Lactiplantibacillus plantarum HY7718 Improves Intestinal Integrity in a DSS-Induced Ulcerative Colitis Mouse Model by Suppressing Inflammation through Modulation of the Gut Microbiota
Abstract
1. Introduction
2. Results
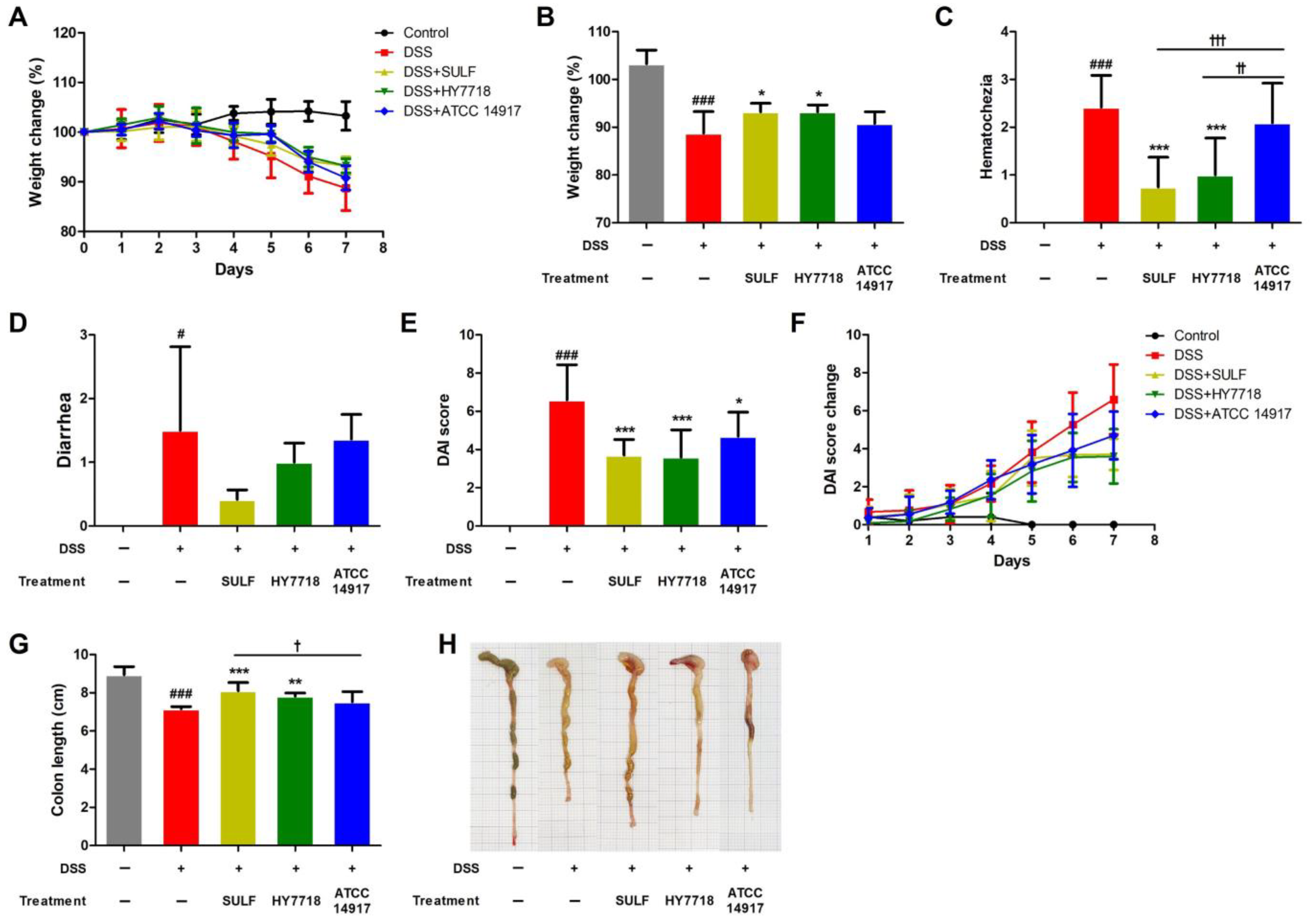
2.1. Effects of HY7718 on Physiological Indicators in Mice with DSS-Induced Colitis
2.2. Effects of HY7718 on the Histological Analysis of Colon Tissues
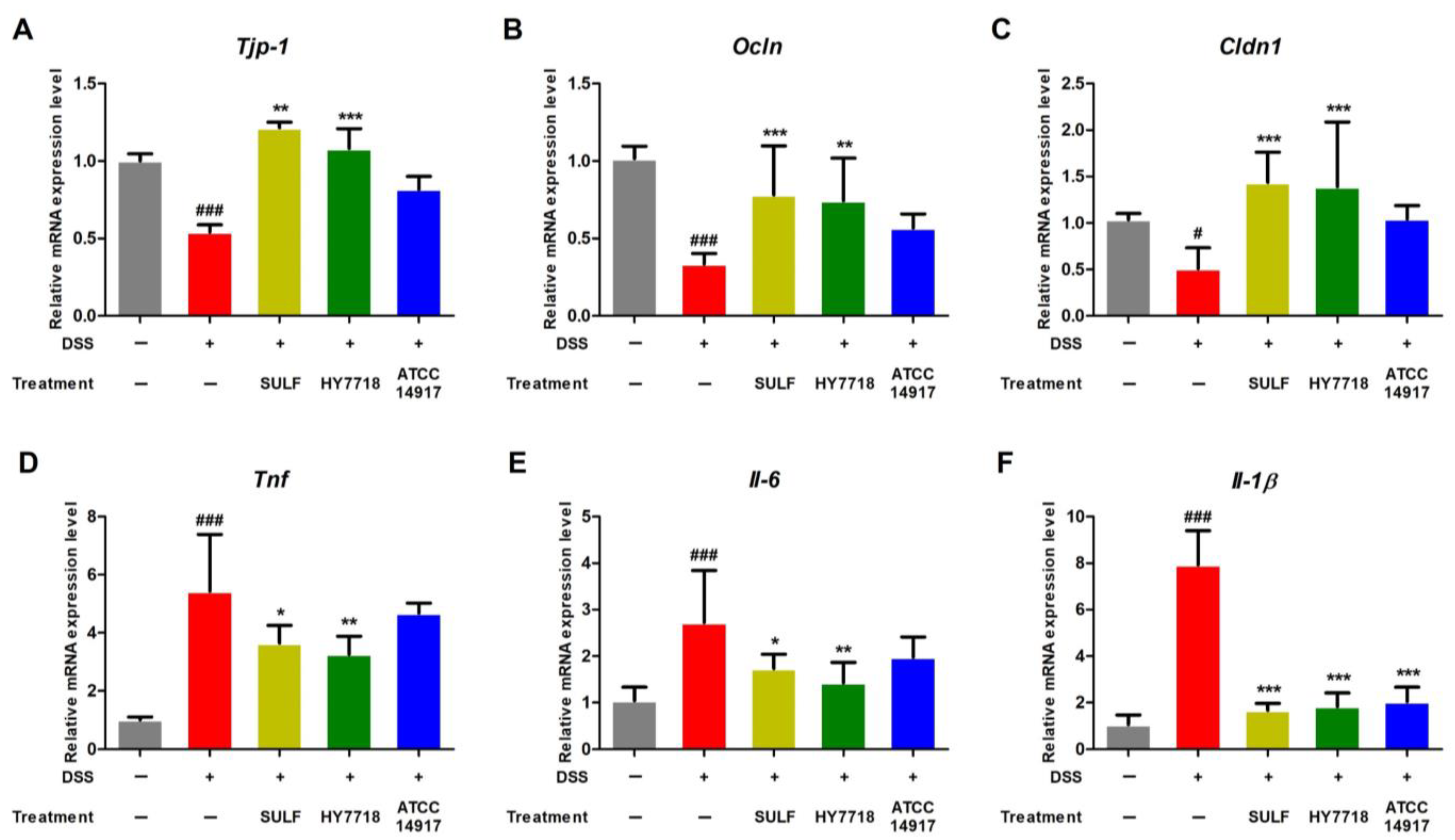
2.3. Effects of HY7718 on TJ- and Pro-Inflammatory Cytokine-Related Gene Expression in Colon Tissues
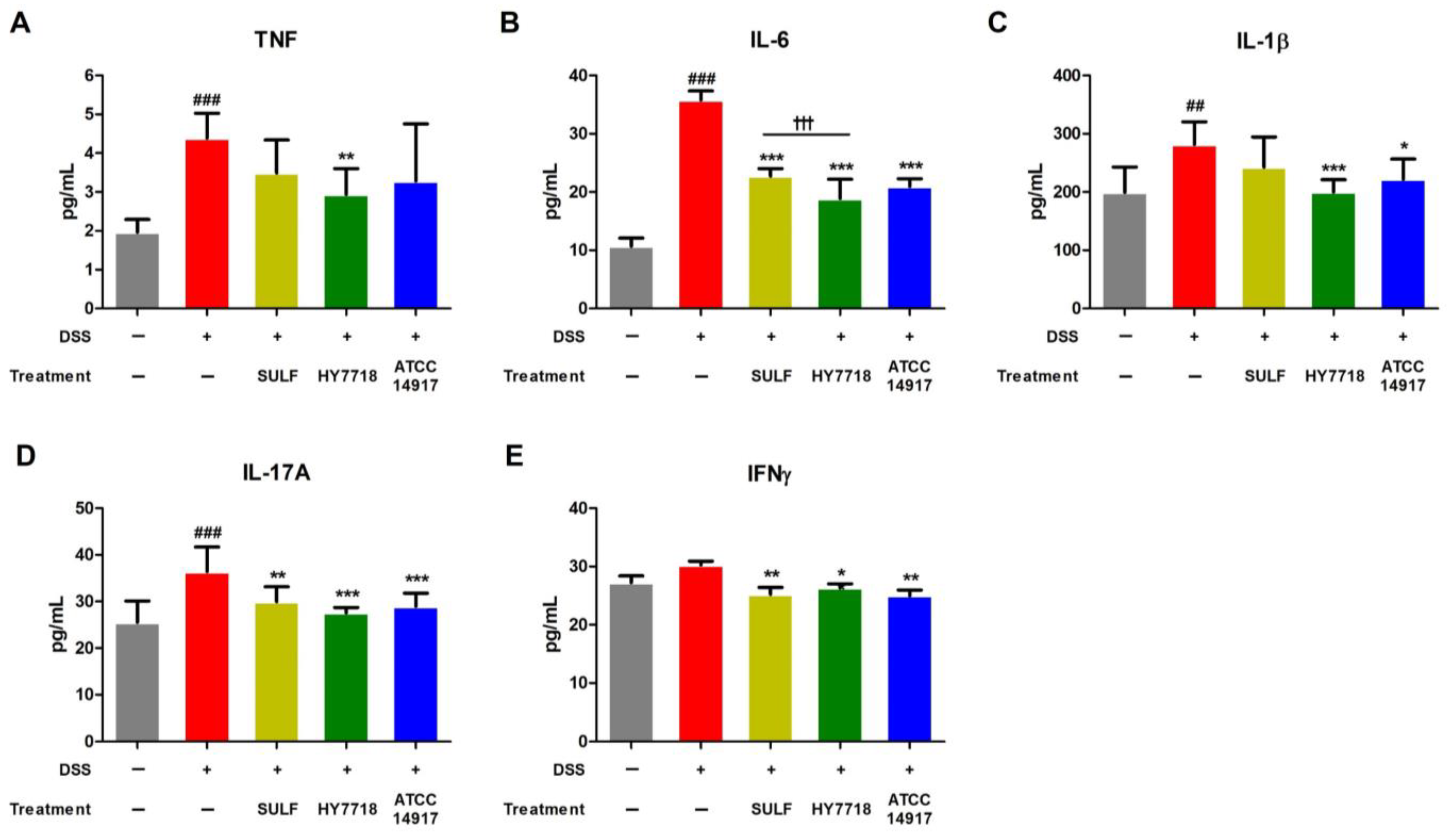
2.4. Effects of HY7718 on the Systemic Secretion of Pro-Inflammatory Cytokines
2.5. Effects of HY7718 on the Expression of TLR/MyD88/NF-κB Signaling Pathway-Related Genes in Colon Tissue
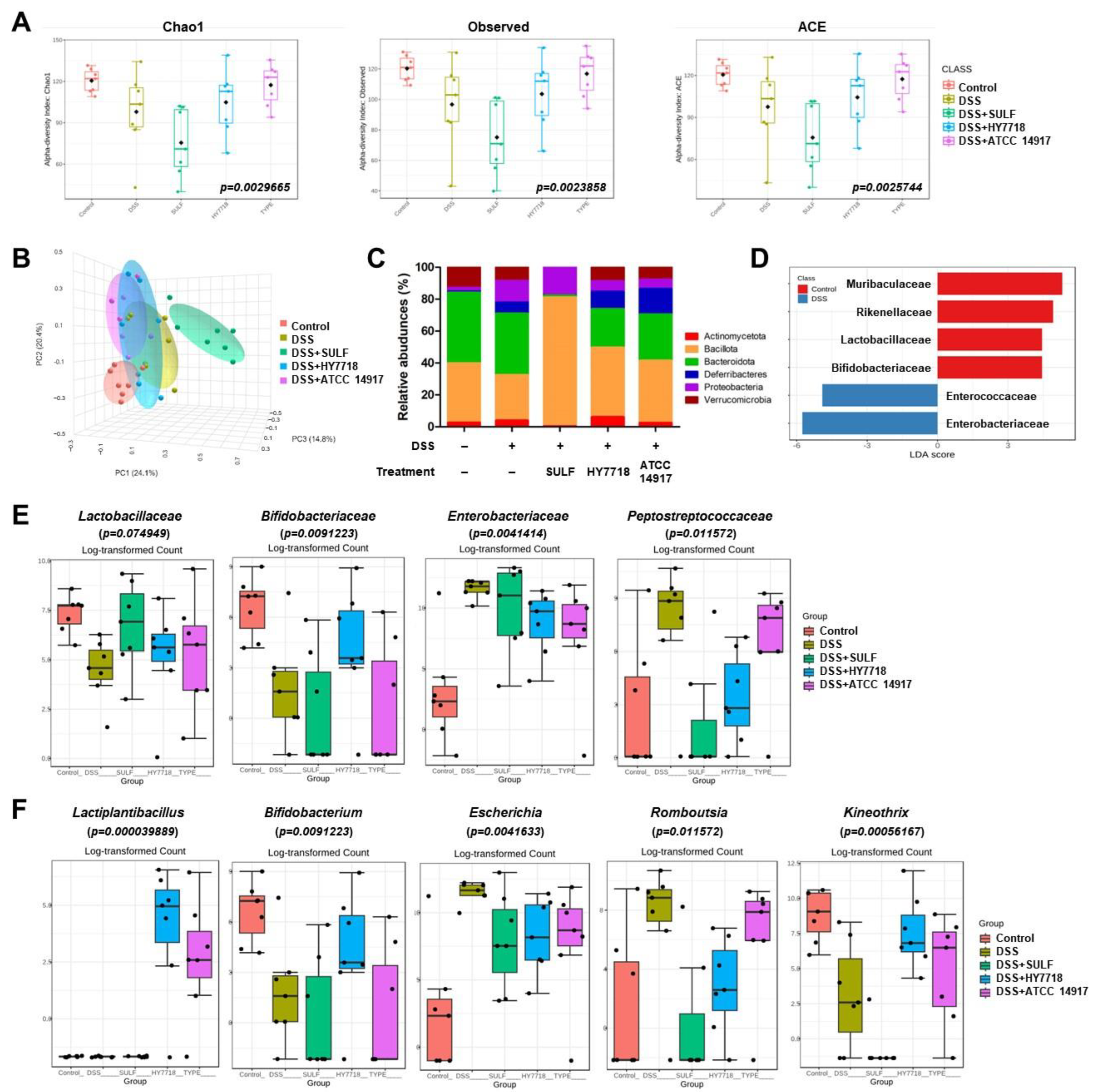
2.6. Effects of HY7718 on the Composition of the Gut Microbiota of Mice with DSS-Induced Colitis
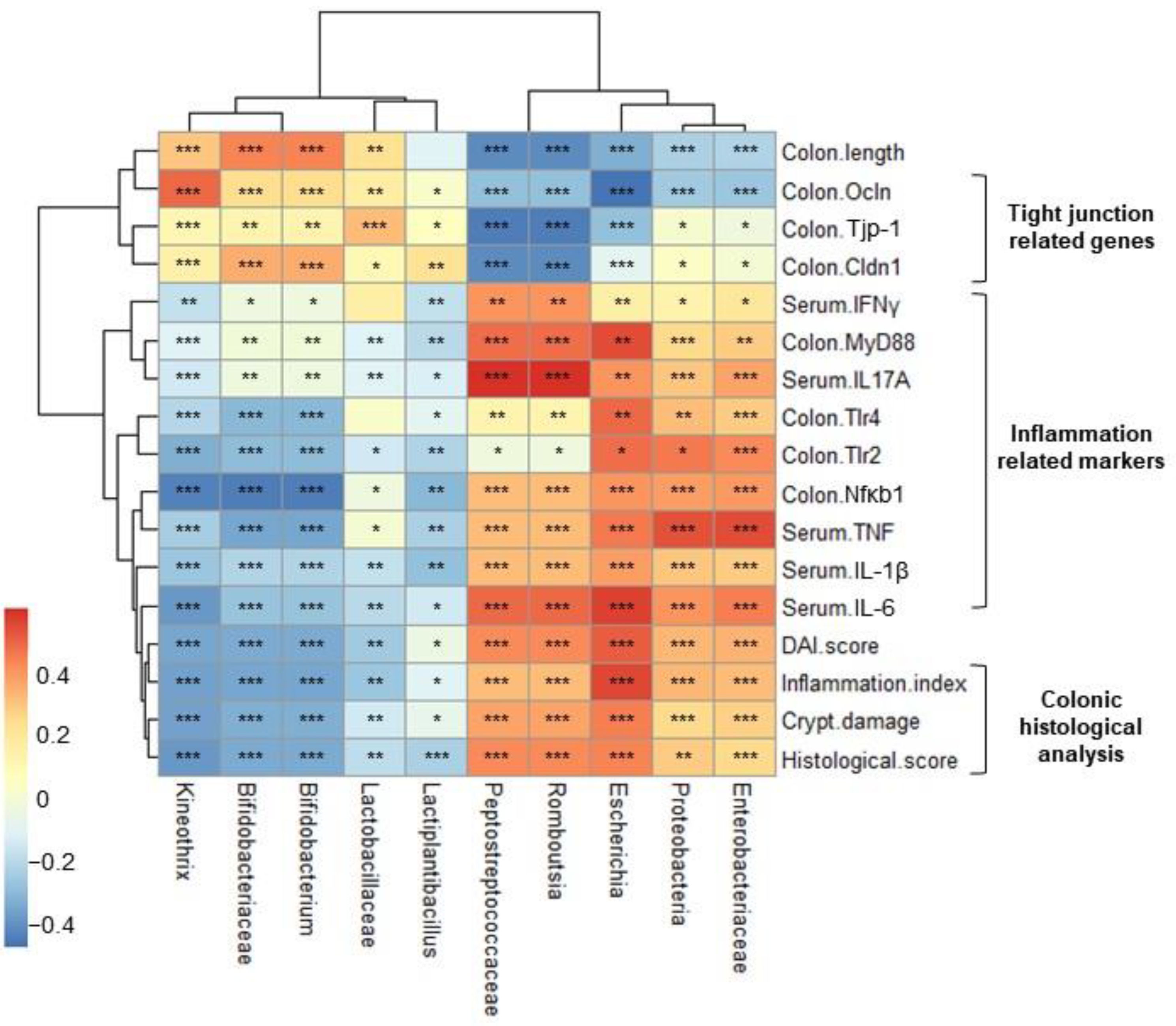
2.7. Correlation Heatmap between Relative Abundance and Biochemical Indicators
3. Discussion
4. Materials and Methods
4.1. Bacterial Culture and Sample Preparation
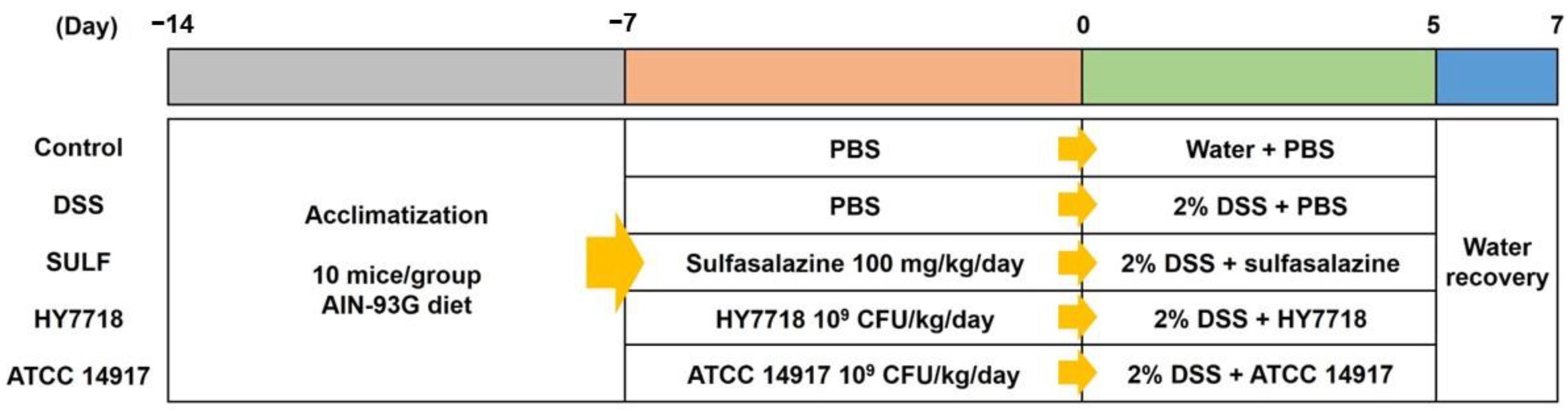
4.2. Animal Experiments
4.3. DAI Scoring
4.4. Histological Analysis
4.5. Extraction of Total RNA and Gene Expression Analysis
4.6. Measurement of Inflammatory Cytokines
4.7. DNA Extraction and Quantification
4.8. Library Construction and Sequencing
- V3-F: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′,
- V4-R: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′
4.9. Bioinformatics Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jang, Y.J.; Kim, W.-K.; Han, D.H.; Lee, K.; Ko, G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes 2019, 10, 696–711. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, C.; Huang, J.; Kuai, X.; Shao, X. The potential therapeutic role of Lactobacillus reuteri for treatment of inflammatory bowel disease. Am. J. Transl. Res. 2020, 12, 1569. [Google Scholar]
- Baumgart, D.C.; Carding, S.R. Inflammatory bowel disease: Cause and immunobiology. Lancet 2007, 369, 1627–1640. [Google Scholar] [CrossRef]
