Contribution of Signal Transducer and Activator of Transcription 3 (STAT3) to Bone Development and Repair
Abstract
:1. Introduction
2. Results
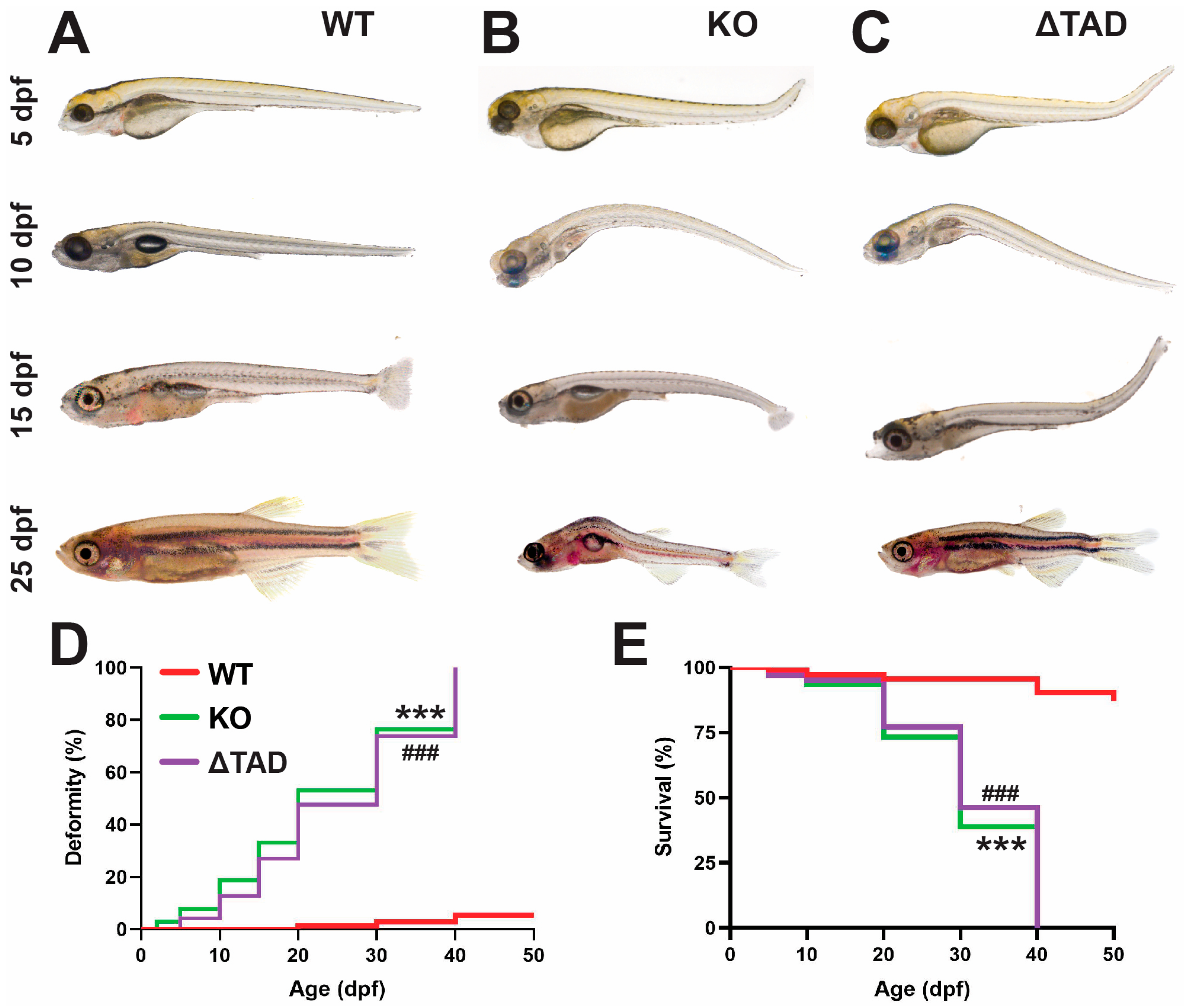
2.1. Stat3 Mutations Affect Development
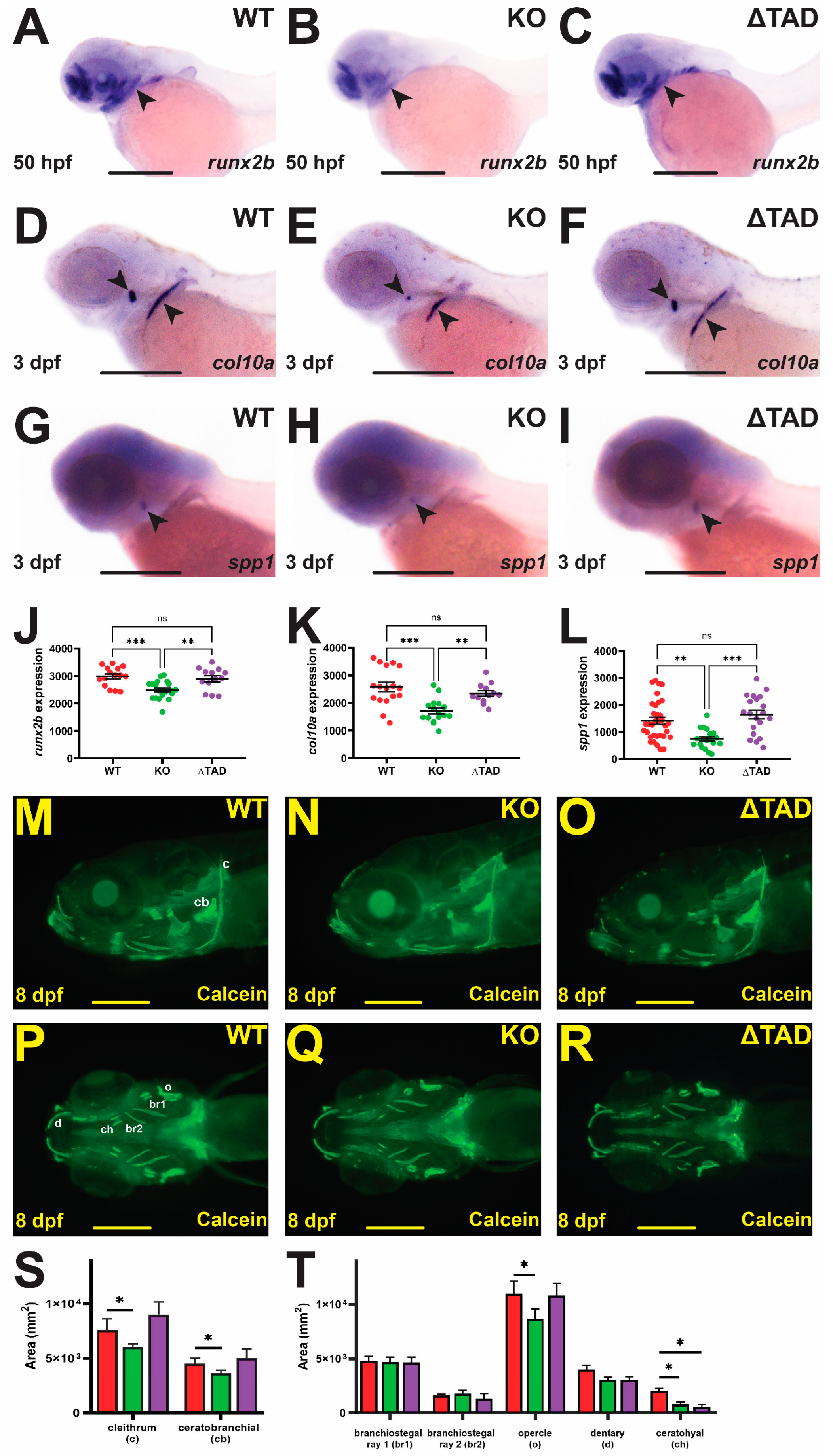
2.2. Stat3 Is Required for Bone Development
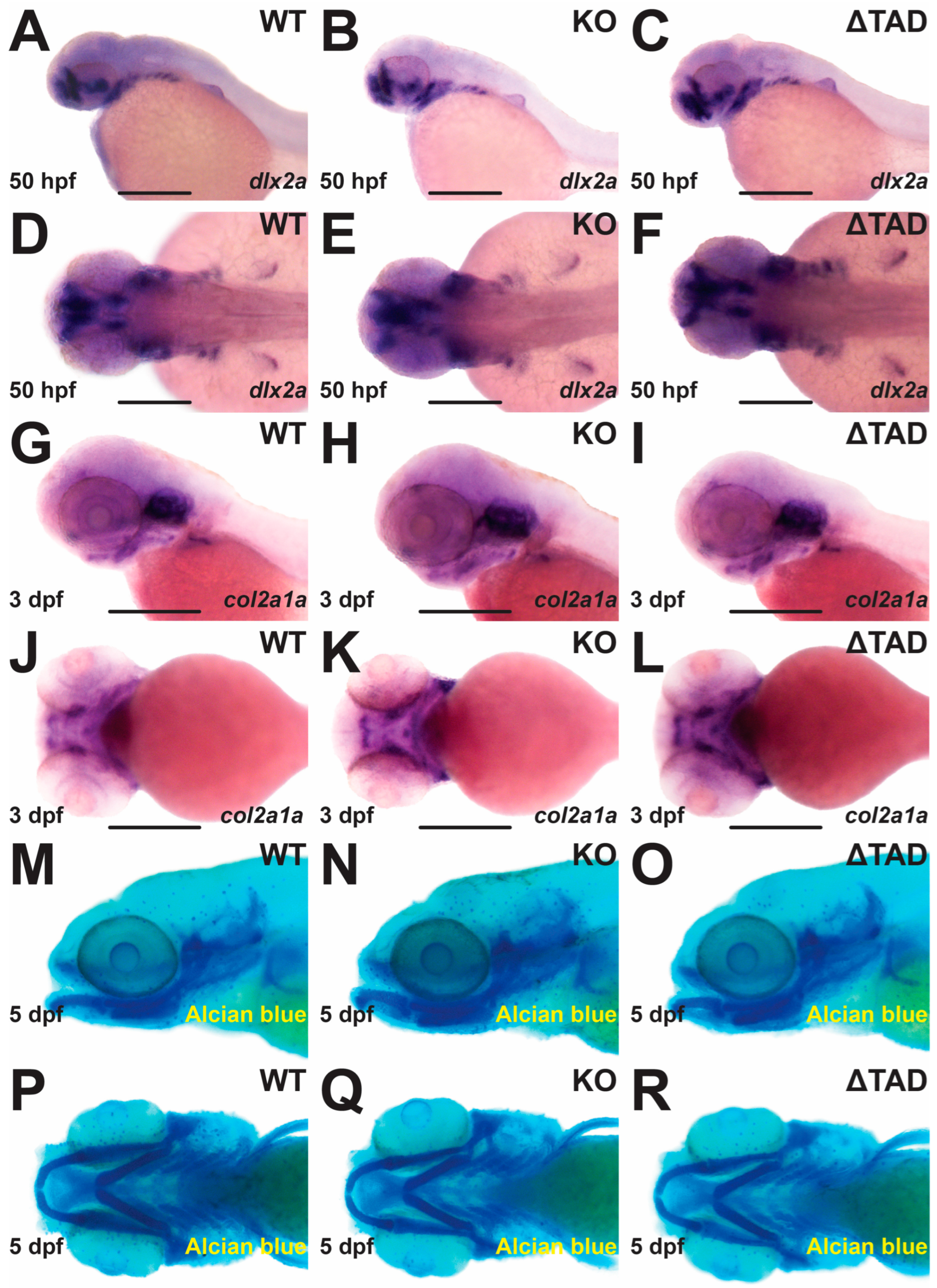
2.3. Stat3 Is Dispensable for Cartilage Formation
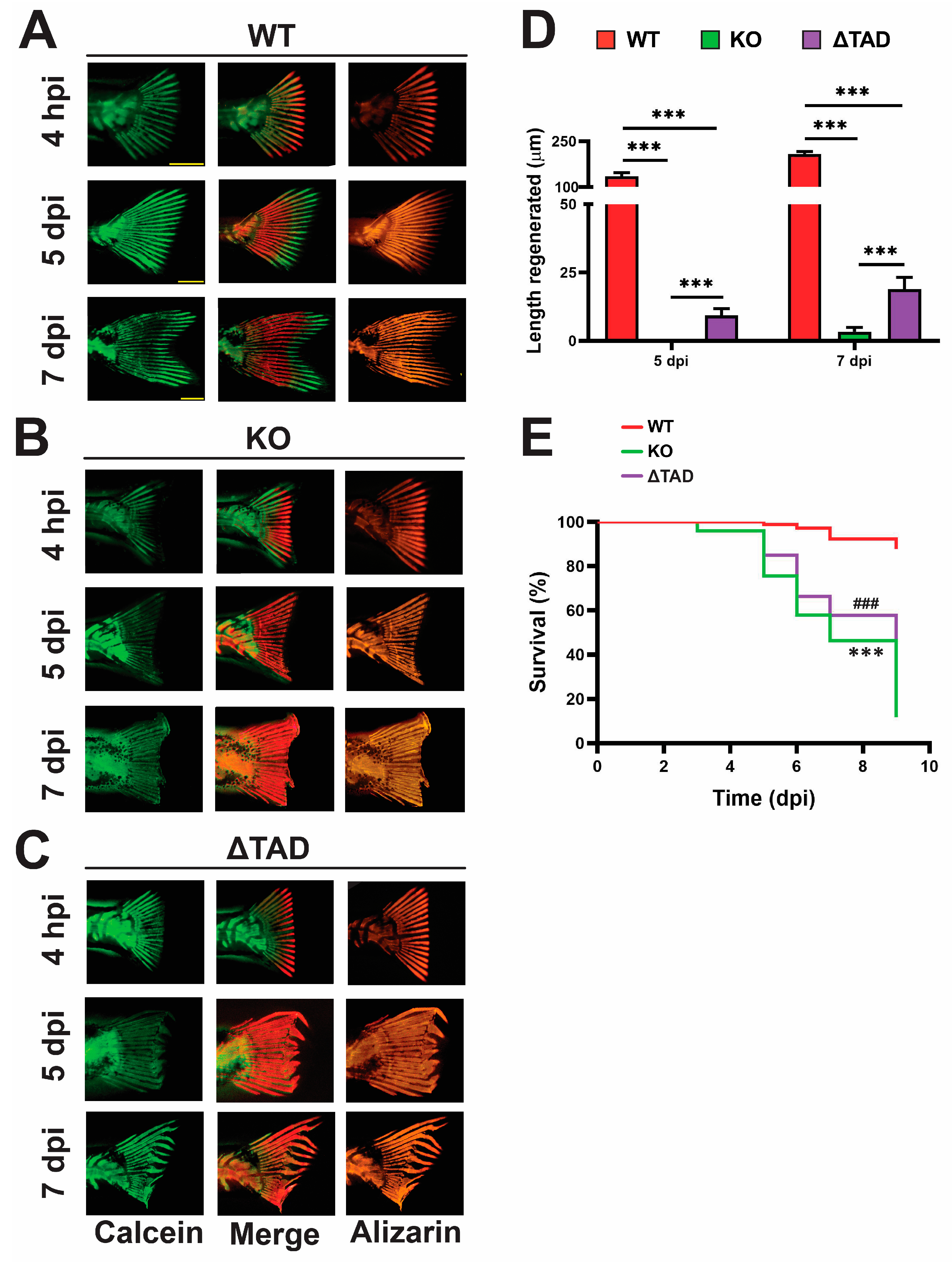
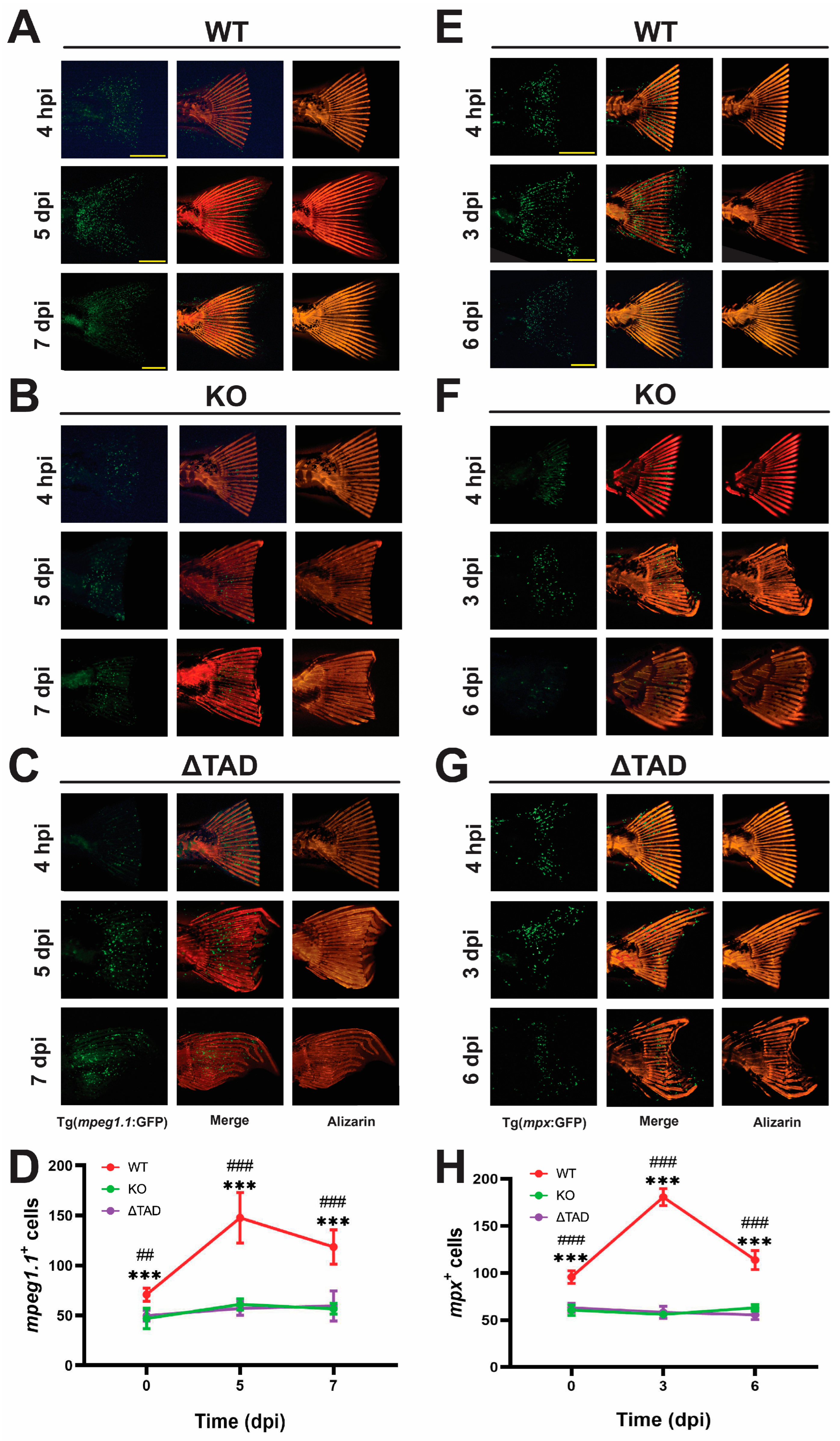
2.4. Stat3 Is Essential for Fin Ray Regeneration and the Associated Innate Immune Response
3. Discussion
4. Materials and Methods
4.1. Zebrafish Husbandry
4.2. Experimental Setup
4.3. Amputation Assay
4.4. Alizarin Red Staining
4.5. Calcein Staining
4.6. Pulse Chase Staining
4.7. Cartilage Staining
4.8. Whole-Mount In Situ Hybridization
4.9. Gene Expression Analysis
4.10. Imaging and Analysis
4.11. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hillmer, E.J.; Zhang, H.; Li, H.S.; Watowich, S.S. STAT3 signaling in immunity. Cytokine Growth Factor Rev. 2016, 31, 1–15. [Google Scholar] [CrossRef]
- Hou, X.; Tian, F. STAT3-mediated osteogenesis and osteoclastogenesis in osteoporosis. Cell Commun. Signal 2022, 20, 112. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Shen, C.; Li, H.; Fan, Q.; Ding, J.; Jin, F.C.; Sha, L. Leptin induces the apoptosis of chondrocytes in an in vitro model of osteoarthritis via the JAK2-STAT3 signaling pathway. Mol. Med. Rep. 2016, 13, 3684–3690. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.Q.; Lin, Y.; Li, L.; Lu, J.; Geng, D.; Zhang, J.; Jashashvili, T.; Buser, Z.; Magallanes, J.; Tassey, J. gp130/STAT3 signaling is required for homeostatic proliferation and anabolism in postnatal growth plate and articular chondrocytes. Commun. Biol. 2022, 5, 64. [Google Scholar]
- Jo, S.; Wang, S.E.; Lee, Y.L.; Kang, S.; Lee, B.; Han, J.; Sung, I.-H.; Park, Y.-S.; Bae, S.-C.; Kim, T.-H. IL-17A induces osteoblast differentiation by activating JAK2/STAT3 in ankylosing spondylitis. Arthritis Res. Ther. 2018, 20, 115. [Google Scholar] [CrossRef]
