, e0120578. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sheng, J.; Hu, S.; Cui, Y.; Xiao, J.; Yu, W.; Peng, J.; Han, W.; He, Q.; Fan, Y.; et al. Estrogen and G protein-coupled estrogen receptor accelerate the progression of benign prostatic hyperplasia by inducing prostatic fibrosis. Cell Death Dis. 2022, 13, 533. [Google Scholar] [CrossRef]
- Aono, Y.; Suzuki, Y.; Horiguchi, R.; Inoue, Y.; Karayama, M.; Hozumi, H.; Furuhashi, K.; Enomoto, N.; Fujisawa, T.; Nakamura, Y.; et al. CD109 on Dendritic Cells Regulates Airway Hyperreactivity and Eosinophilic Airway Inflammation. Am. J. Respir. Cell Mol. Biol. 2023, 68, 201–212. [Google Scholar] [CrossRef]
- Yang, Q.; Underwood, M.J.; Hsin, M.K.; Liu, X.C.; He, G.W. Dysfunction of pulmonary vascular endothelium in chronic obstructive pulmonary disease: Basic considerations for future drug development. Curr. Drug Metab. 2008, 9, 661–667. [Google Scholar] [CrossRef]
- Kyriakopoulos, C.; Chronis, C.; Papapetrou, E.; Tatsioni, A.; Gartzonika, K.; Tsaousi, C.; Gogali, A.; Katsanos, C.; Vaggeli, A.; Tselepi, C.; et al. Prothrombotic state in patients with stable COPD: An observational study. ERJ Open Res. 2021, 7, 00297–2021. [Google Scholar] [CrossRef]
- Lundwall, A.; Dackowski, W.; Cohen, E.; Shaffer, M.; Mahr, A.; Dahlback, B.; Stenflo, J.; Wydro, R. Isolation and sequence of the cDNA for human protein S, a regulator of blood coagulation. Proc. Natl. Acad. Sci. USA 1986, 83, 6716–6720. [Google Scholar] [CrossRef]
- Sauler, M.; Lamontagne, M.; Finnemore, E.; Herazo-Maya, J.D.; Tedrow, J.; Zhang, X.; Morneau, J.E.; Sciurba, F.; Timens, W.; Pare, P.D.; et al. The DNA repair transcriptome in severe COPD. Eur. Respir. J. 2018, 52, 1701994. [Google Scholar] [CrossRef] [PubMed]
- Chu, D.K.; Llop-Guevara, A.; Walker, T.D.; Flader, K.; Goncharova, S.; Boudreau, J.E.; Moore, C.L.; Seunghyun In, T.; Waserman, S.; Coyle, A.J.; et al. IL-33, but not thymic stromal lymphopoietin or IL-25, is central to mite and peanut allergic sensitization. J. Allergy Clin. Immunol. 2013, 131, 187–200.e8. [Google Scholar] [CrossRef] [PubMed]