- Baumgart, D.C.; Sandborn, W.J. Inflammatory bowel disease: Clinical aspects and established and evolving therapies. Lancet 2007, 369, 1641–1657. [Google Scholar] [CrossRef]
- Xavier, R.J.; Podolsky, D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature 2007, 448, 427–434. [Google Scholar] [CrossRef]
- Seyedian, S.S.; Nokhostin, F.; Malamir, M.D. A review of the diagnosis, prevention, and treatment methods of inflammatory bowel disease. J. Med. Life 2019, 12, 113. [Google Scholar] [CrossRef]
- Mitropoulou, M.-A.; Fradelos, E.C.; Lee, K.Y.; Malli, F.; Tsaras, K.; Christodoulou, N.G.; Papathanasiou, I.V.; Lee, K.Y.; Christodoulou, N. Quality of life in patients with inflammatory bowel disease: Importance of psychological symptoms. Cureus 2022, 14, e28502. [Google Scholar] [CrossRef]
- Currò, D.; Ianiro, G.; Pecere, S.; Bibbò, S.; Cammarota, G. Probiotics, fibre and herbal medicinal products for functional and inflammatory bowel disorders. Br. J. Pharmacol. 2017, 174, 1426–1449. [Google Scholar] [CrossRef]
- Osman, N.; Adawi, D.; Molin, G.; Ahrne, S.; Berggren, A.; Jeppsson, B. Bifidobacterium infantis strains with and without a combination of oligofructose and inulin (OFI) attenuate inflammation in DSS-induced colitis in rats. BMC Gastroenterol. 2006, 6, 31. [Google Scholar] [CrossRef]
- Sanders, M.E.; Gibson, G.; Gill, H.S.; Guarner, F. Probiotics: Their potential to impact human health. Counc. Agric. Sci. Technol. Issue Pap. 2007, 36, 1–20. [Google Scholar]
- Amelia, R.; Philip, K.; Pratama, Y.E.; Purwati, E. Characterization and probiotic potential of lactic acid bacteria isolated from dadiah sampled in West Sumatra. Food Sci. Technol. 2020, 41, 746–752. [Google Scholar] [CrossRef]
- Kim, K.J.; Paik, H.-D.; Kim, J.Y. Immune-enhancing effects of Lactiplantibacillus plantarum 200655 isolated from Korean kimchi in a cyclophosphamide-induced immunocompromised mouse model. J. Microbiol. Biotechnol. 2021, 31, 726–732. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, D.; Qi, W.; Hong, T.; Xiong, T.; Wu, T.; Geng, F.; Xie, M.; Nie, S. Exopolysaccharides from Lactiplantibacillus plantarum NCU116 facilitate intestinal homeostasis by modulating intestinal epithelial regeneration and microbiota. J. Agric. Food Chem. 2021, 69, 7863–7873. [Google Scholar] [CrossRef]
