Lipid Metabolism Modulation during SARS-CoV-2 Infection: A Spotlight on Extracellular Vesicles and Therapeutic Prospects
Abstract
1. Introduction
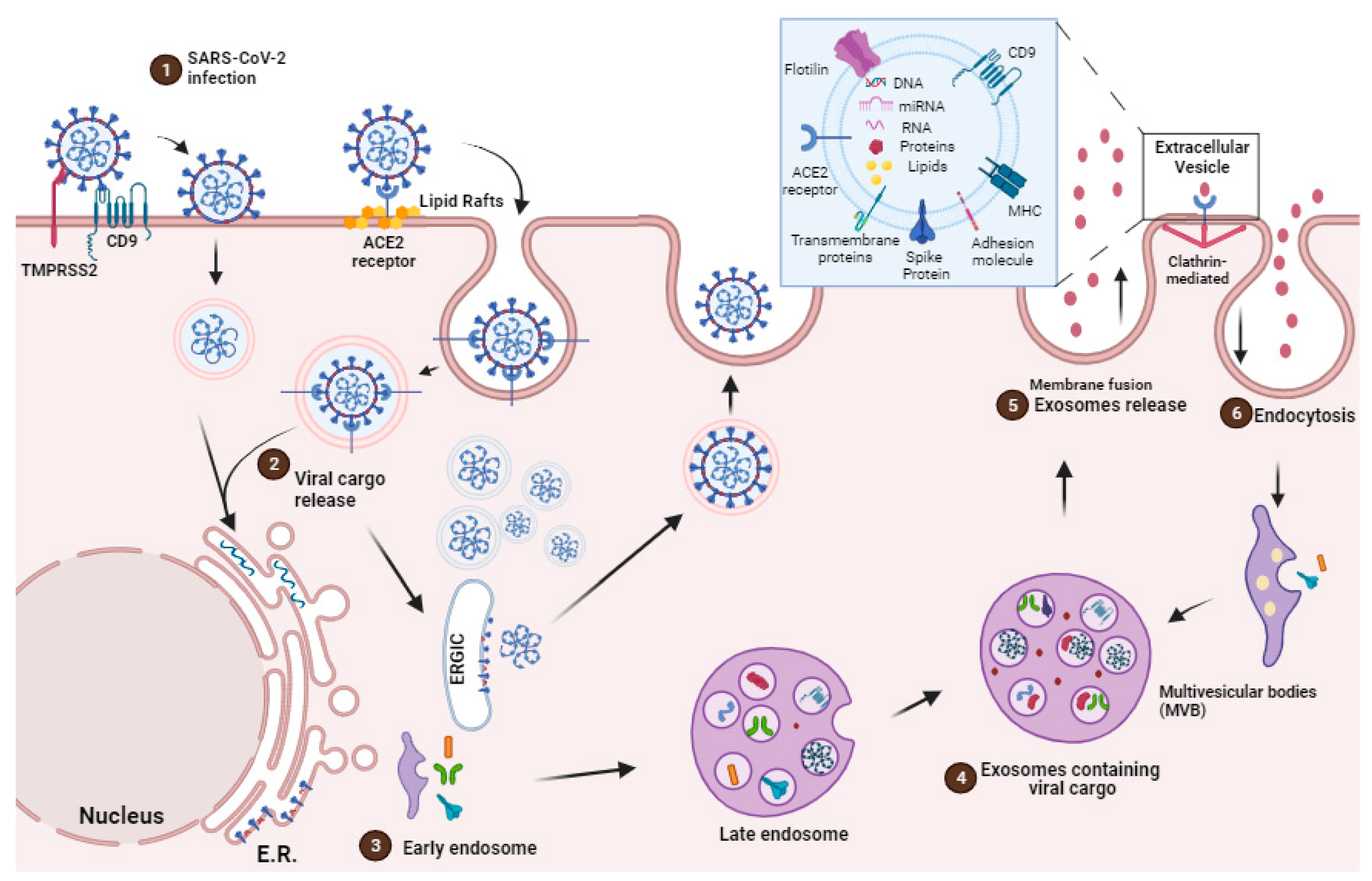
2. EVs Origins, Secretion and Communication
EVs Mediated Cargo Delivery in the Face of SARS-CoV-2 Infection
3. EVs as Potential Role for Immune System Evasion during SARS-CoV-2 Infection
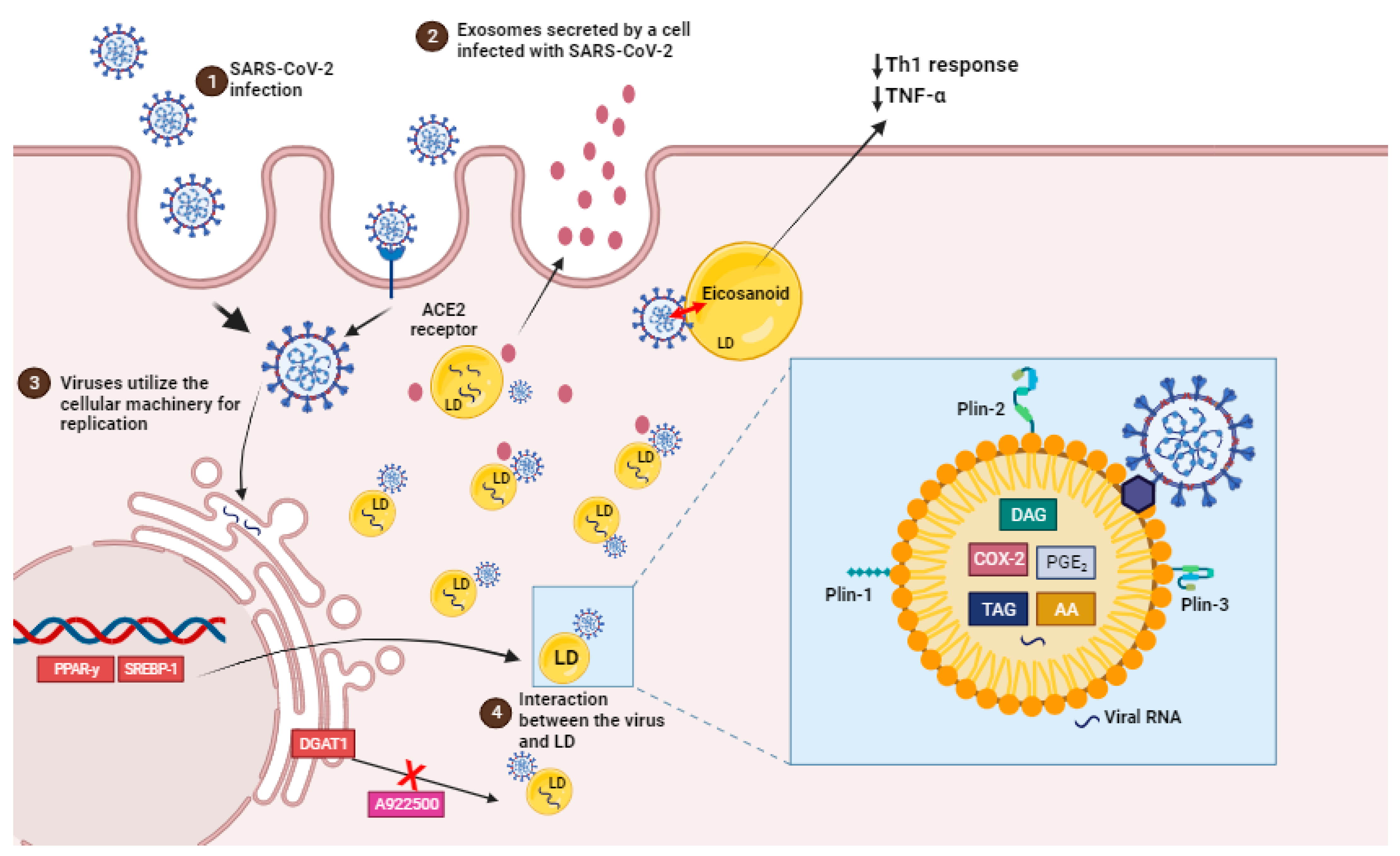
4. Impact of SARS-CoV-2 Infection in the Cellular Lipid Metabolism
Modulation of Cellular Lipid Metabolism as Therapeutic Target on SARS-CoV-2 Infection
5. EVs as Therapeutic Target
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Correction Statement
References
- World Health Organization. Naming the Coronavirus Disease (COVID-19) and the Virus that Causes It. 2020. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it (accessed on 10 October 2023).