- Baines, K.J.; Fu, J.J.; McDonald, V.M.; Gibson, P.G. Airway gene expression of IL-1 pathway mediators predicts exacerbation risk in obstructive airway disease. Int. J. Chronic Obs. Pulmon Dis. 2017, 12, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, M.; Bautista, R.; Larrosa, R.; Cobo, M.A.; Claros, M.G. Biomarker potential of repetitive-element transcriptome in lung cancer. PeerJ 2019, 7, e8277. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Chang, C.Y.; You, R.; Shan, M.; Gu, B.H.; Madison, M.C.; Diehl, G.; Perusich, S.; Song, L.Z.; Cornwell, L.; et al. Cigarette smoke-induced reduction of C1q promotes emphysema. JCI Insight 2019, 4, e124317. [Google Scholar] [CrossRef] [PubMed]
- Draijer, C.; Penke, L.R.K.; Peters-Golden, M. Distinctive Effects of GM-CSF and M-CSF on Proliferation and Polarization of Two Major Pulmonary Macrophage Populations. J. Immunol. 2019, 202, 2700–2709. [Google Scholar] [CrossRef] [PubMed]
- Wick, M.J.; Buesing, E.J.; Wehling, C.A.; Loomis, Z.L.; Cool, C.D.; Zamora, M.R.; Miller, Y.E.; Colgan, S.P.; Hersh, L.B.; Voelkel, N.F.; et al. Decreased neprilysin and pulmonary vascular remodeling in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2011, 183, 330–340. [Google Scholar] [CrossRef]
- Odani, T.; Yasuda, S.; Ota, Y.; Fujieda, Y.; Kon, Y.; Horita, T.; Kawaguchi, Y.; Atsumi, T.; Yamanaka, H.; Koike, T. Up-regulated expression of HLA-DRB5 transcripts and high frequency of the HLA-DRB5*01:05 allele in scleroderma patients with interstitial lung disease. Rheumatology 2012, 51, 1765–1774. [Google Scholar] [CrossRef]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- Rumsaeng, V.; Vliagoftis, H.; Oh, C.K.; Metcalfe, D.D. Lymphotactin gene expression in mast cells following Fc(epsilon) receptor I aggregation: Modulation by TGF-beta, IL-4, dexamethasone, and cyclosporin A. J. Immunol. 1997, 158, 1353–1360. [Google Scholar] [CrossRef]
- Mekori, Y.A.; Metcalfe, D.D. Mast cell-T cell interactions. J. Allergy Clin. Immunol. 1999, 104, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.D.; Fohner, A.; Booker, J.D.; Dong, C.; Krensky, A.M.; Nadeau, K.C. XCL1 enhances regulatory activities of CD4+ CD25high CD127low/− T cells in human allergic asthma. J. Immunol. 2008, 181, 5386–5395. [Google Scholar] [CrossRef] [PubMed]