- Duary, R.K.; Bhausaheb, M.A.; Batish, V.K.; Grover, S. Anti-inflammatory and immunomodulatory efficacy of indigenous probiotic Lactiplantibacillus plantarum Lp91 in colitis mouse model. Mol. Biol. Rep. 2012, 39, 4765–4775. [Google Scholar] [CrossRef]
- Ma, Y.; Fei, Y.; Han, X.; Liu, G.; Fang, J. Lactiplantibacillus plantarum alleviates obesity by altering the composition of the gut microbiota in high-fat diet-fed mice. Front. Nutr. 2022, 9, 947367. [Google Scholar] [CrossRef]
- Li, C.; Nie, S.-P.; Zhu, K.-X.; Ding, Q.; Li, C.; Xiong, T.; Xie, M.-Y. Lactiplantibacillus plantarum NCU116 improves liver function, oxidative stress and lipid metabolism in rats with high fat diet induced non-alcoholic fatty liver disease. Food Funct. 2014, 5, 3216–3223. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, Y.; Park, S.; Lee, D.; Lee, J.; Hlaing, S.P.; Yoo, J.-W.; Rhee, S.H.; Im, E. Lactiplantibacillus plantarum Metabolites Elicit Anticancer Effects by Inhibiting Autophagy-Related Responses. Molecules 2023, 28, 1890. [Google Scholar] [CrossRef]
- Wang, J.; Ji, H.; Wang, S.; Liu, H.; Zhang, W.; Zhang, D.; Wang, Y. Probiotic Lactiplantibacillus plantarum promotes intestinal barrier function by strengthening the epithelium and modulating gut microbiota. Front. Microbiol. 2018, 9, 1953. [Google Scholar]
- Ewaschuk, J.B.; Dieleman, L.A. Probiotics and prebiotics in chronic inflammatory bowel diseases. World J. Gastroenterol. WJG 2006, 12, 5941. [Google Scholar] [CrossRef]
- Li, Q.; Zheng, T.; Ding, H.; Chen, J.; Li, B.; Zhang, Q.; Yang, S.; Zhang, S.; Guan, W. Exploring the Benefits of Probiotics in Gut Inflammation and Diarrhea—From an Antioxidant Perspective. Antioxidants 2023, 12, 1342. [Google Scholar] [CrossRef]
- Chassaing, B.; Aitken, J.D.; Malleshappa, M.; Vijay-Kumar, M. Dextran sulfate sodium (DSS)-induced colitis in mice. Curr. Protoc. Immunol. 2014, 104, 15–25. [Google Scholar] [CrossRef]
- Eichele, D.D.; Kharbanda, K.K. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 2017, 23, 6016. [Google Scholar] [CrossRef]
