Characterization of the Complete Mitochondrial Genome of Agelas nakamurai from the South China Sea
Abstract
1. Introduction
2. Results
2.1. Sample Collection, Sequencing Quality and Assembly Results
2.2. Species Identification
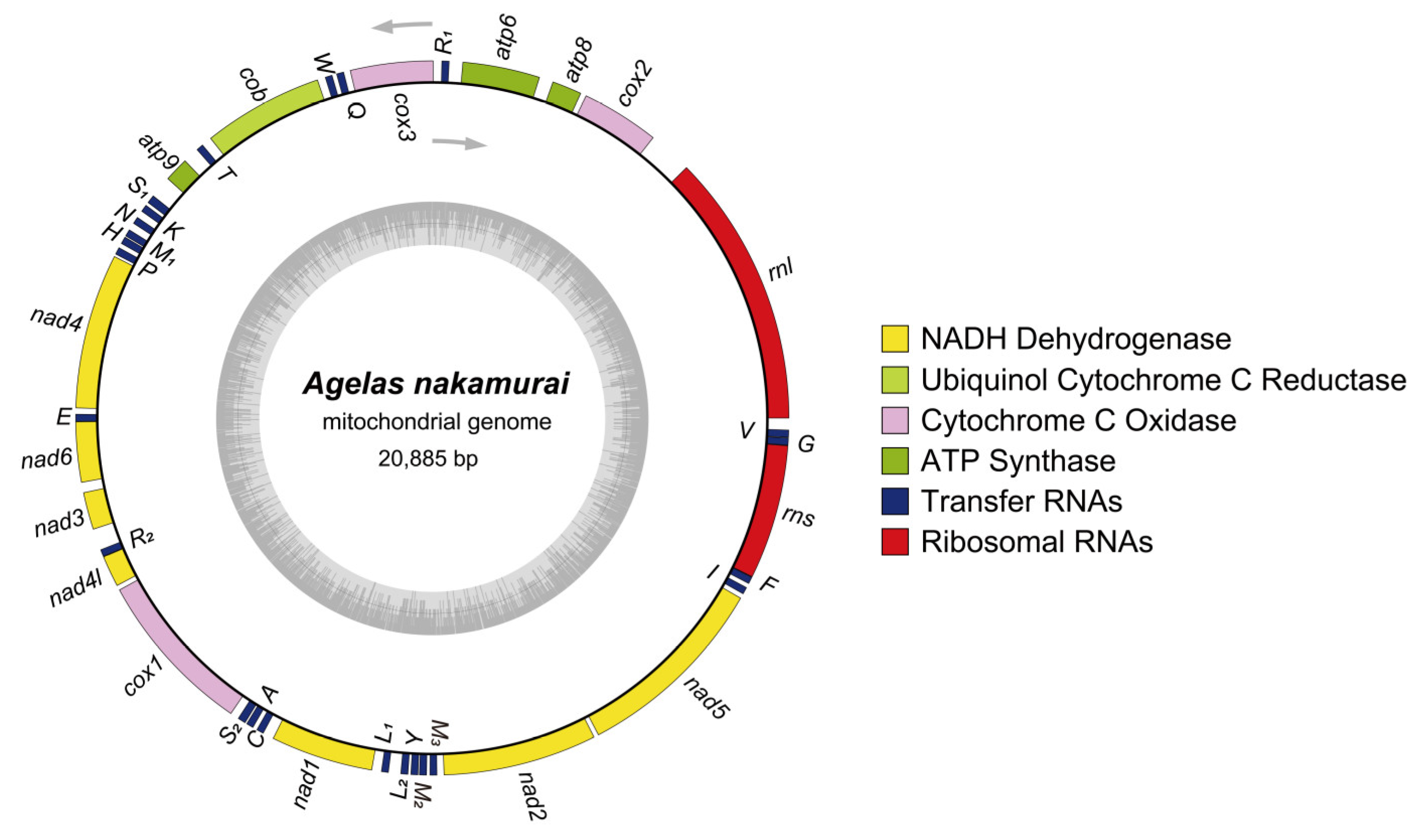
2.3. Mitogenome Composition
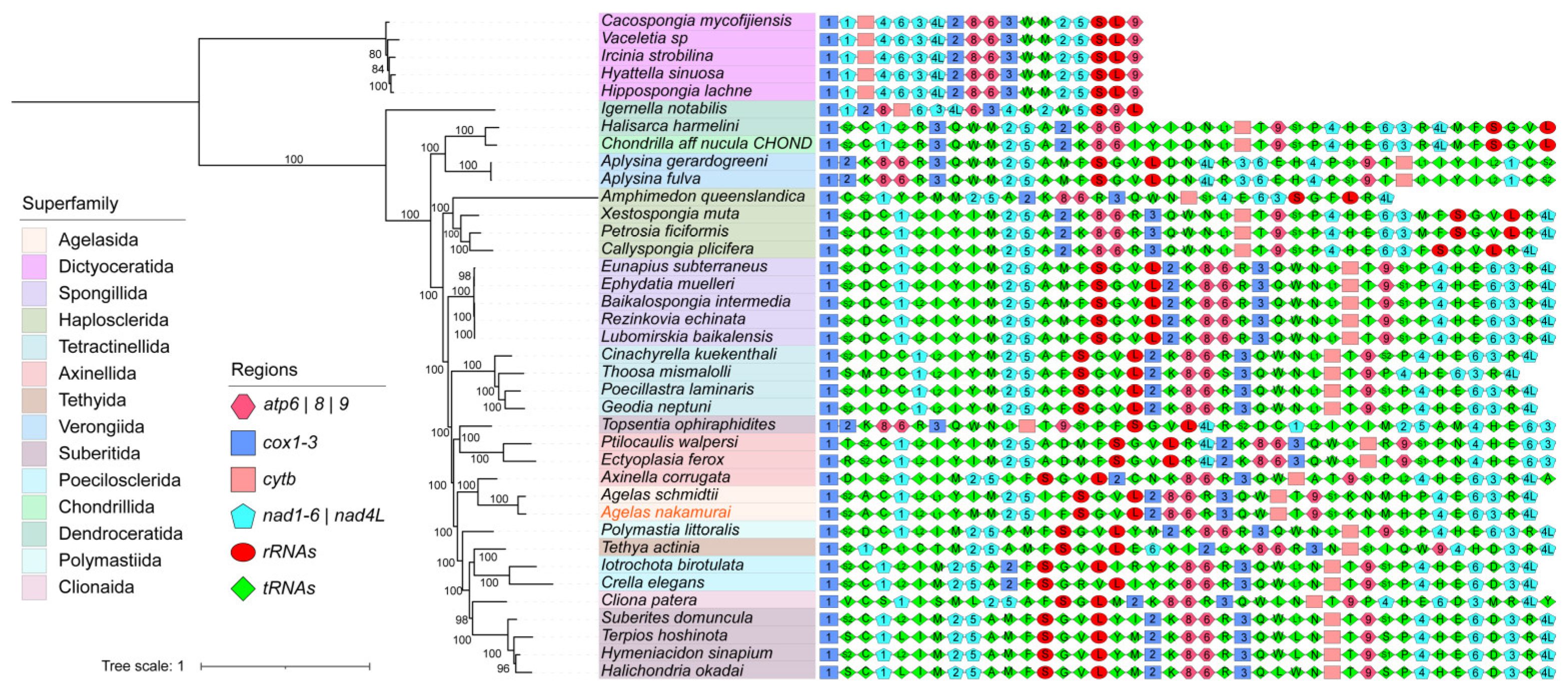
2.4. The Phylogenetic Relationship and Gene Arrangement
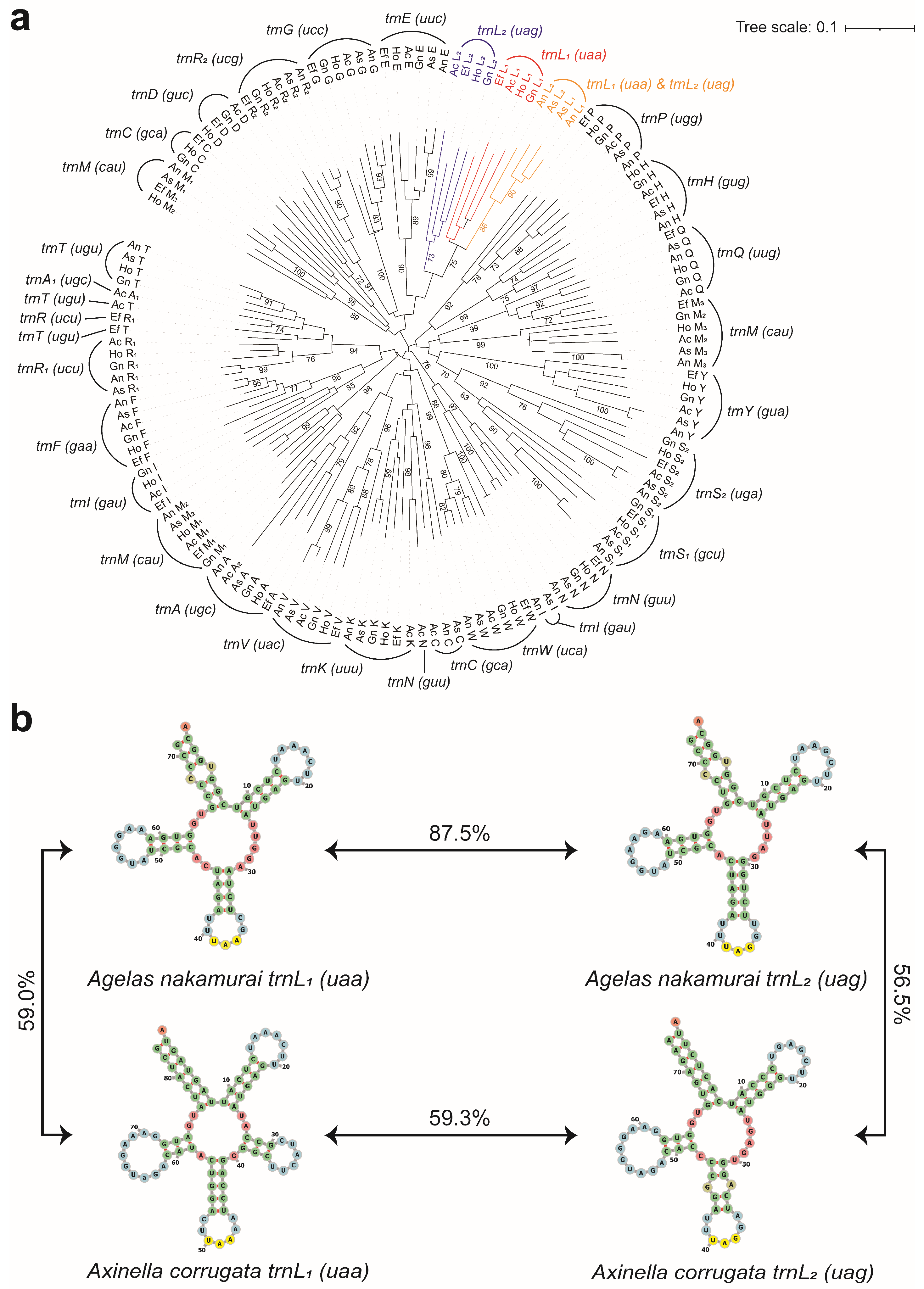
2.5. tRNA Phylogenetic Analysis and Similarity Comparison
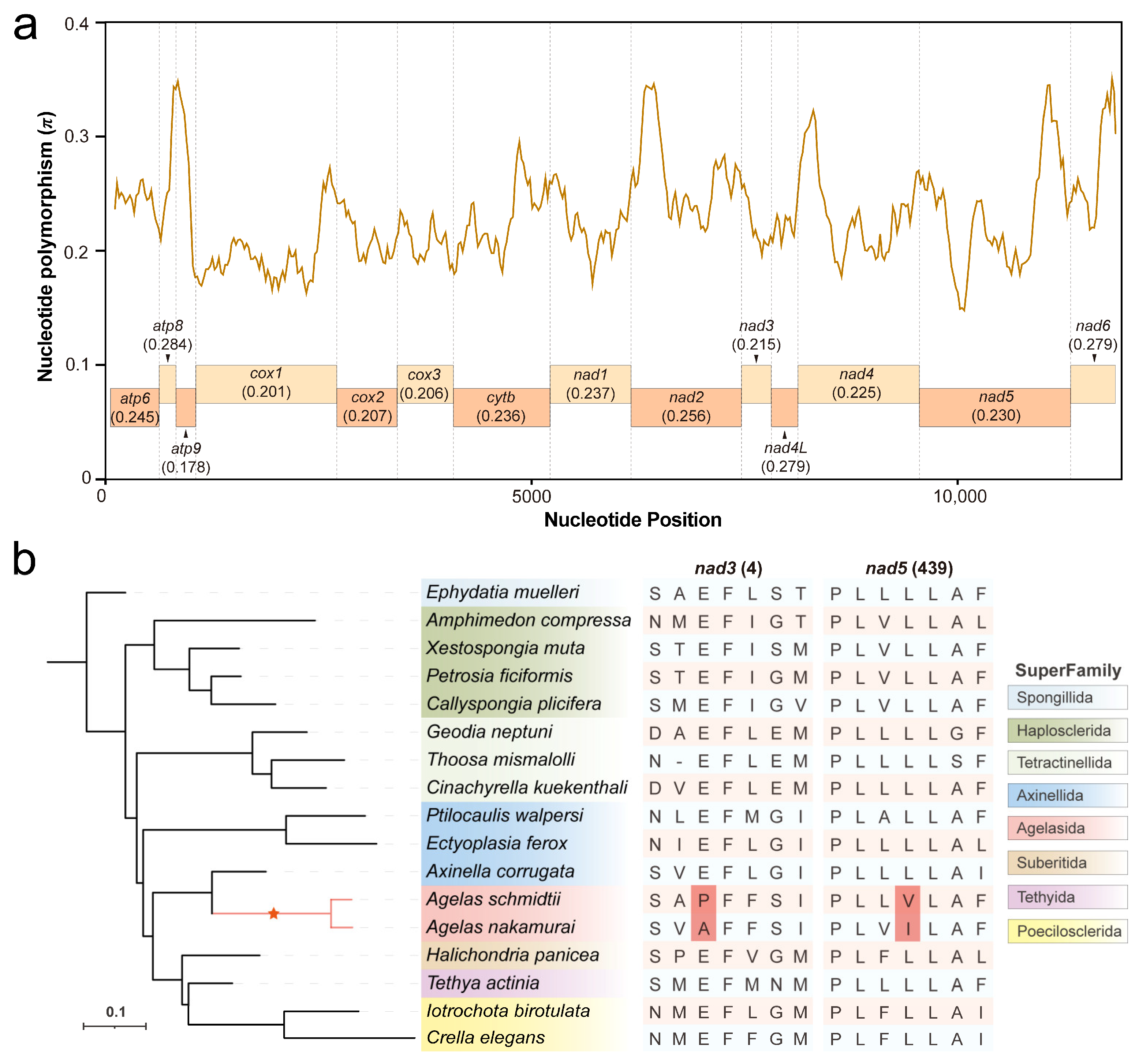
2.6. Nucleotide Diversity and Positive Selection Site Analysis
3. Discussion
4. Materials and Methods
4.1. Sample Collection and DNA Extraction
4.2. Illumina Library Preparation and Sequencing
4.3. Sequence Assembly and Annotation
4.4. Species Identification Based on the cox1 Gene
4.5. Phylogenetic and Gene Arrangement Analysis
4.6. Analysis of tRNA Duplication, and Remoulding
4.7. Nucleotide Diversity and Selection Pressure Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bandaranayake, W.M. The nature and role of pigments of marine invertebrates. Nat. Prod. Rep. 2006, 23, 223–255. [Google Scholar] [CrossRef] [PubMed]
- Parra-Velandia, F.J.; Zea, S.; Van Soest, R.W. Reef sponges of the genus Agelas (Porifera: Demospongiae) from the Greater Caribbean. Zootaxa 2014, 3794, 301–343. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bo, M.; Bertolino, M.; Bavestrello, G.; Canese, S.; Giusti, M.; Angiolillo, M.; Pansini, M.; Taviani, M. Role of deep sponge grounds in the Mediterranean Sea: A case study in southern Italy. Hydrobiologia 2012, 687, 163–177. [Google Scholar] [CrossRef]