- Kumar, D.; Manuel, O.; Natori, Y.; Egawa, H.; Grossi, P.; Han, S.H.; Fernández-Ruiz, M.; Humar, A. COVID-19: A Global Transplant Perspective on Successfully Navigating a Pandemic. Am. J. Transplant. 2020, 20, 1773–1779. [Google Scholar] [CrossRef] [PubMed]
- Prather, B.K.A.; Wang, C.C.; Schooley, R.T. R 1422. Science 2020, 368, 1422–1424. [Google Scholar] [CrossRef] [PubMed]
- van Doremalen, N.; Morris, D.H.; Holbrook, M.G.; Gamble, A.; Williamson, B.N.; Tamin, A.; Harcourt, J.L.; Thornburg, N.J.; Gerber, S.I.; Lloyd-Smith, J.O.; et al. Correspondance Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N. Engl. J. Med. 2020, 382, 1564–1567. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. An Animal Model of Inhaled Vitamin E Acetate and EVALI-like Lung Injury. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- World Health Organization. Tracking SARS-CoV-2 Variants. 2023. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (accessed on 10 October 2023).
- Vitiello, A.; Zovi, A.; Rezza, G. New emerging SARS-CoV-2 variants and antiviral agents. Drug Resist. Update 2023, 70, 100986. [Google Scholar] [CrossRef]
- Liu, Y.J.; Wang, C. A Review of the Regulatory Mechanisms of Extracellular Vesicles-Mediated Intercellular Communication. Cell Commun. Signal. 2023, 21, 77. [Google Scholar] [CrossRef]
- Addetia, A.; Piccoli, L.; Case, J.B.; Park, Y.J.; Beltramello, M.; Guarino, B.; Dang, H.; de Melo, G.D.; Pinto, D.; Sprouse, K.; et al. Neutralization, Effector Function and Immune Imprinting of Omicron Variants. Nature 2023, 621, 592–601. [Google Scholar] [CrossRef]
- Martinelli, S.; Pascucci, D.; Laurenti, P. Humoral Response after a Fourth Dose of SARS-CoV-2 Vaccine in Immunocompromised Patients. Results of a Systematic Review. Front. Public Health 2023, 11, 1108546. [Google Scholar] [CrossRef]
- Bailey, J.; Blankson, J.N.; Wind-Rotolo, M.; Siliciano, R.F. Mechanisms of HIV-1 Escape from Immune Responses and Antiretroviral Drugs. Curr. Opin. Immunol. 2004, 16, 470–476. [Google Scholar] [CrossRef]
- Joyner, A.S.; Willis, J.R.; Crowe, J.E.; Aiken, C. Maturation-Induced Cloaking of Neutralization Epitopes on Hiv-1 Particles. PLoS Pathog. 2011, 7, e1002234. [Google Scholar] [CrossRef]
- Lazarevic, I.; Banko, A.; Miljanovic, D.; Cupic, M. Immune-Escape Hepatitis B Virus Mutations Associated with Viral Reactivation upon Immunosuppression. Viruses 2019, 11, 778. [Google Scholar] [CrossRef] [PubMed]
- Rydell, G.E.; Prakash, K.; Norder, H.; Lindh, M. Hepatitis B Surface Antigen on Subviral Particles Reduces the Neutralizing Effect of Anti-HBs Antibodies on Hepatitis B Viral Particles in Vitro. Virology 2017, 509, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Weber, F.; Haller, O. Viral Suppression of the Interferon System. Biochimie 2007, 89, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W.; Hill, A.B. Viral Interference with Antigen Presentation. Nat. Immunol. 2002, 3, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Dias da Silva Gomes, S.; Soares, V.C.; Ferreira, A.C.; Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Teixeira, L.; da Silva, M.A.N.; Barreto, E.; Mattos, M.; et al. Lipid Droplets Fuel SARS-CoV-2 Replication and Production of Inflammatory Mediators. PLoS Pathog. 2020, 16, e1009127. [Google Scholar] [CrossRef]
- Chen, P.; Wu, M.; He, Y.; Jiang, B.; He, M.L. Metabolic Alterations upon SARS-CoV-2 Infection and Potential Therapeutic Targets against Coronavirus Infection. Signal Transduct. Target. Ther. 2023, 8, 237. [Google Scholar] [CrossRef]
- Altan-Bonnet, N. Lipid Tales on Viral Replication and Transmission Nihal. Trends Cell Biol. 2017, 27, 201–203. [Google Scholar] [CrossRef]
- Akers, J.C.; Gonda, D.; Kim, R.; Carter, B.S.; Chen, C.C. Biogenesis of Extracellular Vesicles (EV): Exosomes, Microvesicles, Retrovirus-like Vesicles, and Apoptotic Bodies. J. Neurooncol. 2013, 1, 1–11. [Google Scholar] [CrossRef]
- Hoen, E.N.; Cremer, T.; Gallo, R.C.; Margolis, L.B. Extracellular Vesicles and Viruses: Are They Close Relatives? Proc. Natl. Acad. Sci. USA 2016, 113, 9155–9161. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes—Vesicular Carriers for Intercellular Communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Mueller, S.K.; Nocera, A.L.; Bleier, B.S. Exosome Function in Aerodigestive Mucosa. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 269–277. [Google Scholar] [CrossRef]
- Kowal, J.; Tkach, M.; Théry, C.; Kowal, J.; Tkach, M.; Biogenesis, C.T. Biogenesis and Secretion of Exosomes To Cite This Version: HAL Id: Inserm-02452742. Curr. Opin. Cell Biol. 2014, 29, 116–125. [Google Scholar] [CrossRef]
- Statello, L.; Maugeri, M.; Garre, E.; Nawaz, M.; Wahlgren, J.; Papadimitriou, A.; Lundqvist, C.; Lindfors, L.; Collén, A.; Sunnerhagen, P.; et al. Identification of RNA-Binding Proteins in Exosomes Capable of Interacting with Different Types of RNA: RBP-Facilitated Transport of RNAs into Exosomes. PLoS ONE 2018, 13, e0195969. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nature Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Gould, S.J.; Booth, A.M.; Hildreth, J.E.K. The Trojan Exosome Hypothesis. Proc. Natl. Acad. Sci. USA 2003, 100, 10592–10597. [Google Scholar] [CrossRef]