- Kanwal, S.; Joseph, T.P.; Aliya, S.; Song, S.; Saleem, M.Z.; Nisar, M.A.; Wang, Y.; Meyiah, A.; Ma, Y.; Xin, Y. Attenuation of DSS induced colitis by Dictyophora indusiata polysaccharide (DIP) via modulation of gut microbiota and inflammatory related signaling pathways. J. Funct. Foods 2020, 64, 103641. [Google Scholar] [CrossRef]
- Zhang, W.-Q.; Quan, K.-Y.; Feng, C.-J.; Zhang, T.; He, Q.-W.; Kwok, L.-Y.; Chen, Y.-F. The Lactobacillus gasseri G098 strain mitigates symptoms of DSS-induced inflammatory bowel disease in mice. Nutrients 2022, 14, 3745. [Google Scholar] [CrossRef]
- Sun, M.-C.; Zhang, F.-C.; Yin, X.; Cheng, B.-J.; Zhao, C.-H.; Wang, Y.-L.; Zhang, Z.-Z.; Hao, H.-W.; Zhang, T.-H.; Ye, H.-Q. Lactobacillus reuteri F-9-35 prevents DSS-Induced colitis by inhibiting proinflammatory gene expression and restoring the gut microbiota in mice. J. Food Sci. 2018, 83, 2645–2652. [Google Scholar] [CrossRef]
- Guo, N.; Lv, L. Mechanistic insights into the role of probiotics in modulating immune cells in ulcerative colitis. Immun. Inflamm. Dis. 2023, 11, e1045. [Google Scholar] [CrossRef]
- Kim, H.; Yoo, M.-S.; Jeon, H.; Shim, J.-J.; Park, W.-J.; Kim, J.-Y.; Lee, J.-L. Probiotic Properties and Safety Evaluation of Lactiplantibacillus plantarum HY7718 with Superior Storage Stability Isolated from Fermented Squid. Microorganisms 2023, 11, 2254. [Google Scholar] [CrossRef]
- Das, K.M. Pharmacotherapy of inflammatory bowel disease. Postgrad. Med. 1983, 74, 141–151. [Google Scholar] [CrossRef]
- Plessas, S.; Nouska, C.; Karapetsas, A.; Kazakos, S.; Alexopoulos, A.; Mantzourani, I.; Chondrou, P.; Fournomiti, M.; Galanis, A.; Bezirtzoglou, E. Isolation, characterization and evaluation of the probiotic potential of a novel Lactobacillus strain isolated from Feta-type cheese. Food Chem. 2017, 226, 102–108. [Google Scholar] [CrossRef]