- Xu, X.; Li, Y.; Shi, L.; He, K.; Sun, Y.; Ding, Y.; Meng, B.; Zhang, J.; Xiang, L.; Dong, J. Myeloid-derived growth factor (MYDGF) protects bone mass through inhibiting osteoclastogenesis and promoting osteoblast differentiation. EMBO Rep. 2022, 23, e53509. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Tang, Y.; Wu, Q.; Ji, Y.C.; Feng, Z.F.; Kang, F.W. HIF-1α facilitates osteocyte-mediated osteoclastogenesis by activating JAK2/STAT3 pathway in vitro. J. Cell Physiol. 2019, 234, 21182–21192. [Google Scholar] [CrossRef]
- Yang, Y.; Chung, M.R.; Zhou, S.; Gong, X.; Xu, H.; Hong, Y.; Jin, A.; Huang, X.; Zou, W.; Dai, Q.; et al. STAT3 controls osteoclast differentiation and bone homeostasis by regulating NFATc1 transcription. J. Biol. Chem. 2019, 294, 15395–15407. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, N.; Liongue, C.; Ward, A.C. STAT proteins: A kaleidoscope of canonical and non-canonical functions in immunity and cancer. J. Hematol. Oncol. 2021, 14, 198. [Google Scholar] [CrossRef]
- Takeda, K.; Noguchi, K.; Shi, W.; Tanaka, T.; Matsumoto, M.; Yoshida, N.; Kishimoto, T.; Akira, S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. USA 1997, 94, 3801–3804. [Google Scholar] [CrossRef]
- Zhou, S.; Dai, Q.; Huang, X.; Jin, A.; Yang, Y.; Gong, X.; Xu, H.; Gao, X.; Jiang, L. STAT3 is critical for skeletal development and bone homeostasis by regulating osteogenesis. Nat. Commun. 2021, 12, 6891. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Welte, T.; Troiano, N.; Maher, S.E.; Fu, X.-Y.; Bothwell, A.L. Osteoporosis with increased osteoclastogenesis in hematopoietic cell-specific STAT3-deficient mice. Biochem. Biophys. Res. Commun. 2005, 328, 800–807. [Google Scholar] [CrossRef] [PubMed]
- Vogel, T.P.; Milner, J.D.; Cooper, M.A. The ying and yang of STAT3 in human disease. J. Clin. Immunol. 2015, 35, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Holland, S.M.; DeLeo, F.R.; Elloumi, H.Z.; Hsu, A.P.; Uzel, G.; Brodsky, N.; Freeman, A.F.; Demidowich, A.; Davis, J.; Turner, M.L.; et al. STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 2007, 357, 1608–1619. [Google Scholar] [CrossRef] [PubMed]