- Kenchington, E.; Power, D.; Koen-Alonso, M. Associations of demersal fish with sponge grounds on the continental slopes of the northwest Atlantic. Mar. Ecol. Prog. Ser. 2013, 477, 217–230. [Google Scholar] [CrossRef]
- Gerovasileiou, V.; Chintiroglou, C.C.; Konstantinou, D.; Voultsiadou, E. Sponges as “living hotels” in Mediterranean marine caves. Sci. Mar. 2016, 80, 279–289. [Google Scholar] [CrossRef]
- Bickmeyer, U.; Drechsler, C.; Kock, M.; Assmann, M. Brominated pyrrole alkaloids from marine Agelas sponges reduce depolarization-induced cellular calcium elevation. Toxicon 2004, 44, 45–51. [Google Scholar] [CrossRef]
- Gazave, E.; Lapebie, P.; Renard, E.; Vacelet, J.; Rocher, C.; Ereskovsky, A.V.; Lavrov, D.V.; Borchiellini, C. Molecular Phylogeny Restores the Supra-Generic Subdivision of Homoscleromorph Sponges (Porifera, Homoscleromorpha). PLoS ONE 2010, 5, 15. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Maikova, O.O.; Pett, W.; Belikov, S.I. Small inverted repeats drive mitochondrial genome evolution in Lake Baikal sponges. Gene 2012, 505, 91–99. [Google Scholar] [CrossRef]
- Plese, B.; Lukic-Bilela, L.; Bruvo-Madaric, B.; Harcet, M.; Imesek, M.; Bilandzija, H.; Cetkovic, H. The mitochondrial genome of stygobitic sponge Eunapius subterraneus: mtDNA is highly conserved in freshwater sponges. Hydrobiologia 2012, 687, 49–59. [Google Scholar] [CrossRef]