- Arellano, K.; Vazquez, J.; Park, H.; Lim, J.; Ji, Y.; Kang, H.J.; Holzapfel, W.H. Safety evaluation and whole-genome annotation of Lactobacillus plantarum strains from different sources with special focus on isolates from green tea. Probiotics Antimicrob. Proteins 2020, 12, 1057–1070. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wu, Z.; Wu, J.; Pan, D.; Zeng, X.; Cheng, K. Effects of salt stress on carbohydrate metabolism of Lactobacillus plantarum ATCC 14917. Curr. Microbiol. 2016, 73, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Ahn, E.-M.; Kim, S.-J. The improving effect of gastrodia elata blume on DSS-induced colitis in mice. Biomed. Sci. Lett. 2018, 24, 168–174. [Google Scholar] [CrossRef]
- Choi, S.; Woo, J.-K.; Jang, Y.-S.; Kang, J.-H.; Jang, J.-E.; Yi, T.-H.; Park, S.-Y.; Kim, S.-Y.; Yoon, Y.-S.; Oh, S.H. Fermented Pueraria Lobata extract ameliorates dextran sulfate sodium-induced colitis by reducing pro-inflammatory cytokines and recovering intestinal barrier function. Lab. Anim. Res. 2016, 32, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Dejban, P.; Nikravangolsefid, N.; Chamanara, M.; Dehpour, A.; Rashidian, A. The role of medicinal products in the treatment of inflammatory bowel diseases (IBD) through inhibition of TLR4/NF-kappaB pathway. Phytother. Res. 2021, 35, 835–845. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Li, X.; Liu, S.; Zhang, Y.; Zhang, D. Toll-like receptors and inflammatory bowel disease. Front. Immunol. 2018, 9, 72. [Google Scholar] [CrossRef]
- Liu, Q.; Jian, W.; Wang, L.; Yang, S.; Niu, Y.; Xie, S.; Hayer, K.; Chen, K.; Zhang, Y.; Guo, Y. Alleviation of DSS-induced colitis in mice by a new-isolated Lactobacillus acidophilus C4. Front. Microbiol. 2023, 14, 1137701. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Ma, Y.; Liu, G.; Yan, W.; Jiang, H.; Fang, J. Lactobacillus brevis alleviates DSS-induced colitis by reprograming intestinal microbiota and influencing serum metabolome in murine model. Front. Physiol. 2019, 10, 1152. [Google Scholar] [CrossRef]
- Kostadinova, F.I.; Baba, T.; Ishida, Y.; Kondo, T.; Popivanova, B.K.; Mukaida, N. Crucial involvement of the CX3CR1-CX3CL1 axis in dextran sulfate sodium-mediated acute colitis in mice. J. Leukoc. Biol. 2010, 88, 133–143. [Google Scholar] [CrossRef]
- Lee, S.H. Intestinal permeability regulation by tight junction: Implication on inflammatory bowel diseases. Intest. Res. 2015, 13, 11–18. [Google Scholar] [CrossRef]
