Systematic Multiomic Analysis of PKHD1L1 Gene Expression and Its Role as a Predicting Biomarker for Immune Cell Infiltration in Skin Cutaneous Melanoma and Lung Adenocarcinoma
Abstract
1. Introduction
2. Results
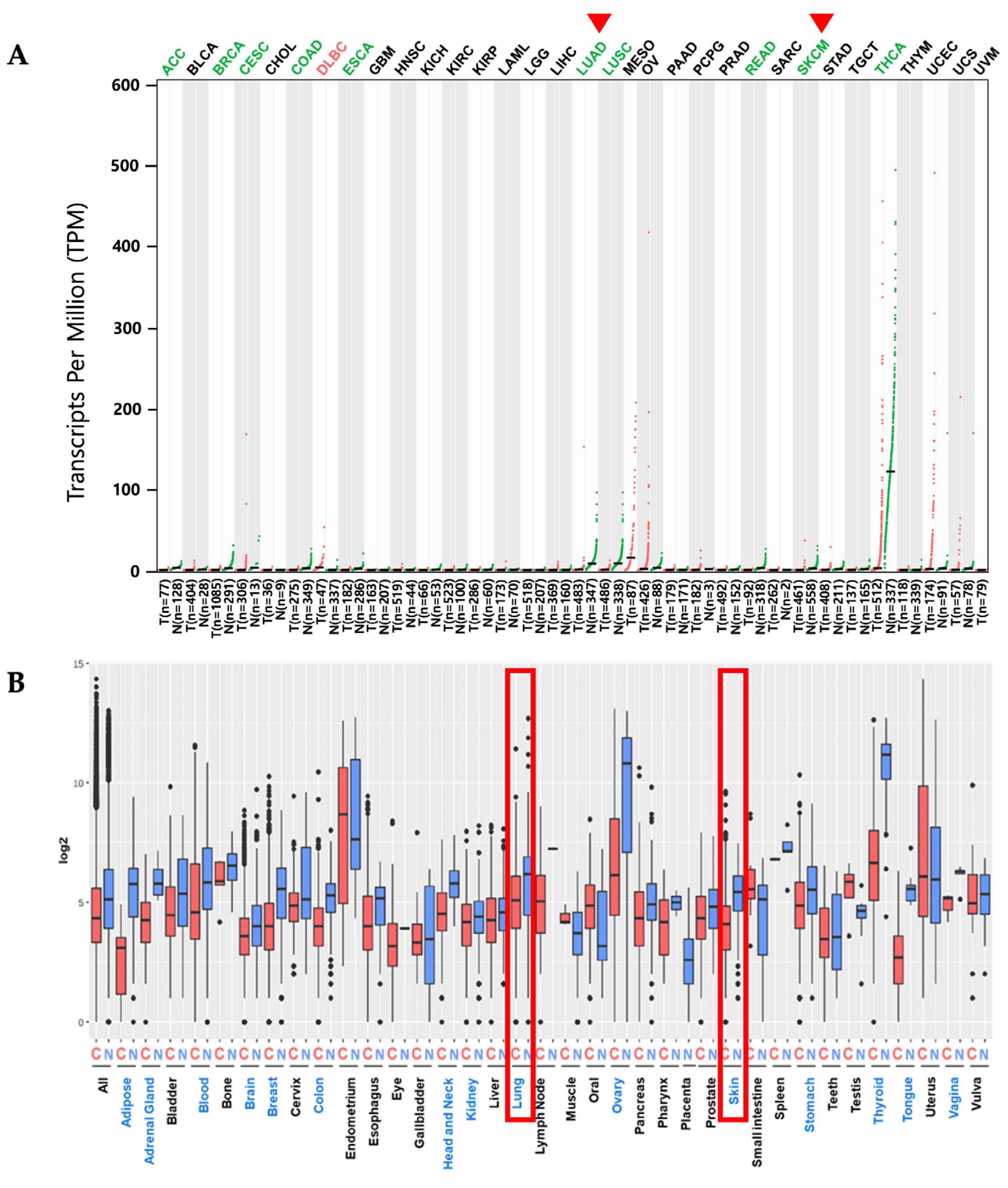
2.1. PKHD1L1 mRNA Expression Levels in Various Cancer Types
2.2. Prognostic Values of PKHD1L1 mRNA Expression Levels in Various Types of Cancers
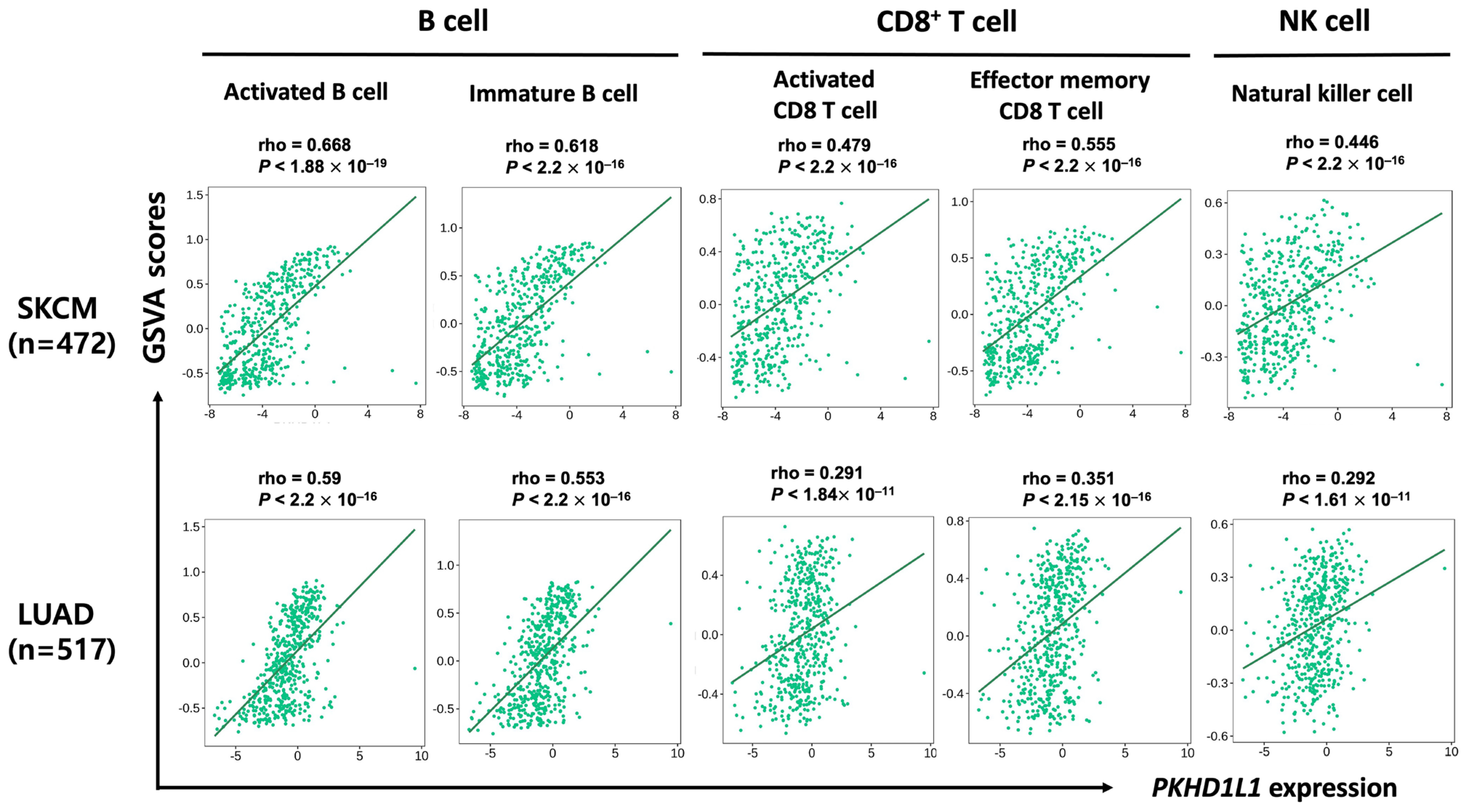
2.3. Correlation of PKHD1L1 Expression and Immune Cell Infiltration
2.4. Expression of PKHD1L1 in Various Types of Immune Cells
2.5. Analysis of Genes Co-Expressed with PKHD1L1
2.6. Prediction of Role of PKHD1L1 Using GeneMANIA and Ontology and Its Co-Expressed Genes
3. Discussion
4. Materials and Methods
4.1. Analysis of PKHD1L1 mRNA Expression Levels in Various Types of Cancers
4.2. Analysis of Relationship between PKHD1L1 Expression Levels and Patient Survival Rates
4.3. Analysis of PKHD1L1 Expression Levels in Various Types of Immune Cells
4.4. Analysis of Correlation of PKHD1L1 Expression Levels with Immune Cell Infiltration
4.5. Identification of Co-Expressed Genes with PKHD1L1
4.6. Cluster Analysis of PKHD1L1 and Its Co-Expressed Genes
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef] [PubMed]
- Thandra, K.C.; Barsouk, A.; Saginala, K.; Aluru, J.S.; Barsouk, A. Epidemiology of lung cancer. Contemp. Oncol. (Pozn) 2021, 25, 45–52. [Google Scholar] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Barsouk, A. Epidemiology of Melanoma. Med. Sci. 2021, 9, 63. [Google Scholar] [CrossRef]
- Kleczko, E.K.; Kwak, J.W.; Schenk, E.L.; Nemenoff, R.A. Targeting the Complement Pathway as a Therapeutic Strategy in Lung Cancer. Front. Immunol. 2019, 10, 954. [Google Scholar] [CrossRef]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghi, A.; Spandidos, D.A.; McCubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef]