- Langlois, A.; Chouinard, F.; Flamand, N.; Ferland, C.; Rola-Pleszczynski, M.; Laviolette, M. Crucial implication of protein kinase C (PKC)-delta, PKC-zeta, ERK-1/2, and p38 MAPK in migration of human asthmatic eosinophils. J. Leukoc. Biol. 2009, 85, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Kytikova, O.Y.; Novgorodtseva, T.P.; Denisenko, Y.K.; Naumov, D.E.; Gvozdenko, T.A.; Perelman, J.M. Thermosensory Transient Receptor Potential Ion Channels and Asthma. Biomedicines 2021, 9, 816. [Google Scholar] [CrossRef] [PubMed]
- Tibbitt, C.A.; Stark, J.M.; Martens, L.; Ma, J.; Mold, J.E.; Deswarte, K.; Oliynyk, G.; Feng, X.; Lambrecht, B.N.; De Bleser, P.; et al. Single-Cell RNA Sequencing of the T Helper Cell Response to House Dust Mites Defines a Distinct Gene Expression Signature in Airway Th2 Cells. Immunity 2019, 51, 169–184.e165. [Google Scholar] [CrossRef] [PubMed]
- Kiwamoto, T.; Brummet, M.E.; Wu, F.; Motari, M.G.; Smith, D.F.; Schnaar, R.L.; Zhu, Z.; Bochner, B.S. Mice deficient in the St3gal3 gene product alpha2,3 sialyltransferase (ST3Gal-III) exhibit enhanced allergic eosinophilic airway inflammation. J. Allergy Clin. Immunol. 2014, 133, 240–247.e3. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Fujinawa, R.; Yoshida, T.; Ueno, M.; Ota, F.; Kizuka, Y.; Hirayama, T.; Korekane, H.; Kitazume, S.; Maeno, T.; et al. A keratan sulfate disaccharide prevents inflammation and the progression of emphysema in murine models. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 312, L268–L276. [Google Scholar] [CrossRef]
- White, S.R.; Dorscheid, D.R.; Rabe, K.F.; Wojcik, K.R.; Hamann, K.J. Role of very late adhesion integrins in mediating repair of human airway epithelial cell monolayers after mechanical injury. Am. J. Respir. Cell Mol. Biol. 1999, 20, 787–796. [Google Scholar] [CrossRef]
- Andersson, C.K.; Weitoft, M.; Rydell-Tormanen, K.; Bjermer, L.; Westergren-Thorsson, G.; Erjefalt, J.S. Uncontrolled asthmatics have increased FceRI+ and TGF-beta-positive MCTC mast cells and collagen VI in the alveolar parenchyma. Clin. Exp. Allergy 2018, 48, 266–277. [Google Scholar] [CrossRef]
- Maskell, N.; British Thoracic Society Pleural Disease Guideline, G. British Thoracic Society Pleural Disease Guidelines--2010 update. Thorax 2010, 65, 667–669. [Google Scholar] [CrossRef]