- Műzes, G.; Molnár, B.; Tulassay, Z.; Sipos, F. Changes of the cytokine profile in inflammatory bowel diseases. World J. Gastroenterol. WJG 2012, 18, 5848. [Google Scholar] [CrossRef]
- Capaldo, C.T.; Farkas, A.E.; Hilgarth, R.S.; Krug, S.M.; Wolf, M.F.; Benedik, J.K.; Fromm, M.; Koval, M.; Parkos, C.; Nusrat, A. Proinflammatory cytokine-induced tight junction remodeling through dynamic self-assembly of claudins. Mol. Biol. Cell 2014, 25, 2710–2719. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadi, R.; Boivin, M.; Ma, T. Mechanism of cytokine modulation of epithelial tight junction barrier. Front. Biosci. A J. Virtual Libr. 2009, 14, 2765. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Liao, M.; Wang, J. TLR4 signaling in the development of colitis-associated cancer and its possible interplay with microRNA-155. Cell Commun. Signal. 2021, 19, 90. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef] [PubMed]
- Frolova, L.; Drastich, P.; Rossmann, P.; Klimesova, K.; Tlaskalova-Hogenova, H. Expression of Toll-like receptor 2 (TLR2), TLR4, and CD14 in biopsy samples of patients with inflammatory bowel diseases: Upregulated expression of TLR2 in terminal ileum of patients with ulcerative colitis. J. Histochem. Cytochem. 2008, 56, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Zhao, Y.; Wang, L.; Li, L. Role of TLR4/MyD88/NF-κB signaling in heart and liver-related complications in a rat model of type 2 diabetes mellitus. J. Int. Med. Res. 2021, 49, 0300060521997590. [Google Scholar] [CrossRef]
- Shan, Y.; Lee, M.; Chang, E.B. The gut microbiome and inflammatory bowel diseases. Annu. Rev. Med. 2022, 73, 455–468. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Ma, L.; Zeng, M.; Liu, Z. Amelioration of dextran sulfate sodium-induced colitis by autoinducer-2-deficient Lactiplantibacillus plantarum is mediated by anti-inflammatory effects and alleviation of dysbiosis of the gut microbiota. Front. Microbiol. 2022, 13, 1013586. [Google Scholar] [CrossRef]