- Al-Shaikhly, T.; Ochs, H.D. Hyper IgE syndromes: Clinical and molecular characteristics. Immunol. Cell Biol. 2019, 97, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Tsilifis, C.; Freeman, A.F.; Gennery, A.R. STAT3 hyper-IgE syndrome—An update and unanswered questions. J. Clin. Immunol. 2021, 41, 864–880. [Google Scholar] [CrossRef]
- Sowerwine, K.J.; Holland, S.M.; Freeman, A.F. Hyper-IgE syndrome update. Ann. N. Y. Acad. Sci. 2012, 1250, 25–32. [Google Scholar] [CrossRef]
- Li, J.; Yin, Z.; Huang, B.; Xu, K.; Su, J. Stat3 signaling pathway: A future therapeutic target for bone-related diseases. Front. Pharmacol. 2022, 13, 897539. [Google Scholar] [CrossRef]
- Latourte, A.; Cherifi, C.; Maillet, J.; Ea, H.-K.; Bouaziz, W.; Funck-Brentano, T.; Cohen-Solal, M.; Hay, E.; Richette, P. Systemic inhibition of IL-6/Stat3 signalling protects against experimental osteoarthritis. Ann. Rheum. Dis. 2017, 76, 748–755. [Google Scholar] [CrossRef]
- Bergen, D.J.; Kague, E.; Hammond, C.L. Zebrafish as an emerging model for osteoporosis: A primary testing platform for screening new osteo-active compounds. Front. Endocrinol. 2019, 10, 6. [Google Scholar] [CrossRef]
- Tonelli, F.; Bek, J.W.; Besio, R.; De Clercq, A.; Leoni, L.; Salmon, P.; Coucke, P.J.; Willaert, A.; Forlino, A. Zebrafish: A resourceful vertebrate model to investigate skeletal disorders. Front. Endocrinol. 2020, 11, 489. [Google Scholar] [CrossRef] [PubMed]
- Oates, A.C.; Wollberg, P.; Pratt, S.J.; Paw, B.H.; Johnson, S.L.; Ho, R.K.; Postlethwait, J.H.; Zon, L.I.; Wilks, A.F. Zebrafish stat3 is expressed in restricted tissues during embryogenesis and stat1 rescues cytokine signaling in a STAT1-deficient human cell line. Dev. Dyn. 1999, 215, 352–370. [Google Scholar] [CrossRef]
- Liongue, C.; Ward, A.C. Evolution of Class I cytokine receptors. BMC Evol. Biol. 2007, 7, 120. [Google Scholar] [CrossRef] [PubMed]
- Liongue, C.; O’Sullivan, L.A.; Trengove, M.C.; Ward, A.C. Evolution of JAK-STAT pathway components: Mechanisms and role in immune system development. PLoS ONE 2012, 7, e32777. [Google Scholar] [CrossRef] [PubMed]
