Traces of Canine Inflammatory Bowel Disease Reflected by Intestinal Organoids
Abstract
1. Introduction
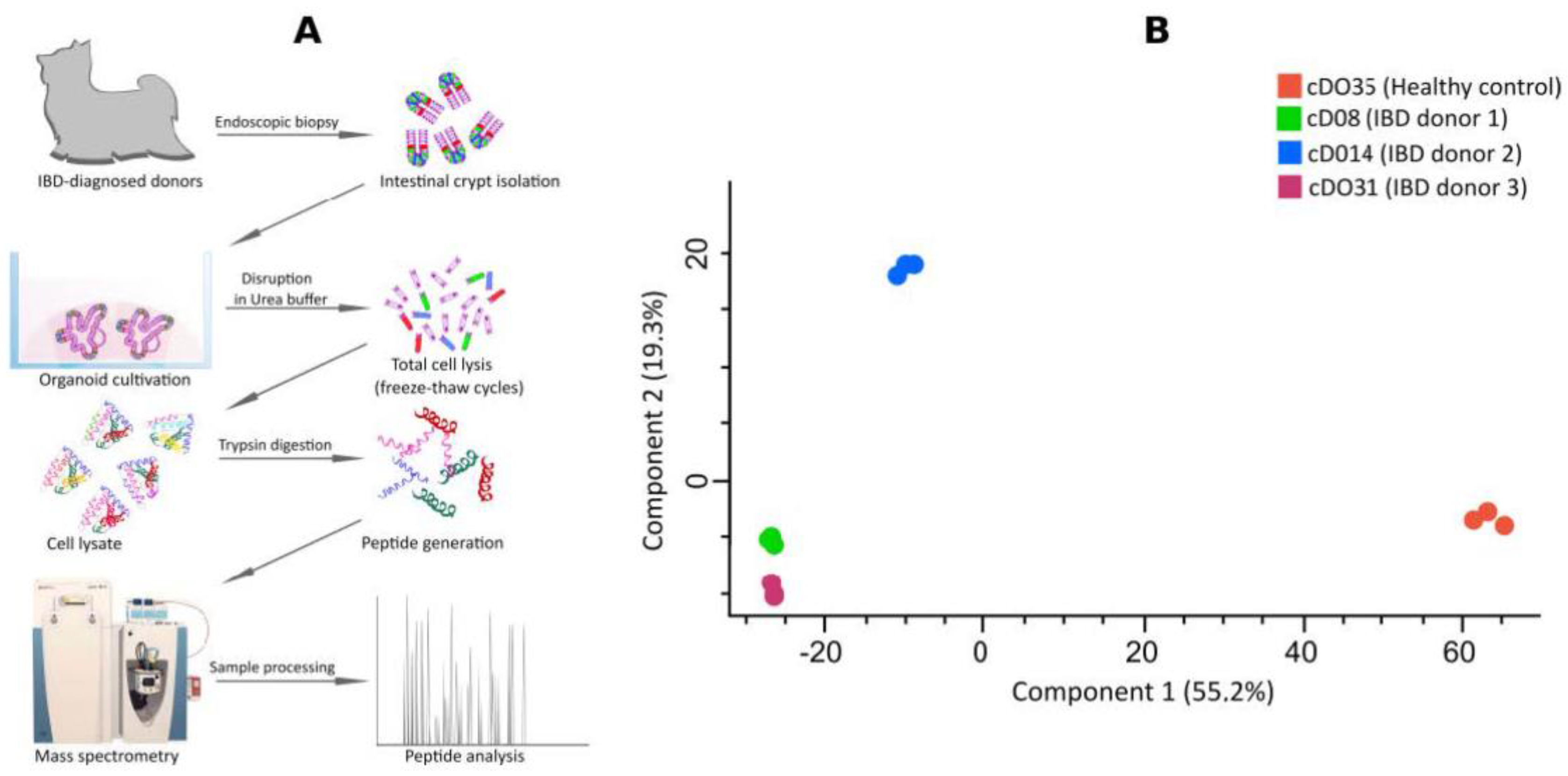
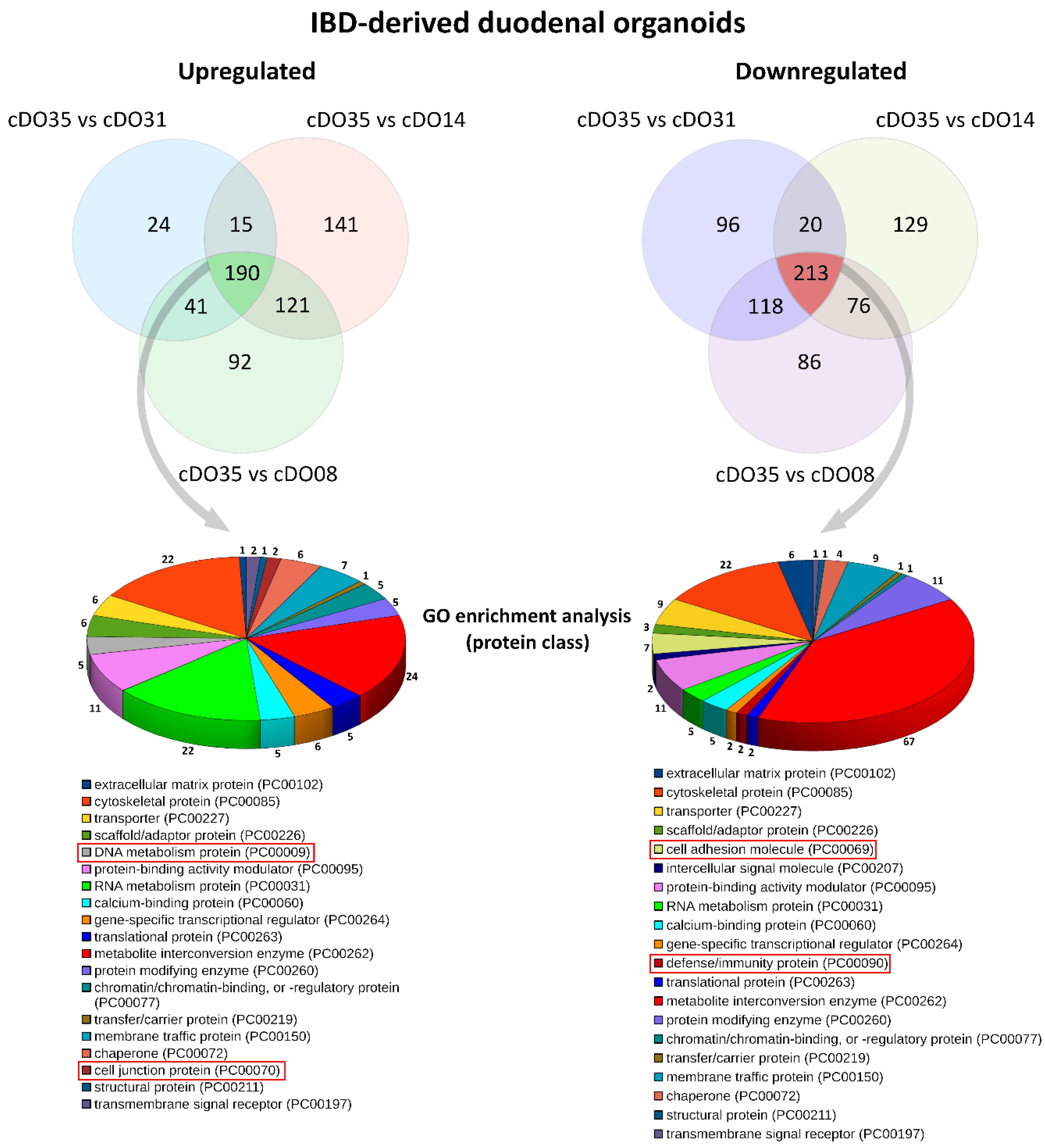
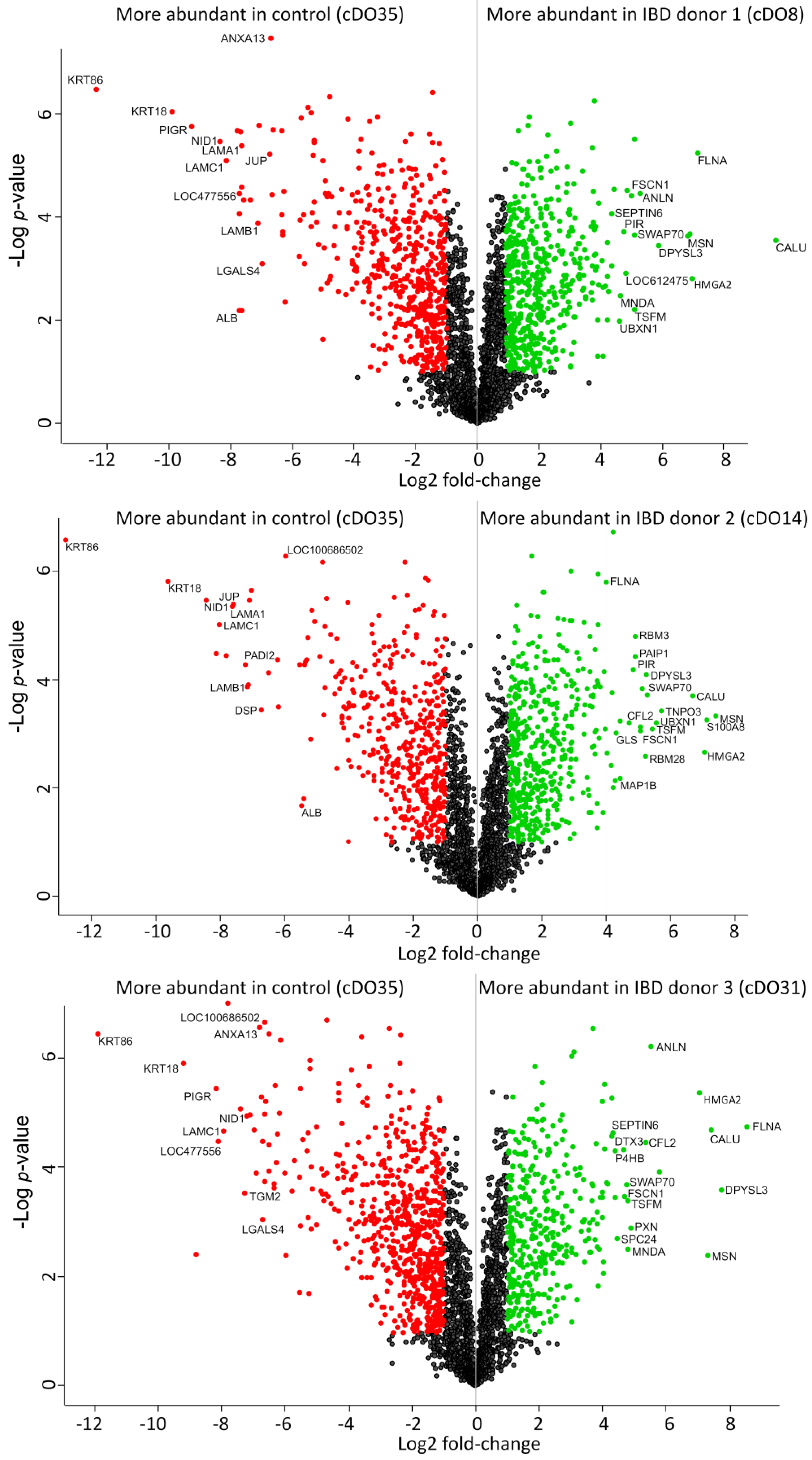
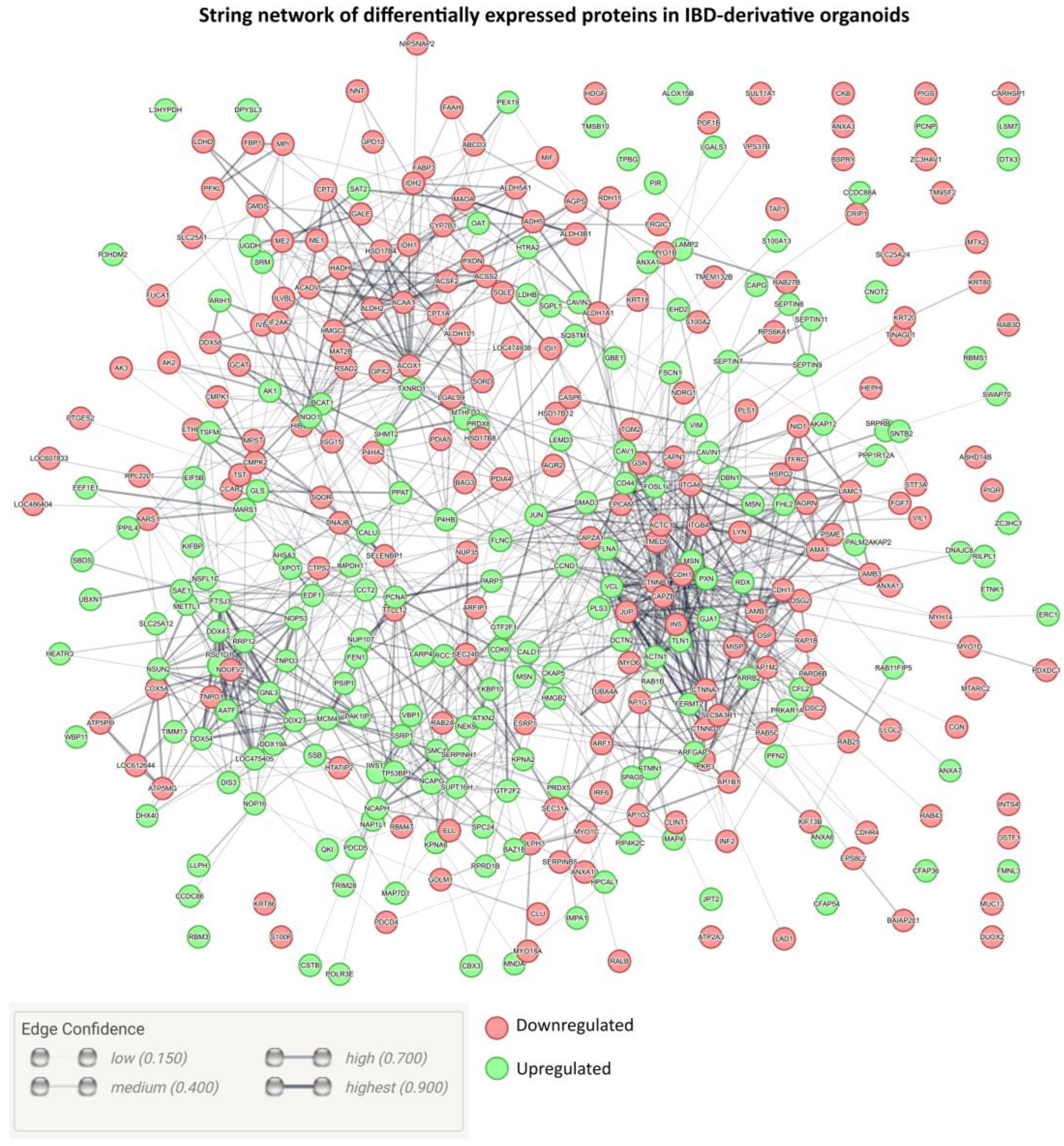
2. Results and Discussion
3. Materials and Methods
3.1. Organoid Cultivation
3.2. Sample Preparation for Liquid Chromatography–Mass Spectrometry
3.3. Sample Analysis
3.4. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The Global, Regional, and National Burden of Inflammatory Bowel Disease in 195 Countries and Territories, 1990–2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Meineri, G.; Martello, E.; Radice, E.; Bruni, N.; Saettone, V.; Atuahene, D.; Armandi, A.; Testa, G.; Ribaldone, D.G. Chronic Intestinal Disorders in Humans and Pets: Current Management and the Potential of Nutraceutical Antioxidants as Alternatives. Animals 2022, 12, 812. [Google Scholar] [CrossRef] [PubMed]
- McDowell, C.; Farooq, U.; Haseeb, M. Inflammatory Bowel Disease; StatPearls: St. Petersburg, FL, USA, 2023. [Google Scholar]
- Kathrani, A.; Werling, D.; Allenspach, K. Canine Breeds at High Risk of Developing Inflammatory Bowel Disease in the South-Eastern UK. Vet. Rec. 2011, 169, 635. [Google Scholar] [CrossRef] [PubMed]
- Greuter, T.; Vavricka, S.R. Extraintestinal Manifestations in Inflammatory Bowel Disease—Epidemiology, Genetics, and Pathogenesis. Expert Rev. Gastroenterol. Hepatol. 2019, 13, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.; Chintanaboina, J. Allergic and Immunologic Perspectives of Inflammatory Bowel Disease. Clin. Rev. Allergy Immunol. 