- Son, S.; Koh, J.; Park, M.; Ryu, S.; Lee, W.; Yun, B.; Lee, J.-H.; Oh, S.; Kim, Y. Effect of the Lactobacillus rhamnosus strain GG and tagatose as a synbiotic combination in a dextran sulfate sodium-induced colitis murine model. J. Dairy Sci. 2019, 102, 2844–2853. [Google Scholar] [CrossRef]
- Pisani, A.; Rausch, P.; Bang, C.; Ellul, S.; Tabone, T.; Marantidis Cordina, C.; Zahra, G.; Franke, A.; Ellul, P. Dysbiosis in the gut microbiota in patients with inflammatory bowel disease during remission. Microbiol. Spectr. 2022, 10, e00616-22. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Lee, T.-S.; Jung, D.-H.; Song, E.-J.; Jang, A.-R.; Park, J.-Y.; Ahn, J.-H.; Seo, I.-S.; Song, S.-J.; Kim, Y.-J. Oral Administration of Lactobacillus sakei CVL-001 Improves Recovery from Dextran Sulfate Sodium-Induced Colitis in Mice by Microbiota Modulation. Microorganisms 2023, 11, 1359. [Google Scholar] [CrossRef] [PubMed]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The role of Enterobacteriaceae in gut microbiota dysbiosis in inflammatory bowel diseases. Microorganisms 2021, 9, 697. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.T.; Amos, G.C.; Murphy, A.R.; Murch, S.; Wellington, E.M.; Arasaradnam, R.P. Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 2020, 12, 1. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.-M.; Guo, H.-X.; Cai, J.-W.; Meng, X.-C. Bifidobacterium breve alleviates DSS-induced colitis in mice by maintaining the mucosal and epithelial barriers and modulating gut microbes. Nutrients 2022, 14, 3671. [Google Scholar] [CrossRef]
- Haas, K.N.; Blanchard, J.L. Kineothrix alysoides, gen. nov., sp. nov., a saccharolytic butyrate-producer within the family Lachnospiraceae. Int. J. Syst. Evol. Microbiol. 2017, 67, 402–410. [Google Scholar] [CrossRef]
- Silva, J.P.; Navegantes-Lima, K.C.; Oliveira, A.L.; Rodrigues, D.V.; Gaspar, S.L.; Monteiro, V.V.; Moura, D.P.; Monteiro, M.C. Protective mechanisms of butyrate on inflammatory bowel disease. Curr. Pharm. Des. 2018, 24, 4154–4166. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, Q.; Zhang, Y.; Ma, W.; Ning, K.; Xiang, J.-Y.; Cui, J.; Xiang, H. Combination of probiotics with different functions alleviate DSS-induced colitis by regulating intestinal microbiota, IL-10, and barrier function. Appl. Microbiol. Biotechnol. 2020, 104, 335–349. [Google Scholar] [CrossRef]