- Bader, J.E.; Voss, K.; Rathmell, J.C. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol. Cell 2020, 78, 1019–1033. [Google Scholar] [CrossRef]
- Katsuta, E.; Rashid, O.M.; Takabe, K. Clinical relevance of tumor microenvironment: Immune cells, vessels, and mouse models. Hum. Cell 2020, 33, 930–937. [Google Scholar] [CrossRef]
- Giraldo, N.A.; Sanchez-Salas, R.; Peske, J.D.; Vano, Y.; Becht, E.; Petitprez, F.; Validire, P.; Ingels, A.; Cathelineau, X.; Fridman, W.H.; et al. The clinical role of the TME in solid cancer. Br. J. Cancer 2019, 120, 45–53. [Google Scholar] [CrossRef]
- Brummel, K.; Eerkens, A.L.; de Bruyn, M.; Nijman, H.W. Tumour-infiltrating lymphocytes: From prognosis to treatment selection. Br. J. Cancer 2023, 128, 451–458. [Google Scholar] [CrossRef]
- Lee, K.H.; Kim, E.Y.; Yun, J.S.; Park, Y.L.; Do, S.I.; Chae, S.W.; Park, C.H. The prognostic and predictive value of tumor-infiltrating lymphocytes and hematologic parameters in patients with breast cancer. BMC Cancer 2018, 18, 938. [Google Scholar] [CrossRef] [PubMed]
- Maibach, F.; Sadozai, H.; Seyed Jafari, S.M.; Hunger, R.E.; Schenk, M. Tumor-Infiltrating Lymphocytes and Their Prognostic Value in Cutaneous Melanoma. Front. Immunol. 2020, 11, 2105. [Google Scholar] [CrossRef] [PubMed]
- Letca, A.F.; Ungureanu, L.; Senila, S.C.; Grigore, L.E.; Pop, S.; Fechete, O.; Vesa, S.C.; Cosgarea, R. Regression and Sentinel Lymph Node Status in Melanoma Progression. Med. Sci. Monit. 2018, 24, 1359–1365. [Google Scholar] [CrossRef] [PubMed]
- de Moll, E.H.; Fu, Y.; Qian, Y.; Perkins, S.H.; Wieder, S.; Gnjatic, S.; Remark, R.; Bernardo, S.G.; Moskalenko, M.; Yao, J.; et al. Immune biomarkers are more accurate in prediction of survival in ulcerated than in non-ulcerated primary melanomas. Cancer Immunol. Immunother. 2015, 64, 1193–1203. [Google Scholar] [CrossRef]