- Aubert, J.D.; Hayashi, S.; Hards, J.; Bai, T.R.; Pare, P.D.; Hogg, J.C. Platelet-derived growth factor and its receptor in lungs from patients with asthma and chronic airflow obstruction. Am. J. Physiol. 1994, 266, L655–L663. [Google Scholar] [CrossRef]
- Bocchino, V.; Bertorelli, G.; D’Ippolito, R.; Castagnaro, A.; Zhuo, X.; Grima, P.; Di Comite, V.; Damia, R.; Olivieri, D. The increased number of very late activation antigen-4-positive cells correlates with eosinophils and severity of disease in the induced sputum of asthmatic patients. J. Allergy Clin. Immunol. 2000, 105, 65–70. [Google Scholar] [CrossRef]
- Bazan-Socha, S.; Zuk, J.; Plutecka, H.; Marcinkiewicz, C.; Zareba, L.; Musial, J. Collagen receptors α1β1 and α2β1 integrins are involved in transmigration of peripheral blood eosinophils, but not mononuclear cells through human microvascular endothelial cells monolayer. J. Physiol. Pharmacol. 2012, 63, 373–379. [Google Scholar]
- Fazlollahi, M.; Lee, T.D.; Andrade, J.; Oguntuyo, K.; Chun, Y.; Grishina, G.; Grishin, A.; Bunyavanich, S. The nasal microbiome in asthma. J. Allergy Clin. Immunol. 2018, 142, 834–843.e832. [Google Scholar] [CrossRef]
- Kai, Y.; Motegi, M.; Suzuki, Y.; Takeuchi, H.; Harada, Y.; Sato, F.; Chiba, Y.; Kamei, J.; Sakai, H. Up-regulation of Rac1 in the bronchial smooth muscle of murine experimental asthma. Basic. Clin. Pharmacol. Toxicol. 2019, 125, 8–15. [Google Scholar] [CrossRef]
- Zeng, L.H.; Barkat, M.Q.; Syed, S.K.; Shah, S.; Abbas, G.; Xu, C.; Mahdy, A.; Hussain, N.; Hussain, L.; Majeed, A.; et al. Hedgehog Signaling: Linking Embryonic Lung Development and Asthmatic Airway Remodeling. Cells 2022, 11, 1774. [Google Scholar] [CrossRef]
- Tam, A.; Osei, E.T.; Cheung, C.Y.; Hughes, M.; Yang, C.X.; McNagny, K.M.; Dorscheid, D.R.; Singhera, G.K.; Hallstrand, T.S.; Warner, S.; et al. Hedgehog Signaling as a Therapeutic Target for Airway Remodeling and Inflammation in Allergic Asthma. Cells 2022, 11, 3016. [Google Scholar] [CrossRef]