| Gene | Gene Name | Catalog Number |
|---|---|---|
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | Mm99999915_g1 |
| Tjp-1 | Tight junction protein 1 | Mm01320638_m1 |
| Ocln | Occludin | Mm00500910_m1 |
| Cldn1 | Claudin-1 | Mm01342184_m1 |
| Tnf | Tumor necrosis factor α | Mm00443258_m1 |
| Il-6 | Interleukin 6 | Mm00446190_m1 |
| Il-1β | Interleukin 1 beta | Mm00434228_m1 |
| Tlr4 | Toll-like receptor 4 | Mm00445273_m1 |
| MyD88 | Myeloid differentiation primary response 88 | Mm00440338_m1 |
| Nfκb1 | Nuclear factor kappa B subunit1 | Mm00476361_m1 |
| Tlr2 | Toll-like receptor 2 | Mm00442346_m1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-J.; Jeon, H.-J.; Kim, J.-Y.; Shim, J.-J.; Lee, J.-H. Lactiplantibacillus plantarum HY7718 Improves Intestinal Integrity in a DSS-Induced Ulcerative Colitis Mouse Model by Suppressing Inflammation through Modulation of the Gut Microbiota. Int. J. Mol. Sci. 2024, 25, 575. https://doi.org/10.3390/ijms25010575
Kim H-J, Jeon H-J, Kim J-Y, Shim J-J, Lee J-H. Lactiplantibacillus plantarum HY7718 Improves Intestinal Integrity in a DSS-Induced Ulcerative Colitis Mouse Model by Suppressing Inflammation through Modulation of the Gut Microbiota. International Journal of Molecular Sciences. 2024; 25(1):575. https://doi.org/10.3390/ijms25010575
Chicago/Turabian StyleKim, Hyeon-Ji, Hye-Jin Jeon, Joo-Yun Kim, Jae-Jung Shim, and Jae-Hwan Lee. 2024. "Lactiplantibacillus plantarum HY7718 Improves Intestinal Integrity in a DSS-Induced Ulcerative Colitis Mouse Model by Suppressing Inflammation through Modulation of the Gut Microbiota" International Journal of Molecular Sciences 25, no. 1: 575. https://doi.org/10.3390/ijms25010575
APA StyleKim, H.-J., Jeon, H.-J., Kim, J.-Y., Shim, J.-J., & Lee, J.-H. (2024). Lactiplantibacillus plantarum HY7718 Improves Intestinal Integrity in a DSS-Induced Ulcerative Colitis Mouse Model by Suppressing Inflammation through Modulation of the Gut Microbiota. International Journal of Molecular Sciences, 25(1), 575. https://doi.org/10.3390/ijms25010575