- Xiong, S.; Wu, J.; Jing, J.; Huang, P.; Li, Z.; Mei, J.; Gui, J.-F. Loss of stat3 function leads to spine malformation and immune disorder in zebrafish. Sci. Bull. 2017, 62, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sepich, D.S.; Solnica-Krezel, L. Stat3/Cdc25a-dependent cell proliferation promotes embryonic axis extension during zebrafish gastrulation. PLoS Genet. 2017, 13, e1006564. [Google Scholar] [CrossRef] [PubMed]
- Khaliq, M.; Ko, S.; Liu, Y.; Wang, H.; Sun, Y.; Solnica-Krezel, L.; Shin, D. Stat3 regulates liver progenitor cell-driven liver regeneration in zebrafish. Gene Expr. 2018, 18, 157–170. [Google Scholar] [CrossRef]
- Sobah, M.L.; Liongue, C.; Ward, A.C. Stat3 regulates hematopoiesis and impacts neutrophil and macrophage function in zebrafish. J. Innate. Immun. 2024; submitted. [Google Scholar]
- Flores, M.V.; Tsang, V.W.K.; Hu, W.; Kalev-Zylinska, M.; Postlethwait, J.; Crosier, P.; Crosier, K.; Fisher, S. Duplicate zebrafish runx2 orthologues are expressed in developing skeletal elements. Gene Expr. Patterns 2004, 4, 573–581. [Google Scholar] [CrossRef]
- Eames, B.F.; Amores, A.; Yan, Y.-L.; Postlethwait, J.H. Evolution of the osteoblast: Skeletogenesis in gar and zebrafish. BMC Evol. Biol. 2012, 12, 27. [Google Scholar] [CrossRef]
- Bergen, D.J.; Tong, Q.; Shukla, A.; Newham, E.; Zethof, J.; Lundberg, M.; Ryan, R.; Youlten, S.E.; Frysz, M.; Croucher, P.I. Regenerating zebrafish scales express a subset of evolutionary conserved genes involved in human skeletal disease. BMC Biol. 2022, 20, 21. [Google Scholar] [CrossRef] [PubMed]
- Talbot, J.C.; Johnson, S.L.; Kimmel, C.B. hand2 and Dlx genes specify dorsal, intermediate and ventral domains within zebrafish pharyngeal arches. Development 2010, 137, 2507–2517. [Google Scholar] [CrossRef] [PubMed]
- Dale, R.M.; Topczewski, J. Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev. Biol. 2011, 357, 518–531. [Google Scholar] [CrossRef] [PubMed]