2019, 57, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Massimino, L.; Lamparelli, L.A.; Houshyar, Y.; D’Alessio, S.; Peyrin-Biroulet, L.; Vetrano, S.; Danese, S.; Ungaro, F. The Inflammatory Bowel Disease Transcriptome and Metatranscriptome Meta-Analysis (IBD TaMMA) Framework. Nat. Comput. Sci. 2021, 1, 511–515. [Google Scholar] [CrossRef]
- Assadsangabi, A.; Evans, C.A.; Corfe, B.M.; Lobo, A. Application of Proteomics to Inflammatory Bowel Disease Research: Current Status and Future Perspectives. Gastroenterol. Res. Pract. 2019, 2019, 1426954. [Google Scholar] [CrossRef] [PubMed]
- Fair, K.L.; Colquhoun, J.; Hannan, N.R.F. Intestinal Organoids for Modelling Intestinal Development and Disease. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170217. [Google Scholar] [CrossRef]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef]
- Csukovich, G.; Pratscher, B.; Burgener, I.A. The World of Organoids: Gastrointestinal Disease Modelling in the Age of 3R and One Health with Specific Relevance to Dogs and Cats. Animals 2022, 12, 2461. [Google Scholar] [CrossRef]
- Kramer, N.; Pratscher, B.; Meneses, A.M.C.; Tschulenk, W.; Walter, I.; Swoboda, A.; Kruitwagen, H.S.; Schneeberger, K.; Penning, L.C.; Spee, B.; et al. Generation of Differentiating and Long-Living Intestinal Organoids Reflecting the Cellular Diversity of Canine Intestine. Cells 2020, 9, 822. [Google Scholar] [CrossRef] [PubMed]
- Allenspach, K.A.; Mochel, J.P.; Du, Y.; Priestnall, S.L.; Moore, F.; Slayter, M.; Rodrigues, A.; Ackermann, M.; Krockenberger, M.; Mansell, J.; et al. Correlating Gastrointestinal Histopathologic Changes to Clinical Disease Activity in Dogs with Idiopathic Inflammatory Bowel Disease. Vet. Pathol. 2019, 56, 435–443. [Google Scholar] [CrossRef] [PubMed]