- Izquierdo-Useros, N.; Lorizate, M.; McLaren, P.J.; Telenti, A.; Kräusslich, H.G.; Martinez-Picado, J. HIV-1 Capture and Transmission by Dendritic Cells: The Role of Viral Glycolipids and the Cellular Receptor Siglec-1. PLoS Pathog. 2014, 10, e1004146. [Google Scholar] [CrossRef]
- Elrashdy, F.; Aljaddawi, A.A.; Redwan, E.M.; Uversky, V.N. On the Potential Role of Exosomes in the COVID-19 Reinfection/Reactivation Opportunity. J. Biomol. Struct. Dyn. 2020, 39, 5831–5842. [Google Scholar] [CrossRef]
- Borowiec, B.M.; Volponi, A.A.; Mozdziak, P.; Kempisty, B.; Dyszkiewicz-Konwińska, M. Small Extracellular Vesicles and Covid19—Using the “Trojan Horse” to Tackle the Giant. Cells 2021, 10, 3383. [Google Scholar] [CrossRef]
- Bello-morales, R.; Praena, B.; Nuez, D.; Rejas, T.; Guerra, M. Role of Microvesicles in the Spread of Herpes Simplex Virus 1 in Oligodendrocytic Cells. J. Virol. 2018, 92, 10–1128. [Google Scholar] [CrossRef]
- Ramakrishnaiah, V.; Thumann, C.; Fofana, I.; Habersetzer, F.; Pan, Q.; De Ruiter, P.E.; Willemsen, R.; Demmers, J.A.A.; Raj, V.S.; Jenster, G.; et al. Exosome-Mediated Transmission of Hepatitis C Virus between Human Hepatoma Huh7.5 Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 13109–13113. [Google Scholar] [CrossRef]
- Feng, Z.; Hensley, L.; Mcknight, K.L.; Hu, F.; Madden, V.; Ping, L.; Jeong, S.; Walker, C.; Lanford, R.E.; Lemon, S.M. Hijacking Cellular Membranes. Nature 2013, 496, 367–371. [Google Scholar] [CrossRef]
- Ota, M.; Serada, S.; Naka, T.; Mori, Y. MHC Class I Molecules Are Incorporated into Human Herpesvirus—6 Viral Particles and Released into the Extracellular Environment. Microbiol. Immunol. 2014, 58, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Cocozza, F.; Névo, N.; Piovesana, E.; Lahaye, X.; Buchrieser, J.; Schwartz, O.; Manel, N.; Tkach, M.; Théry, C.; Martin-Jaular, L. Extracellular Vesicles Containing ACE2 Efficiently Prevent Infection by SARS-CoV-2 Spike Protein-Containing Virus. J. Extracell. Vesicles 2020, 10, e12050. [Google Scholar] [CrossRef] [PubMed]
- El-Shennawy, L.; Hoffmann, A.D.; Dashzeveg, N.K.; McAndrews, K.M.; Mehl, P.J.; Cornish, D.; Yu, Z.; Tokars, V.L.; Nicolaescu, V.; Tomatsidou, A.; et al. Circulating ACE2-Expressing Extracellular Vesicles Block Broad Strains of SARS-CoV-2. Nat. Commun. 2022, 13, 405. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Pan, X.; Luo, R.; Shen, X.; Li, S.; Wang, Y.; Zuo, X.; Wu, Y.; Guo, Y.; Xiao, G.; et al. Extracellular Vesicles Mediate Antibody-Resistant Transmission of SARS-CoV-2. Cell Discov. 2023, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.J.; Atai, N.A.; Cacciottolo, M.; Nice, J.; Salehi, A.; Guo, C.; Sedgwick, A.; Kanagavelu, S.; Gould, S.J. Exosome-Mediated MRNA Delivery in Vivo Is Safe and Can Be Used to Induce SARS-CoV-2 Immunity. J. Biol. Chem. 2021, 297, 101266. [Google Scholar] [CrossRef]
- Troyer, Z.; Alhusaini, N.; Tabler, C.O.; Schlatzer, D.M.; Tilton, J.C.; Sweet, T.; Carias, L.; Inacio, K.; De Carvalho, L.; King, C.L.; et al. Extracellular Vesicles Carry SARS-CoV-2 Spike Protein and Serve as Decoys for Neutralizing Antibodies Zach. J. Extracell. Vesicles 2021, 10, e12112. [Google Scholar] [CrossRef]
- Lam, S.M.; Huang, X.; Shui, G. Neurological Aspects of SARS-CoV-2 Infection: Lipoproteins and Exosomes as Trojan Horses. Trends Endocrinol. Metab. 2022, 33, 554–568. [Google Scholar] [CrossRef]
- Chutipongtanate, S.; Kongsomros, S.; Pongsakul, N.; Panachan, J.; Khowawisetsut, L.; Pattanapanyasat, K.; Hongeng, S.; Thitithanyanont, A. Anti-SARS-CoV-2 Effect of Extracellular Vesicles Released from Mesenchymal Stem Cells. J. Extracell Vesicles 2022, 11, e12201. [Google Scholar] [CrossRef]
- Ning, B.; Huang, Z.; Youngquist, B.M.; Scott, J.W.; Niu, A.; Bojanowski, C.M.; Zwezdaryk, K.J.; Saba, N.S.; Fan, J.; Yin, X.; et al. Liposome-Mediated Detection of SARS-CoV-2 RNA-Positive Extracellular Vesicles in Plasma. Nat. Nanotechnol. 2021, 16, 1039–1044. [Google Scholar] [CrossRef]
- Gould, S.J.; Raposo, G. As We Wait: Coping with an Imperfect Nomenclature for Extracellular Vesicles. J. Extracell. Vesicles 2013, 2, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle Formation during Reticulocyte Maturation. Association of Plasma Membrane Activities with Released Vesicles (Exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef] [PubMed]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome Secretion: Molecular Mechanisms and Roles. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 ( MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 8, 1535750. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Menck, K.; Sönmezer, C.; Worst, T.S.; Schulz, M.; Dihazi, G.H.; Streit, F.; Erdmann, G.; Kling, S.; Boutros, M.; Binder, C.; et al. Neutral Sphingomyelinases Control Extracellular Vesicles Budding from the Plasma Membrane. J. Extracell. Vesicles 2017, 6, 1378056. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Rome, L.H.; Burnette, D.T.; Coffey, R.J.; Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; et al. Reassessment of Exosome Composition Article Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef]
- Hurley, J.H. ESCRTs Are Everywhere. EMBO J. 2015, 34, 2398–2407. [Google Scholar] [CrossRef]