- Petrie, T.A.; Strand, N.S.; Tsung-Yang, C.; Rabinowitz, J.S.; Moon, R.T. Macrophages modulate adult zebrafish tail fin regeneration. Development 2014, 141, 2581–2591. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.S.; Feng, S.; Cong, Q.; Kim, H.; Liu, Y.; Yang, Y. Stat3 loss in mesenchymal progenitors causes Job syndrome-like skeletal defects by reducing Wnt/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2021, 118, e2020100118. [Google Scholar] [CrossRef] [PubMed]
- Peron, M.; Dinarello, A.; Meneghetti, G.; Martorano, L.; Facchinello, N.; Vettori, A.; Licciardello, G.; Tiso, N.; Argenton, F. The stem-like Stat3-responsive cells of zebrafish intestine are Wnt/β-catenin dependent. Development 2020, 147, dev188987. [Google Scholar] [CrossRef] [PubMed]
- Tarasco, M.; Laizé, V.; Cardeira, J.; Cancela, M.L.; Gavaia, P.J. The zebrafish operculum: A powerful system to assess osteogenic bioactivities of molecules with pharmacological and toxicological relevance. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2017, 197, 45–52. [Google Scholar] [CrossRef]
- Mork, L.; Crump, G. Zebrafish craniofacial development: A window into early patterning. Curr. Top. Dev. Biol. 2015, 115, 235–269. [Google Scholar]
- Rosa, J.T.; Tarasco, M.; Gavaia, P.J.; Cancela, M.L.; Laizé, V. Screening of mineralogenic and osteogenic compounds in zebrafish—Tools to improve assay throughput and data accuracy. Pharmaceuticals 2022, 15, 983. [Google Scholar] [CrossRef]
- Lei, L.; Huang, Z.; Feng, J.; Huang, Z.; Tao, Y.; Hu, X.; Zhang, X. Loss of receptor tyrosine kinase-like orphan receptor 2 impairs the osteogenesis of mBMSCs by inhibiting signal transducer and activator of transcription 3. Stem Cell Res. Ther. 2020, 11, 137. [Google Scholar] [CrossRef]
- Dalagiorgou, G.; Piperi, C.; Adamopoulos, C.; Georgopoulou, U.; Gargalionis, A.N.; Spyropoulou, A.; Zoi, I.; Nokhbehsaim, M.; Damanaki, A.; Deschner, J. Mechanosensor polycystin-1 potentiates differentiation of human osteoblastic cells by upregulating Runx2 expression via induction of JAK2/STAT3 signaling axis. Cell Mol. Life Sci. 2017, 74, 921–936. [Google Scholar] [CrossRef] [PubMed]