- Allenspach, K. Diagnosis of Small Intestinal Disorders in Dogs and Cats. Clin. Lab. Med. 2015, 35, 521. [Google Scholar] [CrossRef] [PubMed]
- Chandra, L.; Borcherding, D.C.; Kingsbury, D.; Atherly, T.; Ambrosini, Y.M.; Bourgois-Mochel, A.; Yuan, W.; Kimber, M.; Qi, Y.; Wang, Q.; et al. Derivation of Adult Canine Intestinal Organoids for Translational Research in Gastroenterology. BMC Biol. 2019, 17, 33. [Google Scholar] [CrossRef] [PubMed]
- Powell, R.H.; Behnke, M.S. WRN Conditioned Media Is Sufficient for In Vitro Propagation of Intestinal Organoids from Large Farm and Small Companion Animals. Biol. Open 2017, 6, 698–705. [Google Scholar] [CrossRef] [PubMed]
- Csukovich, G.; Kramer, N.; Pratscher, B.; Gotic, I.; Freund, P.; Hahn, R.; Himmler, G.; Brandt, S.; Burgener, I.A. Neutralising Effects of Different Antibodies on Clostridioides Difficile Toxins TcdA and TcdB in a Translational Approach. Int. J. Mol. Sci. 2023, 24, 3867. [Google Scholar] [CrossRef] [PubMed]
- Csukovich, G.; Huainig, J.; Troester, S.; Pratscher, B.; Burgener, I.A. The Intricacies of Inflammatory Bowel Disease: A Preliminary Study of Redox Biology in Intestinal Organoids. Organoids 2023, 2, 156–164. [Google Scholar] [CrossRef]
- Abdi, H.; Williams, L.J. Principal Component Analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Pearson, K. LIII. On Lines and Planes of Closest Fit to Systems of Points in Space. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1901, 2, 559–572. [Google Scholar] [CrossRef]
- Cangelosi, R.; Goriely, A. Component Retention in Principal Component Analysis with Application to CDNA Microarray Data. Biol. Direct 2007, 2, 2. [Google Scholar] [CrossRef]