- Zhang, J.; Kumar, S.; Jayachandran, M.; Hernandez, L.P.H.; Wang, S.; Wilson, E.M.; Lieske, J.C. Excretion of Urine Extracellular Vesicles Bearing Markers of Activated Immune Cells and Calcium/Phosphorus Physiology Differ between Calcium Kidney Stone Formers and Non-Stone Formers. BMC Nephrol. 2021, 22, 204. [Google Scholar] [CrossRef]
- Jayachandran, M.; Yuzhakov, S.V.; Kumar, S.; Larson, N.B.; Enders, F.T.; Milliner, D.S.; Rule, A.D.; Lieske, J.C. Specific Populations of Urinary Extracellular Vesicles and Proteins Differentiate Type 1 Primary Hyperoxaluria Patients without and with Nephrocalcinosis or Kidney Stones. Orphanet J. Rare Dis. 2020, 15, 319. [Google Scholar] [CrossRef] [PubMed]
- Heijnen, H.F.G.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated Platelets Release Two Types of Membrane Vesicles: Microvesicles by Surface Shedding and Exosomes Derived from Exocytosis of Multivesicular Bodies and α-Granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.L.N.; Breakefield, X.O.; Weaver, A.M.; Hospital, M.G. Extracellular Vesicles: Unique Intercellular Delivery Vehicles. Trends Cell Biol. 2017, 27, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and Release of Arrestin Domain-Containing Protein 1-Mediated Microvesicles (ARMMs) at Plasma Membrane by Recruitment of TSG101 Protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Melino, G.; Shi, Y. Actively or Passively Deacidified Lysosomes Push β-Coronavirus Egress. Cell Death Dis. 2021, 12, 12–14. [Google Scholar] [CrossRef]
- Li, B.; Antonyak, M.A.; Zhang, J.; Cerione, R.A. RhoA Triggers a Specific Signaling Pathway That Generates Transforming Microvesicles in Cancer Cells. Oncogene 2012, 31, 4740–4749. [Google Scholar] [CrossRef]
- Yang, J.; Gould, S.J. The Cis -Acting Signals That Target Proteins to Exosomes and Microvesicles. Biochem. Soc. Trans 2013, 41, 277–282. [Google Scholar] [CrossRef]
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wide-Ranging Implications in Tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef]
- Karnas, E.; Dudek, P.; Zuba-Surma, E.K. Stem Cell-Derived Extracellular Vesicles as New Tools in Regenerative Medicine—Immunomodulatory Role and Future Perspectives. Front. Immunol. 2023, 14, 1120175. [Google Scholar] [CrossRef]
- Buzas, E.I. The Roles of Extracellular Vesicles in the Immune System. Nat. Rev. Immunol. 2023, 23, 236–250. [Google Scholar] [CrossRef]
- Petroni, D.; Fabbri, C.; Babboni, S.; Menichetti, L.; Basta, G.; Del Turco, S. Extracellular Vesicles and Intercellular Communication: Challenges for In Vivo Molecular Imaging and Tracking. Pharmaceutics 2023, 15, 1639. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and Mechanisms of Extracellular Vesicle Uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [PubMed]
- French, K.C.; Antonyak, M.A.; Cerione, R.A. Extracellular Vesicle Docking at the Cellular Port: Extracellular Vesicle Binding and Uptake. Semin. Cell Dev. Biol. 2017, 67, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of Immune Responses by Extracellular Vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef]
- De Carvalho, J.V.; De Castro, R.O.; Da Silva, E.Z.M.; Silveira, P.P.; Da Silva-Januário, M.E.; Arruda, E.; Jamur, M.C.; Oliver, C.; Aguiar, R.S.; DaSilva, L.L.P. Nef Neutralizes the Ability of Exosomes from CD4+ T Cells to Act as Decoys during HIV-1 Infection. PLoS ONE 2014, 9, e113691. [Google Scholar] [CrossRef]
- Keller, M.D.; Ching, K.L.; Liang, F.X.; Dhabaria, A.; Tam, K.; Ueberheide, B.M.; Unutmaz, D.; Torres, V.J.; Cadwell, K. Decoy Exosomes Provide Protection against Bacterial Toxins. Nature 2020, 579, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Pocsfalvi, G.; Mammadova, R.; Ramos Juarez, A.P.; Bokka, R.; Trepiccione, F.; Capasso, G. COVID-19 and Extracellular Vesicles: An Intriguing Interplay. Kidney Blood Press. Res. 2020, 45, 661–670. [Google Scholar] [CrossRef]
- Urbanelli, L.; Buratta, S.; Tancini, B.; Sagini, K.; Delo, F.; Porcellati, S.; Emiliani, C. The Role of Extracellular Vesicles in Viral Infection and Transmission. Vaccines 2019, 7, 102. [Google Scholar] [CrossRef]
- Wurdinger, T.; Gatson, N.N.; Balaj, L.; Kaur, B.; Breakefield, X.O.; Pegtel, D.M. Extracellular Vesicles and Their Convergence with Viral Pathways. Adv. Virol. 2012, 2012, 767694. [Google Scholar] [CrossRef]
- Meckes, D.G. Exosomal Communication Goes Viral. J. Virol. 2015, 89, 5200–5203. [Google Scholar] [CrossRef]
- Van Dongen, H.M.; Masoumi, N.; Witwer, K.W.; Michiel, D. Extracellular Vesicles Exploit Viral Entry Routes for Cargo Delivery. Microbiol. Mol. Biol. Rev. 2016, 80, 369–386. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Yuan, P.; Zheng, J.C. Emerging Roles of Extracellular Vesicles in COVID—Edged Sword? Immunology 2021, 163, 416–430. [Google Scholar] [CrossRef] [PubMed]
- Fozouni, P.; Son, S.; Díaz de León Derby, M.; Knott, G.J.; Gray, C.N.; D’Ambrosio, M.V.; Zhao, C.; Switz, N.A.; Kumar, G.R.; Stephens, S.I.; et al. Amplification-Free Detection of SARS-CoV-2 with CRISPR-Cas13a and Mobile Phone Microscopy. Cell 2021, 184, 323–333. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Lv, P.; Jiang, J.; Liu, Y.; Yan, R.; Shu, S.; Hu, B.; Xiao, H.; Cai, K.; Yuan, S.; et al. Advances in Developing ACE2 Derivatives against SARS-CoV-2. Lancet Microbe 2023, 4, e369–e378. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20. [Google Scholar] [CrossRef]