- Acampora, D.; Merlo, G.R.; Paleari, L.; Zerega, B.; Postiglione, M.P.; Mantero, S.; Bober, E.; Barbieri, O.; Simeone, A.; Levi, G. Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development 1999, 126, 3795–3809. [Google Scholar] [CrossRef] [PubMed]
- Heude, E.; Shaikho, S.; Ekker, M. The dlx5a/dlx6a genes play essential roles in the early development of zebrafish median fin and pectoral structures. PLoS ONE 2014, 9, e98505. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.D.; Murray, C.A.; Valdez, M.J.; Perantoni, A.O. Mesoderm-specific Stat3 deletion affects expression of Sox9 yielding Sox9-dependent phenotypes. PLoS Genet. 2017, 13, e1006610. [Google Scholar] [CrossRef] [PubMed]
- Gensel, J.C.; Nakamura, S.; Guan, Z.; van Rooijen, N.; Ankeny, D.P.; Popovich, P.G. Macrophages promote axon regeneration with concurrent neurotoxicity. J. Neurosci. 2009, 29, 3956–3968. [Google Scholar] [CrossRef] [PubMed]
- Bohaud, C.; Johansen, M.D.; Jorgensen, C.; Ipseiz, N.; Kremer, L.; Djouad, F. The role of macrophages during zebrafish injury and tissue regeneration under infectious and non-infectious conditions. Front. Immunol. 2021, 12, 707824. [Google Scholar] [CrossRef] [PubMed]
- Vi, L.; Baht, G.S.; Whetstone, H.; Ng, A.; Wei, Q.; Poon, R.; Mylvaganam, S.; Grynpas, M.; Alman, B.A. Macrophages promote osteoblastic differentiation in vivo: Implications in fracture repair and bone homeostasis. J. Bone Miner. Res. 2015, 30, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, T.; Hall, C.J.; Crosier, P.S.; Abe, G.; Kawakami, K.; Kudo, A.; Kawakami, A. Transient inflammatory response mediated by interleukin-1beta is required for proper regeneration in zebrafish fin fold. eLife 2017, 6, e22716. [Google Scholar] [CrossRef]
- Peiseler, M.; Kubes, P. More friend than foe: The emerging role of neutrophils in tissue repair. J. Clin. Investig. 2019, 129, 2629–2639. [Google Scholar] [CrossRef]
- Li, L.; Yan, B.; Shi, Y.Q.; Zhang, W.Q.; Wen, Z.L. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J. Biol. Chem. 2012, 287, 25353–25360. [Google Scholar] [CrossRef] [PubMed]