- Nielsen, J.L.; Graziano, S.L.; Seitz, A.C. Fine-scale population genetic structure in Alaskan Pacific halibut (Hippoglossus stenolepis). Conserv. Genet. 2010, 11, 999–1012. [Google Scholar] [CrossRef]
- Rokas, A.; Holland, P.W. Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 2000, 15, 454–459. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V. Key transitions in animal evolution: A mitochondrial DNA perspective. Integr. Comp. Biol. 2007, 47, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lavrov, D.V. Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the Demospongiae. PLoS ONE 2008, 3, e2723. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Pett, W.; Voigt, O.; Wörheide, G.; Forget, L.; Lang, B.F.; Kayal, E. Mitochondrial DNA of Clathrina clathrus (Calcarea, Calcinea): Six linear chromosomes, fragmented rRNAs, tRNA editing, and a novel genetic code. Mol. Biol. Evol. 2013, 30, 865–880. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Pett, W. Animal Mitochondrial DNA as We Do Not Know It: Mt-Genome Organization and Evolution in Nonbilaterian Lineages. Genome Biol. Evol. 2016, 8, 2896–2913. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Adamski, M.; Chevaldonne, P.; Adamska, M. Extensive Mitochondrial mRNA Editing and Unusual Mitochondrial Genome Organization in Calcaronean Sponges. Curr. Biol. 2016, 26, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Formaggioni, A.; Luchetti, A.; Plazzi, F. Mitochondrial genomic landscape: A portrait of the mitochondrial genome 40 years after the first complete sequence. Life 2021, 11, 663. [Google Scholar] [CrossRef]