- Crescenzi, M.; Giuliani, A. The Main Biological Determinants of Tumor Line Taxonomy Elucidated by a Principal Component Analysis of Microarray Data. FEBS Lett. 2001, 507, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Alter, O.; Brown, P.O.; Botstein, D. Singular Value Decomposition for Genome-Wide Expression Data Processing and Modeling. Proc. Natl. Acad. Sci. USA 2000, 97, 10101–10106. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Gutierrez, J.; Kim, E.M.; Batth, T.S.; Cho, N.; Hu, Q.; Chan, L.J.G.; Petzold, C.J.; Hillson, N.J.; Adams, P.D.; Keasling, J.D.; et al. Principal Component Analysis of Proteomics (PCAP) as a Tool to Direct Metabolic Engineering. Metab. Eng. 2015, 28, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus Computational Platform for Comprehensive Analysis of (Prote)Omics Data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER Version 14: More Genomes, a New PANTHER GO-Slim and Improvements in Enrichment Analysis Tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Turula, H.; Wobus, C.E. The Role of the Polymeric Immunoglobulin Receptor and Secretory Immunoglobulins during Mucosal Infection and Immunity. Viruses 2018, 10, 237. [Google Scholar] [CrossRef] [PubMed]
- Pabst, O.; Slack, E. IgA and the Intestinal Microbiota: The Importance of Being Specific. Mucosal Immunol. 2019, 13, 12–21. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut Microbiota and IBD: Causation or Correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef]
- Corfe, B.M.; Majumdar, D.; Assadsangabi, A.; Marsh, A.M.R.; Cross, S.S.; Connolly, J.B.; Evans, C.A.; Lobo, A.J. Inflammation Decreases Keratin Level in Ulcerative Colitis; Inadequate Restoration Associates with Increased Risk of Colitis-Associated Cancer. BMJ Open Gastroenterol. 2015, 2, e000024. [Google Scholar] [CrossRef]