- Tomecka, M.J.; Ethiraj, L.P.; Sánchez, L.M.; Roehl, H.H.; Carney, T.J. Clinical pathologies of bone fracture modelled in zebrafish. Dis. Models Mech. 2019, 12, dmm037630. [Google Scholar] [CrossRef] [PubMed]
- Bastian, O.W.; Koenderman, L.; Alblas, J.; Leenen, L.P.; Blokhuis, T.J. Neutrophils contribute to fracture healing by synthesizing fibronectin+ extracellular matrix rapidly after injury. Clin. Immunol. 2016, 164, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Houseright, R.A.; Miskolci, V.; Mulvaney, O.; Bortnov, V.; Mosher, D.F.; Rindy, J.; Bennin, D.A.; Huttenlocher, A. Myeloid-derived growth factor regulates neutrophil motility in interstitial tissue damage. J. Cell Biol. 2021, 220, e202103054. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T. IL-6 in inflammation, autoimmunity and cancer. Int. Immunol. 2021, 33, 127–148. [Google Scholar] [CrossRef] [PubMed]
- Weigele, J.; Franz-Odendaal, T.A. Functional bone histology of zebrafish reveals two types of endochondral ossification, different types of osteoblast clusters and a new bone type. J. Anat. 2016, 229, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Ellett, F.; Pase, L.; Hayman, J.W.; Andrianopoulos, A.; Lieschke, G.J. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 2011, 117, e49–e56. [Google Scholar] [CrossRef] [PubMed]
- Buchan, K.D.; Prajsnar, T.K.; Ogryzko, N.V.; de Jong, N.W.M.; van Gent, M.; Kolata, J.; Foster, S.J.; van Strijp, J.A.G.; Renshaw, S.A. A transgenic zebrafish line for in vivo visualisation of neutrophil myeloperoxidase. PLoS ONE 2019, 14, e0215592. [Google Scholar] [CrossRef]
- Dobrzycki, T.; Krecsmarik, M.; Monteiro, R. Genotyping and quantification of in situ hybridization staining in zebrafish. J. Vis. Exp. 2020, e59956. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book: A Guide for The Laboratory Use of Zebrafish; University of Oregon: Eugene, OR, USA, 2000. [Google Scholar]