- Avanzato, V.A.; Seifert, S.N.; De Wit, E. Case Study: Prolonged Infectious SARS-CoV-2 Shedding from an Asymptomatic Immunocompromised Individual with Cancer. Cell 2020, 183, 1901–1912. [Google Scholar] [CrossRef]
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474. [Google Scholar] [CrossRef]
- Perrier, A.; Bonnin, A.; Desmarets, L.; Danneels, A.; Goffard, A.; Rouillé, Y.; Dubuisson, J.; Belouzard, X.S. Cro The C-Terminal Domain of the MERS Coronavirus M Protein Contains a Trans -Golgi Network Localization Signal. J. Biol. Chem. 2019, 294, 14406–14421. [Google Scholar] [CrossRef]
- Senapati, S.; Banerjee, P.; Bhagavatula, S.; Kushwaha, P.P.; Kumar, S. Contributions of Human ACE2 and TMPRSS2 in Determining Host–Pathogen Interaction of COVID-19. J. Genet. 2021, 100, 12. [Google Scholar] [CrossRef]
- Cabal, A.B.S.; Wu, T.Y. Recombinant Protein Technology in the Challenging Era of Coronaviruses. Processes 2022, 10, 946. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, S.; Bihl, J. Exosome-Mediated Transfer of ACE2 (Angiotensin-Converting Enzyme 2) from Endothelial Progenitor Cells Promotes Survival and Function of Endothelial Cell. Oxid. Med. Cell. Longev. 2020, 2020, 4213541. [Google Scholar] [CrossRef] [PubMed]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 Infects Cells after Viral Entry via Clathrin-Mediated Endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef] [PubMed]
- Earnest, J.T.; Hantak, M.P.; Li, K.; Mccray, P.B.; Perlman, S.; Gallagher, T. The Tetraspanin CD9 Facilitates MERS- Coronavirus Entry by Scaffolding Host Cell Receptors and Proteases. PLoS Pathog. 2017, 13, e1006546. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2: A Master of Immune Evasion. Biomedicines 2022, 10, 1339. [Google Scholar] [CrossRef] [PubMed]
- Horn, M.D.; MacLean, A.G. Extracellular Vesicles as a Means of Viral Immune Evasion, CNS Invasion, and Glia-Induced Neurodegeneration. Front. Cell. Neurosci. 2021, 15, 695899. [Google Scholar] [CrossRef]
- Pesce, E.; Manfrini, N.; Cordiglieri, C.; Santi, S.; Bandera, A.; Gobbini, A.; Gruarin, P.; Favalli, A.; Bombaci, M.; Cuomo, A.; et al. Exosomes Recovered From the Plasma of COVID-19 Patients Expose SARS-CoV-2 Spike-Derived Fragments and Contribute to the Adaptive Immune Response. Front. Immunol. 2022, 12, 785941. [Google Scholar] [CrossRef]
- Barberis, E.; Vanella, V.V.; Falasca, M.; Caneapero, V.; Cappellano, G.; Raineri, D.; Ghirimoldi, M.; De Giorgis, V.; Puricelli, C.; Vaschetto, R.; et al. Circulating Exosomes Are Strongly Involved in SARS-CoV-2 Infection. Front. Mol. Biosci. 2021, 8, 632290. [Google Scholar] [CrossRef]
- Popowski, K.D.; Dinh, P.C.; Cheng, K. Exosome therapeutics for COVID-19 and respiratory viruses. View 2021, 2, 20200186. [Google Scholar] [CrossRef]
- Machhi, J.; Shahjin, F.; Das, S.; Patel, M.; Abdelmoaty, M.M.; Cohen, J.D.; Singh, P.A.; Baldi, A.; Bajwa, N.; Kumar, R.; et al. A Role for Extracellular Vesicles in SARS-CoV-2 Therapeutics and Prevention. J. Neuroimmune Pharmacol. 2021, 16, 270–288. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, F.A.; Razeeth, M.; Mohammed, S.; Alkarim, S.; Azhar, E.I.; El-magd, M.A.; Hawsawi, Y.; Abdulaal, W.H.; Yusuf, A.; Alhatmi, A.; et al. Untargeted Metabolic Profiling of Extracellular Vesicles of SARS-CoV-2-Infected Patients Shows Presence of Potent Anti-Inflammatory Metabolites. Int. J. Mol. Sci. 2021, 22, 10467. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Brindisi, M.; Shahabi, D.; Chapman, M.E.; Mesecar, A.D. Drug Development and Medicinal Chemistry Efforts toward SARS-Coronavirus and COVID-19 Therapeutics. ChemMedChem 2020, 15, 907–932. [Google Scholar] [CrossRef] [PubMed]
- Villareal, V.A.; Rodgers, M.A.; Costello, D.A.; Yang, P.L. Targeting Host Lipid Synthesis and Metabolism to Inhibit Dengue and Hepatitis C Viruses. Antivir. Res. 2015, 124, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Lyn, R.K.; Hope, G.; Sherratt, A.R.; McLauchlan, J.; Pezacki, J.P. Bidirectional Lipid Droplet Velocities Are Controlled by Differential Binding Strengths of HCV Core DII Protein. PLoS ONE 2013, 8, e78065. [Google Scholar] [CrossRef]
- Filipe, A.; McLauchlan, J. Hepatitis C Virus and Lipid Droplets: Finding a Niche. Trends Mol. Med. 2015, 21, 34–42. [Google Scholar] [CrossRef]
- Cheung, W.; Gill, M.; Esposito, A.; Kaminski, C.F.; Courousse, N.; Chwetzoff, S.; Trugnan, G.; Keshavan, N.; Lever, A.; Desselberger, U. Rotaviruses Associate with Cellular Lipid Droplet Components To Replicate in Viroplasms, and Compounds Disrupting or Blocking Lipid Droplets Inhibit Viroplasm Formation and Viral Replication. J. Virol. 2010, 84, 6782–6798. [Google Scholar] [CrossRef]
- Coffey, C.M.; Sheh, A.; Kim, I.S.; Chandran, K.; Nibert, M.L.; Parker, J.S.L. Reovirus Outer Capsid Protein Μ1 Induces Apoptosis and Associates with Lipid Droplets, Endoplasmic Reticulum, and Mitochondria. J. Virol. 2006, 80, 8422–8438. [Google Scholar] [CrossRef]