- Nedeljkovic, I.; Lahousse, L.; Carnero-Montoro, E.; Faiz, A.; Vonk, J.M.; de Jong, K.; van der Plaat, D.A.; van Diemen, C.C.; van den Berge, M.; Obeidat, M.; et al. COPD GWAS variant at 19q13.2 in relation with DNA methylation and gene expression. Hum. Mol. Genet. 2018, 27, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.Y.W.; Nguyen, N.; Peh, H.Y.; Shanmugasundaram, M.; Chandna, R.; Tee, J.H.; Ong, C.B.; Hossain, M.Z.; Venugopal, S.; Zhang, T.; et al. ISM1 protects lung homeostasis via cell-surface GRP78-mediated alveolar macrophage apoptosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2019161119. [Google Scholar] [CrossRef]
- Krane, C.M.; Deng, B.; Mutyam, V.; McDonald, C.A.; Pazdziorko, S.; Mason, L.; Goldman, S.; Kasaian, M.; Chaudhary, D.; Williams, C.; et al. Altered regulation of aquaporin gene expression in allergen and IL-13-induced mouse models of asthma. Cytokine 2009, 46, 111–118. [Google Scholar] [CrossRef][Green Version]
- Agusti, A.; Calverley, P.M.; Celli, B.; Coxson, H.O.; Edwards, L.D.; Lomas, D.A.; MacNee, W.; Miller, B.E.; Rennard, S.; Silverman, E.K.; et al. Characterisation of COPD heterogeneity in the ECLIPSE cohort. Respir. Res. 2010, 11, 122. [Google Scholar] [CrossRef] [PubMed]
- Hillus, D.; Schwarz, T.; Tober-Lau, P.; Vanshylla, K.; Hastor, H.; Thibeault, C.; Jentzsch, S.; Helbig, E.T.; Lippert, L.J.; Tscheak, P.; et al. Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost immunisation with ChAdOx1 nCoV-19 and BNT162b2: A prospective cohort study. Lancet Respir. Med. 2021, 9, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Boschetto, P.; Miniati, M.; Miotto, D.; Braccioni, F.; De Rosa, E.; Bononi, I.; Papi, A.; Saetta, M.; Fabbri, L.M.; Mapp, C.E. Predominant emphysema phenotype in chronic obstructive pulmonary disease patients. Eur. Respir. J. 2003, 21, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Castaldi, P.J.; Dy, J.; Ross, J.; Chang, Y.; Washko, G.R.; Curran-Everett, D.; Williams, A.; Lynch, D.A.; Make, B.J.; Crapo, J.D.; et al. Cluster analysis in the COPDGene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax 2014, 69, 415–422. [Google Scholar] [CrossRef]
- Cerveri, I.; Corsico, A.G.; Grosso, A.; Albicini, F.; Ronzoni, V.; Tripon, B.; Imberti, F.; Galasso, T.; Klersy, C.; Luisetti, M.; et al. The rapid FEV1 decline in chronic obstructive pulmonary disease is associated with predominant emphysema: A longitudinal study. COPD 2013, 10, 55–61. [Google Scholar] [CrossRef]