- Sakata-Haga, H.; Uchishiba, M.; Shimada, H.; Tsukada, T.; Mitani, M.; Arikawa, T.; Shoji, H.; Hatta, T. A rapid and nondestructive protocol for whole-mount bone staining of small fish and Xenopus. Sci. Rep. 2018, 8, 7453. [Google Scholar] [CrossRef]
- Du, S.J.; Frenkel, V.; Kindschi, G.; Zohar, Y. Visualizing normal and defective bone development in zebrafish embryos using the fluorescent chromophore calcein. Dev. Biol. 2001, 238, 239–246. [Google Scholar] [CrossRef] [PubMed]
- McGowan, L.M.; Kague, E.; Vorster, A.; Newham, E.; Cross, S.; Hammond, C.L. Wnt16 elicits a protective effect against fractures and supports bone repair in zebrafish. JBMR Plus 2021, 5, e10461. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.; Kimmel, C. A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 2007, 82, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Thisse, B.; Thisse, C. In situ hybridization on whole-mount zebrafish embryos and young larvae. In In Situ Hybridization Protocols; Springer: Berlin/Heidelberg, Germany, 2014; pp. 53–67. [Google Scholar]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2^(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobah, M.L.; Liongue, C.; Ward, A.C. Contribution of Signal Transducer and Activator of Transcription 3 (STAT3) to Bone Development and Repair. Int. J. Mol. Sci. 2024, 25, 389. https://doi.org/10.3390/ijms25010389
Sobah ML, Liongue C, Ward AC. Contribution of Signal Transducer and Activator of Transcription 3 (STAT3) to Bone Development and Repair. International Journal of Molecular Sciences. 2024; 25(1):389. https://doi.org/10.3390/ijms25010389
Chicago/Turabian StyleSobah, Mohamed L., Clifford Liongue, and Alister C. Ward. 2024. "Contribution of Signal Transducer and Activator of Transcription 3 (STAT3) to Bone Development and Repair" International Journal of Molecular Sciences 25, no. 1: 389. https://doi.org/10.3390/ijms25010389
APA StyleSobah, M. L., Liongue, C., & Ward, A. C. (2024). Contribution of Signal Transducer and Activator of Transcription 3 (STAT3) to Bone Development and Repair. International Journal of Molecular Sciences, 25(1), 389. https://doi.org/10.3390/ijms25010389