- Samsa, M.M.; Mondotte, J.A.; Iglesias, N.G.; Assunção-Miranda, I.; Barbosa-Lima, G.; Da Poian, A.T.; Bozza, P.T.; Gamarnik, A.V. Dengue Virus Capsid Protein Usurps Lipid Droplets for Viral Particle Formation. PLoS Pathog. 2009, 5, e1000632. [Google Scholar] [CrossRef]
- Soares, V.C.; Dias, S.S.G.; Santos, J.C.; Azevedo-Quintanilha, I.G.; Moreira, I.B.G.; Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; da Silva, M.A.N.; Barreto-Vieira, D.F.; et al. Inhibition of the SREBP Pathway Prevents SARS-CoV-2 Replication and Inflammasome Activation. Life Sci. Alliance 2023, 6, e202302049. [Google Scholar] [CrossRef]
- Li, Y.; Xu, S.; Jiang, B.; Cohen, R.A.; Zang, M. Activation of Sterol Regulatory Element Binding Protein and NLRP3 Inflammasome in Atherosclerotic Lesion Development in Diabetic Pigs. PLoS ONE 2013, 8, e67532. [Google Scholar] [CrossRef] [PubMed]
- Fintelman-Rodrigues, N.; Sacramento, C.Q.; Lima, C.R.; da Silva, F.S.; Ferreira, A.; Mattos, M.; de Freitas, C.S.; Soares, V.C.; da Silva Gomes Dias, S.; Temerozo, J.R.; et al. Atazanavir Inhibits SARS-CoV-2 Replication and pro-Inflammatory Cytokine Production. bioRxiv 2020. [Google Scholar] [CrossRef]
- Coperchini, F.; Chiovato, L.; Croce, L.; Magri, F.; Rotondi, M. The Cytokine Storm in COVID-19: An Overview of the Involvement of the Chemokine/Chemokine-Receptor System. Cytokine Growth Factor Rev. 2020, 53, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Ricciotti, E.; Fitzgerald, G.A. Prostaglandins and Inflammation. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 986–1000. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tan, L.; Wang, Q.; Zhang, D.; Ding, J.; Huang, Q.; Tang, Y.Q.; Wang, Q.; Miao, H. Lymphopenia Predicts Disease Severity of COVID-19: A Descriptive and Predictive Study. Signal Transduct. Target. Ther. 2020, 5, 16–18. [Google Scholar] [CrossRef]
- Kong, M.; Zhang, H.; Cao, X.; Mao, X.; Lu, Z. Higher Level of Neutrophil-to-Lymphocyte Is Associated with Severe COVID-19. Epidemiol. Infect. 2020, 148, e139. [Google Scholar] [CrossRef]
- Wilk, A.J.; Rustagi, A.; Zhao, N.Q.; Roque, J.; Martínez-Colón, G.J.; McKechnie, J.L.; Ivison, G.T.; Ranganath, T.; Vergara, R.; Hollis, T.; et al. A Single-Cell Atlas of the Peripheral Immune Response in Patients with Severe COVID-19. Nat. Med. 2020, 26, 1070–1076. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-Cell Landscape of Bronchoalveolar Immune Cells in Patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Huang, L.; Zhao, A.; Wong, F.; Ayala, J.M.; Struthers, M.; Ujjainwalla, F.; Wright, S.D.; Springer, M.S.; Evans, J.; Cui, J. Leukotriene B4 Strongly Increases Monocyte Chemoattractant Protein-1 in Human Monocytes. Arterioscler. Thromb. Vasc. Biol. 2004, 24, 1783–1788. [Google Scholar] [CrossRef]
- Archambault, A.S.; Zaid, Y.; Rakotoarivelo, V.; Turcotte, C.; Doré, É.; Dubuc, I.; Martin, C.; Flamand, O.; Amar, Y.; Cheikh, A.; et al. High Levels of Eicosanoids and Docosanoids in the Lungs of Intubated COVID-19 Patients. FASEB J. 2021, 35, e21666. [Google Scholar] [CrossRef]
- Chatel-Chaix, L.; Bartenschlager, R. Dengue Virus- and Hepatitis C Virus-Induced Replication and Assembly Compartments: The Enemy Inside—Caught in the Web. J. Virol. 2014, 88, 5907–5911. [Google Scholar] [CrossRef]
- Lee, J.Y.; Cortese, M.; Haselmann, U.; Tabata, K.; Romero-Brey, I.; Funaya, C.; Schieber, N.L.; Qiang, Y.; Bartenschlager, M.; Kallis, S.; et al. Spatiotemporal Coupling of the Hepatitis C Virus Replication Cycle by Creating a Lipid Droplet- Proximal Membranous Replication Compartment. Cell Rep. 2019, 27, 3602–3617.e5. [Google Scholar] [CrossRef] [PubMed]
- Miyanari, Y.; Atsuzawa, K.; Usuda, N.; Watashi, K.; Hishiki, T.; Zayas, M.; Bartenschlager, R.; Wakita, T.; Hijikata, M.; Shimotohno, K. The Lipid Droplet Is an Important Organelle for Hepatitis C Virus Production. Nat. Cell Biol. 2007, 9, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Laufman, O.; Perrino, J.; Andino, R. Viral Generated Inter-Organelle Contacts Redirect Lipid Flux for Genome Replication. Cell 2019, 178, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, S.; Guarino, A.M.; Giaquinto, L.; Polishchuk, E.V.; Santoro, M.; Di Tullio, G.; Wilson, C.; Panariello, F.; Soares, V.C.; Dias, S.S.G.; et al. The Role of NSP6 in the Biogenesis of the SARS-CoV-2 Replication Organelle. Nature 2022, 606, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Tan, Q.; Ma, Y.; Wang, S.; Zhong, H.; Liao, Y.; Huang, Q.; Xiao, W.; Xia, H.; Tan, X.; et al. Proteomics of Extracellular Vesicles in Plasma Reveals the Characteristics and Residual Traces of COVID-19 Patients without Underlying Diseases after 3 Months of Recovery. Cell Death Dis. 2021, 12, 541. [Google Scholar] [CrossRef]
- Lu, Y.; Zhou, T.; Xu, C.; Wang, R.; Feng, D.; Li, J.; Wang, X.; Kong, Y.; Hu, G.; Kong, X.; et al. Occludin Is a Target of Src Kinase and Promotes Lipid Secretion by Binding to BTN1a1 and XOR. PLoS Biol. 2022, 20, e3001518. [Google Scholar] [CrossRef]
- Collot, M.; Fam, T.K.; Ashokkumar, P.; Faklaris, O.; Galli, T.; Danglot, L.; Klymchenko, A.S. Ultrabright and Fluorogenic Probes for Multicolor Imaging and Tracking of Lipid Droplets in Cells and Tissues. J. Am. Chem. Soc. 2018, 140, 5401–5411. [Google Scholar] [CrossRef]