- Miller, P.G.; Qiao, D.; Rojas-Quintero, J.; Honigberg, M.C.; Sperling, A.S.; Gibson, C.J.; Bick, A.G.; Niroula, A.; McConkey, M.E.; Sandoval, B.; et al. Association of clonal hematopoiesis with chronic obstructive pulmonary disease. Blood 2022, 139, 357–368. [Google Scholar] [CrossRef]


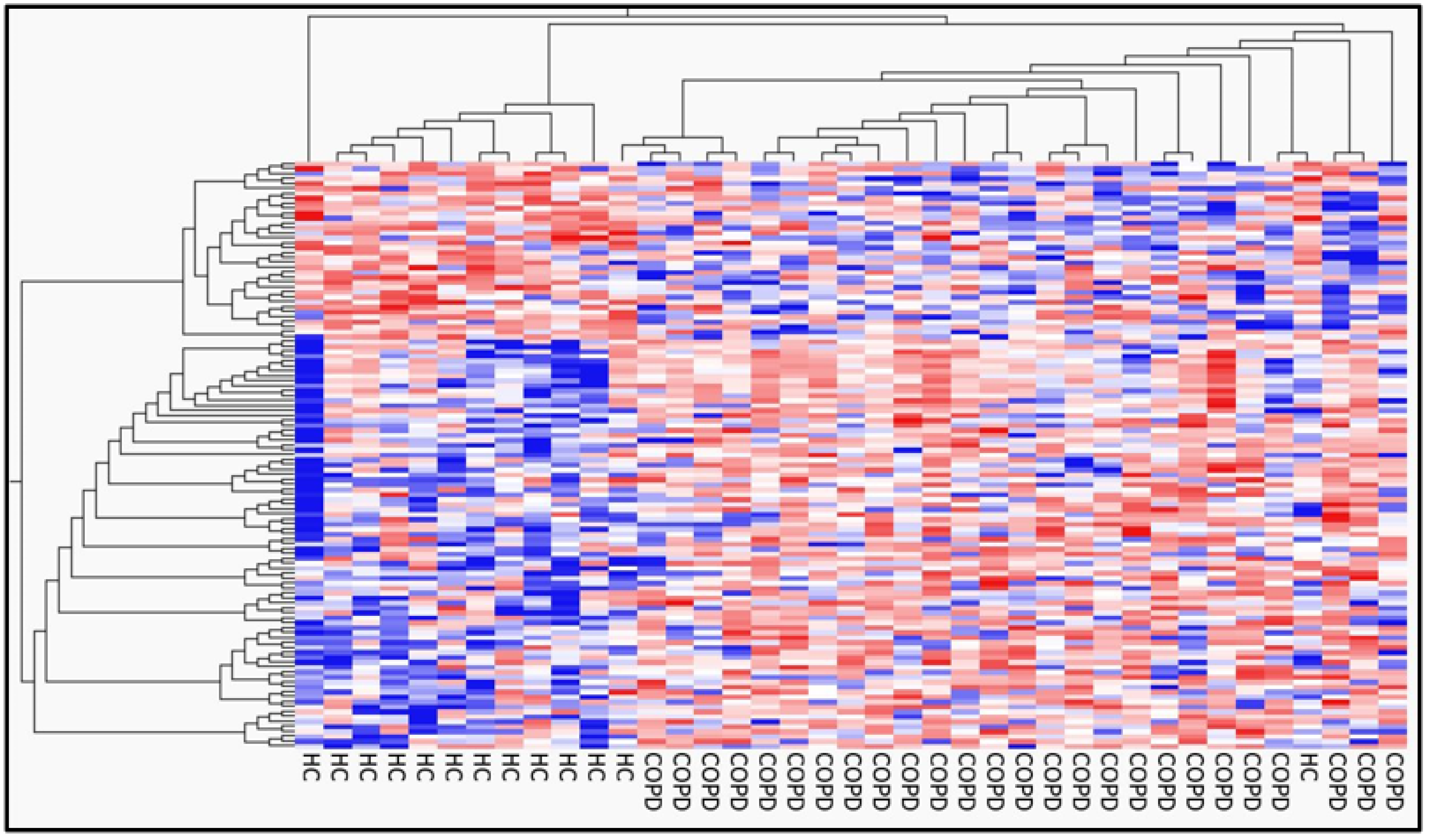
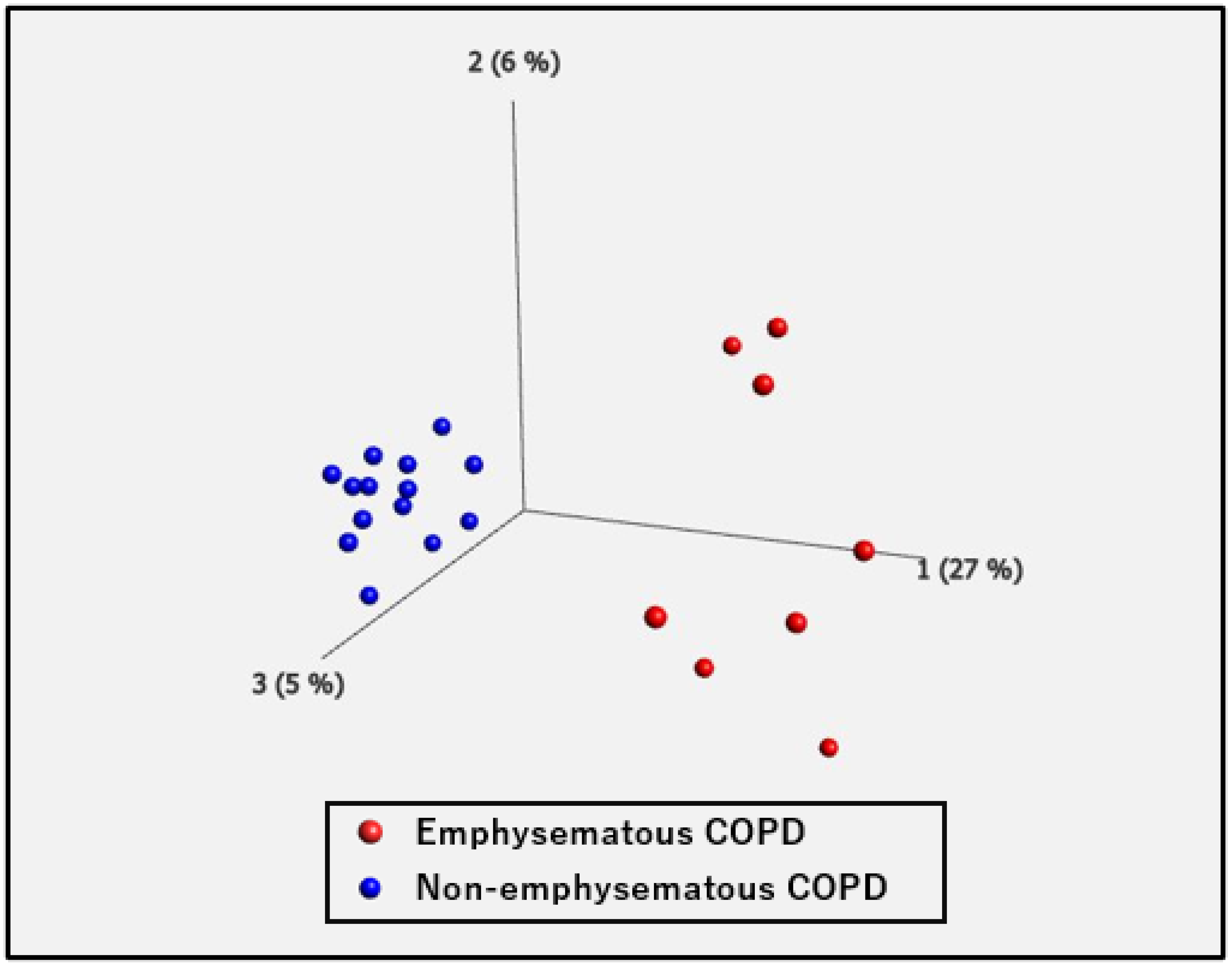
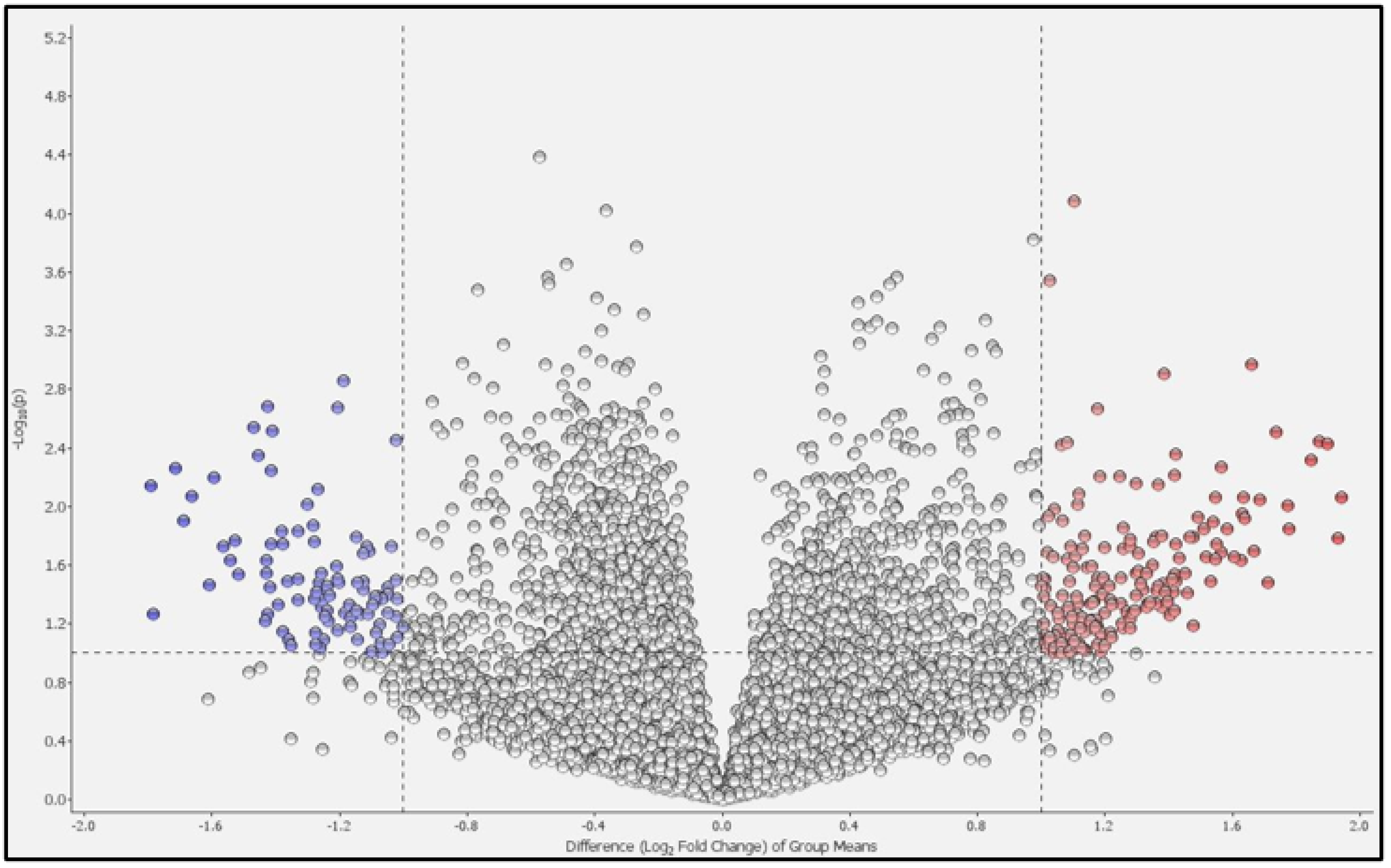

| COPD | Healthy Controls | p-Value | |
|---|---|---|---|
| Number of participants | 26 | 13 | N/A |
| Sex (male/female) | 22/4 | 10/3 | p = 0.51 |
| Age | 68.0 ± 0.3 | 66.5 ± 0.6 | p = 0.54 |
| BMI (kg/m2) | 23.5 ± 3.7 | N/A | N/A |
| Current smokers, number (%) | 6 (23.1) | 1 (7.7) | p = 0.39 |
| Treatment | Number (%) |
|---|---|
| ICS | 2 (7.7) |
| ICS/LABA | 2 (7.7) |
| ICS/LAMA/LABA | 2 (7.7) |
| A. Gene Ontology (biological process): Relevant terms were excerpted | ||
| Term (Gene Ontology: biological process) with upregulated genes | p-value | DEGs |
| Regulation of chromatin binding (GO:0035561) | 0.00074 | CDT1, DDX11 |
| Positive regulation of ERBB signaling pathway (GO:1901186) | 0.00443 | NEU3, GPER1 |
| Acute inflammatory response (GO:0002526) | 0.00518 | EHHADH, ELANE |
| Negative regulation of wound healing (GO:0061045) | 0.00600 | CD109, TFPI |
| Positive regulation of epidermal growth factor receptor signaling (GO:0042058) | 0.00827 | NEU3, GPER1 |
| Positive regulation of cell development (GO:0010720) | 0.00926 | MME, GPER1 |
| Interstrand cross-link repair (GO:0036297) | 0.00978 | DCLRE1A, NEIL3 |
| Term (Gene Ontology: biological process) with downregulated genes | p-value | DEGs |
| Ameboidal-type cell migration (GO:0001667) | 0.00059 | PKN3, AMOT |
| Epithelial cell migration (GO:0010631) | 0.00310 | PKN3, LOXL2 |
| Negative regulation of blood vessel morphogenesis (GO:2000181) | 0.00847 | COL4A3, AMOT |
| Positive regulation of Wnt signaling pathway, planar cell (GO:0060071) | 0.00897 | NKD1 |
| Negative regulation of morphogenesis of an epithelium (GO:1905331) | 0.00897 | NKD1 |
| B. KEGG pathway: Relevant terms were excerpted | ||
| Term (KEGG pathway) with upregulated genes | p-value | DEGs |
| Hematopoietic cell lineage | 3.50 × 10−6 | HLA-DRB5, CSF1, MME, IL1R1, IL1R2, CD1A |