- Lavrov, D.V. Rapid proliferation of repetitive palindromic elements in mtDNA of the endemic Baikalian sponge Lubomirskia baicalensis. Mol. Biol. Evol. 2010, 27, 757–760. [Google Scholar] [CrossRef]
- Schuster, A.; Vargas, S.; Knapp, I.S.; Pomponi, S.A.; Toonen, R.J.; Erpenbeck, D.; Wörheide, G. Divergence times in demosponges (Porifera): First insights from new mitogenomes and the inclusion of fossils in a birth-death clock model. BMC Evol. Biol. 2018, 18, 114. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Wang, X.; Kelly, M. Reconstructing ordinal relationships in the Demospongiae using mitochondrial genomic data. Mol. Phylogenet. Evol. 2008, 49, 111–124. [Google Scholar] [CrossRef]
- Borchiellini, C.; Chombard, C.; Manuel, M.; Alivon, E.; Vacelet, J.; Boury-Esnault, N. Molecular phylogeny of Demospongiae: Implications for classification and scenarios of character evolution. Mol. Phylogenet. Evol. 2004, 32, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Pett, W.; Ryan, J.F.; Pang, K.; Mullikin, J.C.; Martindale, M.Q.; Baxevanis, A.D.; Lavrov, D.V. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: Insight from mtDNA and the nuclear genome. Mitochondrial DNA 2011, 22, 130–142. [Google Scholar] [CrossRef] [PubMed]