- Flaherty, S.E., III; Grijalva, A.; Xu, X.; Ables, E.; Nomani, A.; Ferrante, A.W., Jr. A Lipase-Independent Pathway of Lipid Release and Immune Modulation by Adipocytes. Science 2019, 363, 989–993. [Google Scholar] [CrossRef]
- Amarasinghe, I.; Phillips, W.; Hill, A.F.; Cheng, L.; Helbig, K.J.; Willms, E.; Monson, E.A. Cellular Communication through extracellular vesicles and lipid droplets. J. Extracell Biol. 2023, 2, e77. [Google Scholar] [CrossRef]
- Kowalska, K.; Sabatowska, Z.; Forycka, J.; Młynarska, E.; Franczyk, B.; Rysz, J. The Influence of SARS-CoV-2 Infection on Lipid Metabolism—The Potential Use of Lipid-Lowering Agents in COVID-19 Management. Biomedicines 2022, 10, 2320. [Google Scholar] [CrossRef] [PubMed]
- Grassmé, H.; Jekle, A.; Riehle, A.; Schwarz, H.; Berger, J.; Sandhoff, K.; Kolesnick, R.; Gulbins, E. CD95 Signaling via Ceramide-Rich Membrane Rafts. J. Biol. Chem. 2001, 276, 20589–20596. [Google Scholar] [CrossRef] [PubMed]
- Tam, C.; Idone, V.; Devlin, C.; Fernandes, M.C.; Flannery, A.; He, X.; Schuchman, E.; Tabas, I.; Andrews, N.W. Exocytosis of Acid Sphingomyelinase by Wounded Cells Promotes Endocytosis and Plasma Membrane Repair. J. Cell Biol. 2010, 189, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Ermini, L.; Farrell, A.; Alahari, S.; Ausman, J.; Park, C.; Sallais, J.; Melland-Smith, M.; Porter, T.; Edson, M.; Nevo, O.; et al. Ceramide-Induced Lysosomal Biogenesis and Exocytosis in Early-Onset Preeclampsia Promotes Exosomal Release of SMPD1 Causing Endothelial Dysfunction. Front. Cell Dev. Biol. 2021, 9, 652651. [Google Scholar] [CrossRef] [PubMed]
- Bianco, F.; Perrotta, C.; Novellino, L.; Francolini, M.; Riganti, L.; Menna, E.; Saglietti, L.; Schuchman, E.H.; Furlan, R.; Clementi, E.; et al. Acid Sphingomyelinase Activity Triggers Microparticle Release from Glial Cells. EMBO J. 2009, 28, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, F.S.; Abrami, L.; Sergeeva, O.; Turelli, P.; Qing, E.; Kunz, B.; Raclot, C.; Paz Montoya, J.; Abriata, L.A.; Gallagher, T.; et al. S-Acylation Controls SARS-CoV-2 Membrane Lipid Organization and Enhances Infectivity. Dev. Cell 2021, 56, 2790–2807. [Google Scholar] [CrossRef] [PubMed]
- Timilsina, U.; Umthong, S.; Ivey, E.B.; Waxman, B.; Stavrou, S. SARS-CoV-2 ORF7a Potently Inhibits the Antiviral Effect of the Host Factor SERINC5. Nat. Commun. 2022, 13, 2935. [Google Scholar] [CrossRef]
- Lorizate, M.; Kräusslich, H.G. Role of Lipids in Virus Replication. Cold Spring Harb. Perspect. Biol. 2011, 3, a004820. [Google Scholar] [CrossRef]
- Sureda, A.; Alizadeh, J.; Fazel, S.; Berindan-neagoe, I.; Mohammad, S.; Ghavami, S. Endoplasmic Reticulum as a Potential Therapeutic Target for COVID-19 Infection Management? Eur. J. Pharmacol. 2020, 882, 173288. [Google Scholar] [CrossRef]
- Pol, A.; Gross, S.P.; Parton, R.G. Biogenesis of the Multifunctional Lipid Droplet: Lipids, Proteins, and Sites. J. Cell Biol. 2014, 204, 635–646. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Carvalho, P. Dynamics and Functions of Lipid Droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Yan, B.; Cao, J.; Ye, Z.W.; Liang, R.; Tang, K.; Luo, C.; Cai, J.; Chu, H.; Chung, T.W.H.; et al. SARS-CoV-2 Exploits Host DGAT and ADRP for Efficient Replication. Cell Discov. 2021, 7, 100. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.G.; Jureka, A.S.; Silvas, J.A.; Nicolini, A.M.; Chvatal, S.A.; Carlson-Stevermer, J.; Oki, J.; Holden, K.; Basler, C.F. Inhibitors of VPS34 and Fatty-Acid Metabolism Suppress SARS-CoV-2 Replication. Cell Rep. 2021, 36, 109479. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.; Xing, C.; Du, Y.; Duan, T.; Liu, S.; Zhang, P.; Cheng, C.; Henley, J.; Liu, X.; Qian, C.; et al. Pharmacological Inhibition of Fatty Acid Synthesis Blocks SARS-CoV-2 Replication. Nat. Metab. 2021, 3, 1466–1475. [Google Scholar] [CrossRef] [PubMed]
- Casari, I.; Manfredi, M.; Metharom, P.; Falasca, M. Dissecting Lipid Metabolism Alterations in SARS-CoV-2. Prog. Lipid Res. J. 2021, 82, 101092. [Google Scholar] [CrossRef]
- Jiang, W.; Hu, J.W.; He, X.R.; Jin, W.L.; He, X.Y. Statins: A Repurposed Drug to Fight Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 241. [Google Scholar] [CrossRef]
- Geiger, N.; Kersting, L.; Schlegel, J.; Stelz, L.; Fähr, S.; Diesendorf, V.; Roll, V.; Sostmann, M.; König, E.M.; Reinhard, S.; et al. The Acid Ceramidase Is a SARS-CoV-2 Host Factor. Cells 2022, 11, 2532. [Google Scholar] [CrossRef]
- Vitner, E.B.; Avraham, R.; Politi, B.; Melamed, S.; Israely, T. Elevation in Sphingolipid upon SARS-CoV-2 Infection: Possible Implications for COVID-19 Pathology. Life Sci. Alliance 2022, 5, e202101168. [Google Scholar] [CrossRef]
- Yan, B.; Yuan, S.; Cao, J.; Fung, K.; Lai, P.M.; Yin, F.; Sze, K.H.; Qin, Z.; Xie, Y.; Ye, Z.W.; et al. Phosphatidic Acid Phosphatase 1 Impairs SARS-CoV-2 Replication by Affecting the Glycerophospholipid Metabolism Pathway. Int. J. Biol. Sci. 2022, 18, 4744–4755. [Google Scholar] [CrossRef]
- Kuate, S.; Cinatl, J.; Doerr, H.W.; Überla, K. Exosomal Vaccines Containing the S Protein of the SARS Coronavirus Induce High Levels of Neutralizing Antibodies. Virology 2007, 362, 26–37. [Google Scholar] [CrossRef]