- Uryvaev, A.; Passhak, M.; Hershkovits, D.; Sabo, E.; Bar-Sela, G. The role of tumor-infiltrating lymphocytes (TILs) as a predictive biomarker of response to anti-PD1 therapy in patients with metastatic non-small cell lung cancer or metastatic melanoma. Med. Oncol. 2018, 35, 25. [Google Scholar] [CrossRef]
- Eberst, G.; Vernerey, D.; Laheurte, C.; Meurisse, A.; Kaulek, V.; Cuche, L.; Jacoulet, P.; Almotlak, H.; Lahourcade, J.; Gainet-Brun, M.; et al. Prognostic value of CD4+ T lymphopenia in non-small cell lung Cancer. BMC Cancer 2022, 22, 529. [Google Scholar] [CrossRef]
- Giatromanolaki, A.; Anestopoulos, I.; Panayiotidis, M.I.; Mitrakas, A.; Pappa, A.; Koukourakis, M.I. Prognostic Relevance of the Relative Presence of CD4, CD8 and CD20 Expressing Tumor Infiltrating Lymphocytes in Operable Non-small Cell Lung Cancer Patients. Anticancer Res. 2021, 41, 3989–3995. [Google Scholar] [CrossRef]
- Wang, S.S.; Liu, W.; Ly, D.; Xu, H.; Qu, L.; Zhang, L. Tumor-infiltrating B cells: Their role and application in anti-tumor immunity in lung cancer. Cell. Mol. Immunol. 2019, 16, 6–18. [Google Scholar] [CrossRef]
- Laumont, C.M.; Banville, A.C.; Gilardi, M.; Hollern, D.P.; Nelson, B.H. Tumour-infiltrating B cells: Immunological mechanisms, clinical impact and therapeutic opportunities. Nat. Rev. Cancer 2022, 22, 414–430. [Google Scholar] [CrossRef]
- Leong, T.L.; Bryant, V.L. B cells in lung cancer-not just a bystander cell: A literature review. Transl. Lung Cancer Res. 2021, 10, 2830–2841. [Google Scholar] [CrossRef]
- Ma, C.; Kang, W.; Yu, L.; Yang, Z.; Ding, T. AUNIP Expression Is Correlated with Immune Infiltration and Is a Candidate Diagnostic and Prognostic Biomarker for Hepatocellular Carcinoma and Lung Adenocarcinoma. Front. Oncol. 2020, 10, 590006. [Google Scholar] [CrossRef] [PubMed]
- Ventola, C.L. Cancer Immunotherapy, Part 3: Challenges and Future Trends. Pharm. Ther. 2017, 42, 514–521. [Google Scholar]