- Moll, R.; Divo, M.; Langbein, L. The Human Keratins: Biology and Pathology. Histochem. Cell Biol. 2008, 129, 705–733. [Google Scholar] [CrossRef]
- Mun, J.; Hur, W.; Ku, N.O. Roles of Keratins in Intestine. Int. J. Mol. Sci. 2022, 23, 8051. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zikusoka, M.; Trindade, A.; Dassopoulos, T.; Harris, M.L.; Bayless, T.M.; Brant, S.R.; Chakravarti, S.; Kwon, J.H. MicroRNAs Are Differentially Expressed in Ulcerative Colitis and Alter Expression of Macrophage Inflammatory Peptide-2α. Gastroenterology 2008, 135, 1624–1635.e24. [Google Scholar] [CrossRef] [PubMed]
- Galamb, O.; Györffy, B.; Sipos, F.; Spisák, S.; Németh, A.M.; Miheller, P.; Tulassay, Z.; Dinya, E.; Molnár, B. Inflammation, Adenoma and Cancer: Objective Classification of Colon Biopsy Specimens with Gene Expression Signature. Dis. Markers 2008, 25, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nasrabadi, P.N.; Nayeri, Z.; Gharib, E.; Salmanipour, R.; Masoomi, F.; Mahjoubi, F.; Zomorodipour, A. Establishment of a CALU, AURKA, and MCM2 Gene Panel for Discrimination of Metastasis from Primary Colon and Lung Cancers. PLoS ONE 2020, 15, e0233717. [Google Scholar] [CrossRef]
- Zada, A.; Zhao, Y.; Halim, D.; Windster, J.; van der Linde, H.C.; Glodener, J.; Overkleeft, S.; de Graaf, B.M.; Verdijk, R.M.; Brooks, A.S.; et al. The Long Filamin-A Isoform Is Required for Intestinal Development and Motility: Implications for Chronic Intestinal Pseudo-Obstruction. Hum. Mol. Genet. 2023, 32, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Marcin, W.; Michael, S. Genetics and Epigenetics of Inflammatory Bowel Disease. Swiss Med. Wkly. 2018, 148, w14671. [Google Scholar] [CrossRef]