| Protein digestion and absorption | 0.00905 | CPA3, MME, COL24A1 |
| Complement and coagulation cascades | 0.04867 | PROS1, TFPI |
| NF-kappa B signaling pathway | 0.06947 | IL1R1, BLNK |
| Th17 cell differentiation | 0.07298 | HLA-DRB5, IL1R1 |
| Term (KEGG pathway) with downregulated genes | p-value | DEGs |
| Hippo signaling pathway | 0.03470 | NKD1, AMOT |
| Focal adhesion | 0.05065 | MYLPF, COL4A3 |
| MAPK signaling pathway | 0.09795 | PLA2G4C, CACNA2D2 |
| Emphysematous COPD | Non-Emphysematous COPD | p-Value | |
|---|---|---|---|
| Number of participants | 14 | 8 | N/A |
| Sex (male/female) | 13/1 | 6/2 | NS |
| Age (years) | 69.3 ± 1.0 | 67.8 ± 0.6 | NS |
| BMI (kg/m2) | 22.7 ± 8.0 | 23.1 ± 6.2 | NS |
| Current smokers, N (%) | 4 (28.6) | 2 (25.0) | NS |
| FEV1/FVC (%) | 42.0 ± 2.8 | 61.2 ± 2.3 | p = 0.0009 |
| %FEV1 (%) | 50.7 ± 1.7 | 76.7 ± 1.0 | p = 0.0003 |
| B.I. | 1191.9 ± 71.5 | 943.6 ± 40.8 | NS |
| Eosinophils (/μL) | 293 ± 179.7 | 251.3 ± 245.4 | NS |
| Eosinophils (%) | 3.8 ± 2.4 | 3.8 ± 3.4 | NS |
| Serum IgE (IU/mL) | 197.0 ± 342.0 (n = 7) | 188.9 ± 205.8 (n = 12) | NS |
| FeNO (ppb) | 16.5 ± 5.5 (n = 6) | 26.4 ± 32.4 (n = 10) | NS |
| A. Gene Ontology (biological process): relevant terms were excerpted | ||
| Terms (Gene Ontology: biological process) with upregulated genes | p-value | DEGs |
| Positive regulation of T-helper 2 cell cytokine production (GO:2000553) | 0.00075 | XCL1, PRKCZ |
| Regulation of T-helper 2 Cell cytokine production (GO:2000551) | 0.00158 | XCL1, PRKCZ |
| Positive regulation of Type 2 immune Response (GO:0002830) | 0.00271 | XCL1, PRKCZ |
| Keratan sulfate biosynthetic process (GO:0018146) | 0.00362 | B3GNT7, ST3GAL3 |
| Keratan sulfate metabolic process (GO:0042339) | 0.00465 | B3GNT7, ST3GAL3 |
| Cellular response to inorganic substance (GO:0071241) | 0.00521 | DDI2, AQP1 |
| Regulation of T cell cytokine production (GO:0002724) | 0.00521 | XCL1, TRPM4 |
| Positive regulation of T cell cytokine production (GO:0002726) | 0.00707 | XCL1, PRKCZ |
| Regulation of angiogenesis (GO:0045765) | 0.00821 | SERPINF1, ISM1, HSPG2, AQP1, PAK4 |
| Regulation of T cell migration (GO:2000404) | 0.00845 | CD200R1, TMEM102 |