- Haen, K.M.; Pett, W.; Lavrov, D.V. Parallel Loss of Nuclear-Encoded Mitochondrial Aminoacyl-tRNA Synthetases and mtDNA-Encoded tRNAs in Cnidaria. Mol. Biol. Evol. 2010, 27, 2216–2219. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, T.A.; Collins, T.M.; Bieler, R. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proc. Natl. Acad. Sci. USA 2003, 100, 15700–15705. [Google Scholar] [CrossRef] [PubMed]
- Romanova, E.V.; Bukin, Y.S.; Mikhailov, K.V.; Logacheva, M.D.; Aleoshin, V.V.; Sherbakov, D.Y. Hidden cases of tRNA gene duplication and remolding in mitochondrial genomes of amphipods. Mol. Phylogenet. Evol. 2020, 144, 106710. [Google Scholar] [CrossRef] [PubMed]
- Da Fonseca, R.R.; Johnson, W.E.; O’brien, S.J.; Ramos, M.J.; Antunes, A. The adaptive evolution of the mammalian mitochondrial genome. BMC Genom. 2008, 9, 119. [Google Scholar] [CrossRef]
- Brandt, U. Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 2006, 75, 69–92. [Google Scholar] [CrossRef]
- Nakamaru-Ogiso, E.; Han, H.; Matsuno-Yagi, A.; Keinan, E.; Sinha, S.C.; Yagi, T.; Ohnishi, T. The ND2 subunit is labeled by a photoaffinity analogue of asimicin, a potent complex I inhibitor. FEBS Lett. 2010, 584, 883–888. [Google Scholar] [CrossRef]
- Sharma, L.K.; Lu, J.; Bai, Y. Mitochondrial respiratory complex I: Structure, function and implication in human diseases. Curr. Med. Chem. 2009, 16, 1266–1277. [Google Scholar] [CrossRef]