- Ashar, H.R.; Chouinard, R.A.; Dokur, M.; Chada, K. In Vivo Modulation of HMGA2 Expression. Biochim. Biophys. Acta 2010, 1799, 55–61. [Google Scholar] [CrossRef]
- Brandt, M.; Grazioso, T.P.; Fawal, M.A.; Tummala, K.S.; Torres-Ruiz, R.; Rodriguez-Perales, S.; Perna, C.; Djouder, N. MTORC1 Inactivation Promotes Colitis-Induced Colorectal Cancer but Protects from APC Loss-Dependent Tumorigenesis. Cell Metab. 2018, 27, 118–135.e8. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Ferguson, L.R. Chronic Inflammation and Mutagenesis. Mutat. Res. 2010, 690, 3–11. [Google Scholar] [CrossRef]
- Li, Y. Modern Epigenetics Methods in Biological Research. Methods 2021, 187, 104. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Powell, D.R.; Curtis, D.J.; Wong, N.C. From Reads to Insight: A Hitchhiker’s Guide to ATAC-Seq Data Analysis. Genome Biol. 2020, 21, 22. [Google Scholar] [CrossRef] [PubMed]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Galler, A.I.; Suchodolski, J.S.; Steiner, J.M.; Sung, C.-H.; Hittmair, K.M.; Richter, B.; Burgener, I.A. Microbial Dysbiosis and Fecal Metabolomic Perturbations in Yorkshire Terriers with Chronic Enteropathy. Sci. Rep. 2022, 12, 12977. [Google Scholar] [CrossRef] [PubMed]
- Galler, A.I.; Klavins, K.; Burgener, I.A. A Preliminary Metabolomic Study of Yorkshire Terrier Enteropathy. Metabolites 2022, 12, 264. [Google Scholar] [CrossRef] [PubMed]
- Doulidis, P.G.; Galler, A.I.; Hausmann, B.; Berry, D.; Rodríguez-Rojas, A.; Burgener, I.A. Gut Microbiome Signatures of Yorkshire Terrier Enteropathy during Disease and Remission. Sci. Rep. 2023, 13, 4337. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.G.; Snippert, H.J.; Van De Wetering, M.; Barker, N.; Stange, D.E.; Van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures In Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for Micro-Purification, Enrichment, Pre-Fractionation and Storage of Peptides for Proteomics Using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Carlson, A.; Sinitcyn, P.; Mann, M.; Cox, J. Visualization of LC-MS/MS Proteomics Data in MaxQuant. Proteomics 2015, 15, 1453–1456. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pratscher, B.; Kuropka, B.; Csukovich, G.; Doulidis, P.G.; Spirk, K.; Kramer, N.; Freund, P.; Rodríguez-Rojas, A.; Burgener, I.A. Traces of Canine Inflammatory Bowel Disease Reflected by Intestinal Organoids. Int. J. Mol. Sci. 2024, 25, 576. https://doi.org/10.3390/ijms25010576
Pratscher B, Kuropka B, Csukovich G, Doulidis PG, Spirk K, Kramer N, Freund P, Rodríguez-Rojas A, Burgener IA. Traces of Canine Inflammatory Bowel Disease Reflected by Intestinal Organoids. International Journal of Molecular Sciences. 2024; 25(1):576. https://doi.org/10.3390/ijms25010576
Chicago/Turabian StylePratscher, Barbara, Benno Kuropka, Georg Csukovich, Pavlos G. Doulidis, Katrin Spirk, Nina Kramer, Patricia Freund, Alexandro Rodríguez-Rojas, and Iwan A. Burgener. 2024. "Traces of Canine Inflammatory Bowel Disease Reflected by Intestinal Organoids" International Journal of Molecular Sciences 25, no. 1: 576. https://doi.org/10.3390/ijms25010576
APA StylePratscher, B., Kuropka, B., Csukovich, G., Doulidis, P. G., Spirk, K., Kramer, N., Freund, P., Rodríguez-Rojas, A., & Burgener, I. A. (2024). Traces of Canine Inflammatory Bowel Disease Reflected by Intestinal Organoids. International Journal of Molecular Sciences, 25(1), 576. https://doi.org/10.3390/ijms25010576