| Positive regulation of T cell migration (GO:2000406) | 0.00994 | TMEM102, XCL1 |
| Terms (Gene Ontology: biological process) with downregulated genes | p-value | DEGs |
| Regulation of vasoconstriction (GO:0019229) | 0.00292 | KCNMB4, ADM |
| Peptidyl-proline modification (GO:0018208) | 0.00316 | P3H2, FKBP7 |
| B. KEGG pathway: relevant terms were excerpted | ||
| Terms (KEGG pathway) with upregulated genes | p-value | DEGs |
| ECM-receptor interaction | 0.01633 | ITGA3, COL6A1, HSPG2 |
| Focal adhesion | 0.03390 | PDGFRB, ITGA3, COL6A1, PAK4 |
| Hedgehog signaling pathway | 0.04523 | GPR161, EFCAB7 |
| Terms (KEGG pathway) with downregulated genes | p-value | DEGs |
| Vascular smooth muscle contraction | 0.00817 | KCNMB4, ADM, MYLK4 |
| cGMP-PKG signaling pathway | 0.09475 | KCNMB4, MYLK4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imamoto, T.; Kawasaki, T.; Sato, H.; Tatsumi, K.; Ishii, D.; Yoshioka, K.; Hasegawa, Y.; Ohara, O.; Suzuki, T. Different Transcriptome Features of Peripheral Blood Mononuclear Cells in Non-Emphysematous Chronic Obstructive Pulmonary Disease. Int. J. Mol. Sci. 2024, 25, 66. https://doi.org/10.3390/ijms25010066
Imamoto T, Kawasaki T, Sato H, Tatsumi K, Ishii D, Yoshioka K, Hasegawa Y, Ohara O, Suzuki T. Different Transcriptome Features of Peripheral Blood Mononuclear Cells in Non-Emphysematous Chronic Obstructive Pulmonary Disease. International Journal of Molecular Sciences. 2024; 25(1):66. https://doi.org/10.3390/ijms25010066
Chicago/Turabian StyleImamoto, Takuro, Takeshi Kawasaki, Hironori Sato, Koichiro Tatsumi, Daisuke Ishii, Keiichiro Yoshioka, Yoshinori Hasegawa, Osamu Ohara, and Takuji Suzuki. 2024. "Different Transcriptome Features of Peripheral Blood Mononuclear Cells in Non-Emphysematous Chronic Obstructive Pulmonary Disease" International Journal of Molecular Sciences 25, no. 1: 66. https://doi.org/10.3390/ijms25010066
APA StyleImamoto, T., Kawasaki, T., Sato, H., Tatsumi, K., Ishii, D., Yoshioka, K., Hasegawa, Y., Ohara, O., & Suzuki, T. (2024). Different Transcriptome Features of Peripheral Blood Mononuclear Cells in Non-Emphysematous Chronic Obstructive Pulmonary Disease. International Journal of Molecular Sciences, 25(1), 66. https://doi.org/10.3390/ijms25010066