- Catalano, M.; O’Driscoll, L. Inhibiting Extracellular Vesicles Formation and Release: A Review of EV Inhibitors. J. Extracell. Vesicles 2020, 9, 1703244. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, W.; Yao, Q.; Zhang, H.; Dong, G.; Zhang, M.; Liu, Y.; Chen, J.K.; Dong, Z. Exosome Production and Its Regulation of EGFR during Wound Healing in Renal Tubular Cells. Am. J. Physiol. -Ren. Physiol. 2017, 312, F963–F970. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, H.; Sheokand, N.; Kumar, S.; Chauhan, A.S.; Kumar, M.; Jakhar, P.; Boradia, V.M.; Raje, C.I.; Raje, M. Exosomes: Tunable Nano Vehicles for Macromolecular Delivery of Transferrin and Lactoferrin to Specific Intracellular Compartment. J. Biomed. Nanotechnol. 2016, 12, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.J.; Speth, J.M.; Penke, L.R.; Wettlaufer, S.H.; Swanson, J.A.; Peters-Golden, M. Mechanisms and Modulation of Microvesicle Uptake in a Model of Alveolar Cell Communication. J. Biol. Chem. 2017, 292, 20897–20910. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Berumen, G.; Bunn, K.E.; Pua, H.H.; Rafat, M. Extracellular Vesicles: Mediators of Intercellular Communication in Tissue Injury and Disease. Cell Commun. Signal. 2021, 19, 104. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Tu, C.; Zhang, J.; Wang, J. Inhibition of Multiple Myeloma-derived Exosomes Uptake Suppresses the Functional Response in Bone Marrow Stromal Cell. Int. J. Oncol. 2019, 54, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Devhare, P.B.; Ray, R.B. A Novel Role of Exosomes in the Vaccination Approach. Ann. Transl. Med. 2017, 5, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Thapa, N. Exosome-Based COVID-19 Vaccine. Methods Mol Biol. 2023, 268, 301–311. [Google Scholar] [CrossRef]
- Rosell, A.; Havervall, S.; Von Meijenfeldt, F.; Hisada, Y.; Aguilera, K.; Grover, S.P.; Lisman, T.; MacKman, N.; Thålin, C. Patients With COVID-19 Have Elevated Levels of Circulating Extracellular Vesicle Tissue Factor Activity That Is Associated With Severity and Mortality—Brief Report. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 878–882. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Z.; Dinh, P.U.C.; Zhu, D.; Popowski, K.D.; Lutz, H.; Hu, S.; Lewis, M.G.; Cook, A.; Andersen, H.; et al. Cell-Mimicking Nanodecoys Neutralize SARS-CoV-2 and Mitigate Lung Injury in a Non-Human Primate Model of COVID-19. Nat. Nanotechnol. 2021, 16, 942–951. [Google Scholar] [CrossRef]
- Gilligan, K.E.; Dwyer, R.M. Engineering Exosomes for Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 1122. [Google Scholar] [CrossRef] [PubMed]
- Cooper, L.F.; Ravindran, S.; Huang, C.C.; Kang, M. A Role for Exosomes in Craniofacial Tissue Engineering and Regeneration. Front. Physiol. 2020, 10, 1569. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Zhou, S.; Li, S.; Kuang, L.; Chen, H.; Luo, X.; Ouyang, J.; He, M.; Du, X.; Chen, L. Exosomes: Roles and Therapeutic Potential in Osteoarthritis. Bone Res. 2020, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Petrik, J. Immunomodulatory Effects of Exosomes Produced by Virus-Infected Cells. Transfus. Apher. Sci. 2016, 55, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Fan, J.; Lyon, C.; Wan, M.; Hu, Y. Role of Extracellular Vesicles in Viral and Bacterial Infections: Pathogenesis, Diagnostics, and Therapeutics. Theranostics 2018, 8, 2709–2721. [Google Scholar] [CrossRef]
- Börger, V.; Weiss, D.J.; Anderson, J.D.; Borràs, F.E.; Bussolati, B.; Carter, D.R.F.; Dominici, M.; Falcón-Pérez, J.M.; Gimona, M.; Hill, A.F.; et al. International Society for Extracellular Vesicles and International Society for Cell and Gene Therapy Statement on Extracellular Vesicles from Mesenchymal Stromal Cells and Other Cells: Considerations for Potential Therapeutic Agents to Suppress Coronavirus Disease-19. Cytotherapy 2020, 22, 482–485. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Avila, H.; Lima, C.N.R.; Rampinelli, P.G.; Mateus, L.C.O.; Sousa Silva, R.V.d.; Correa, J.R.; Almeida, P.E.d. Lipid Metabolism Modulation during SARS-CoV-2 Infection: A Spotlight on Extracellular Vesicles and Therapeutic Prospects. Int. J. Mol. Sci. 2024, 25, 640. https://doi.org/10.3390/ijms25010640
D’Avila H, Lima CNR, Rampinelli PG, Mateus LCO, Sousa Silva RVd, Correa JR, Almeida PEd. Lipid Metabolism Modulation during SARS-CoV-2 Infection: A Spotlight on Extracellular Vesicles and Therapeutic Prospects. International Journal of Molecular Sciences. 2024; 25(1):640. https://doi.org/10.3390/ijms25010640
Chicago/Turabian StyleD’Avila, Heloisa, Claudia Natércia Rocha Lima, Pollianne Garbero Rampinelli, Laiza Camila Oliveira Mateus, Renata Vieira de Sousa Silva, José Raimundo Correa, and Patrícia Elaine de Almeida. 2024. "Lipid Metabolism Modulation during SARS-CoV-2 Infection: A Spotlight on Extracellular Vesicles and Therapeutic Prospects" International Journal of Molecular Sciences 25, no. 1: 640. https://doi.org/10.3390/ijms25010640
APA StyleD’Avila, H., Lima, C. N. R., Rampinelli, P. G., Mateus, L. C. O., Sousa Silva, R. V. d., Correa, J. R., & Almeida, P. E. d. (2024). Lipid Metabolism Modulation during SARS-CoV-2 Infection: A Spotlight on Extracellular Vesicles and Therapeutic Prospects. International Journal of Molecular Sciences, 25(1), 640. https://doi.org/10.3390/ijms25010640