- Gish, W.; States, D.J. Identification of protein coding regions by database similarity search. Nat. Genet. 1993, 3, 266–272. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.; Boguski, M.; Lipman, D.; Ostell, J. The National Center for Biotechnology Information. Genomics 1990, 6, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D. bold: The Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Greiner, S.; Lehwark, P.; Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019, 47, W59–W64. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Kerpedjiev, P.; Hammer, S.; Hofacker, I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 2015, 31, 3377–3379. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-Delbarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, Z.; Lin, Q.; Zhang, H. Characterization of the Complete Mitochondrial Genome of Agelas nakamurai from the South China Sea. Int. J. Mol. Sci. 2024, 25, 357. https://doi.org/10.3390/ijms25010357
Lu Z, Lin Q, Zhang H. Characterization of the Complete Mitochondrial Genome of Agelas nakamurai from the South China Sea. International Journal of Molecular Sciences. 2024; 25(1):357. https://doi.org/10.3390/ijms25010357
Chicago/Turabian StyleLu, Zijian, Qiang Lin, and Huixian Zhang. 2024. "Characterization of the Complete Mitochondrial Genome of Agelas nakamurai from the South China Sea" International Journal of Molecular Sciences 25, no. 1: 357. https://doi.org/10.3390/ijms25010357
APA StyleLu, Z., Lin, Q., & Zhang, H. (2024). Characterization of the Complete Mitochondrial Genome of Agelas nakamurai from the South China Sea. International Journal of Molecular Sciences, 25(1), 357. https://doi.org/10.3390/ijms25010357