- Lauriola, A.; Davalli, P.; Marverti, G.; Santi, S.; Caporali, A.; D’Arca, D. Targeting the Interplay of Independent Cellular Pathways and Immunity: A Challenge in Cancer Immunotherapy. Cancers 2023, 15, 3009. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Heo, Y.J.; Han, D.K. New opportunities for nanoparticles in cancer immunotherapy. Biomater. Res. 2018, 22, 24. [Google Scholar] [CrossRef] [PubMed]
- UniprotKB_PKHD1L1. Available online: https://www.uniprot.org/uniprot/Q86WI1 (accessed on 15 January 2023).
- Wu, X.; Ivanchenko, M.V.; Al Jandal, H.; Cicconet, M.; Indzhykulian, A.A.; Corey, D.P. PKHD1L1 is a coat protein of hair-cell stereocilia and is required for normal hearing. Nat. Commun. 2019, 10, 3801. [Google Scholar] [CrossRef] [PubMed]
- Bork, P.; Doerks, T.; Springer, T.A.; Snel, B. Domains in plexins: Links to integrins and transcription factors. Trends Biochem. Sci. 1999, 24, 261–263. [Google Scholar] [CrossRef] [PubMed]
- He, Q.Y.; Liu, X.H.; Li, Q.; Studholme, D.J.; Li, X.W.; Liang, S.P. G8: A novel domain associated with polycystic kidney disease and non-syndromic hearing loss. Bioinformatics 2006, 22, 2189–2191. [Google Scholar] [CrossRef]
- Jenkins, J.; Mayans, O.; Pickersgill, R. Structure and evolution of parallel beta-helix proteins. J. Struct. Biol. 1998, 122, 236–246. [Google Scholar] [CrossRef]
- Saravia, C.H.; Flores, C.; Schwarz, L.J.; Bravo, L.; Zavaleta, J.; Araujo, J.; Neciosup, S.; Pinto, J.A. Patterns of Mutation Enrichment in Metastatic Triple-Negative Breast Cancer. Clin. Med. Insights Oncol. 2019, 13, 1179554919868482. [Google Scholar] [CrossRef]
- Zheng, C.; Quan, R.; Xia, E.J.; Bhandari, A.; Zhang, X. Original tumour suppressor gene polycystic kidney and hepatic disease 1-like 1 is associated with thyroid cancer cell progression. Oncol. Lett. 2019, 18, 3227–3235. [Google Scholar] [CrossRef]
- Hogan, M.C.; Griffin, M.D.; Rossetti, S.; Torres, V.E.; Ward, C.J.; Harris, P.C. PKHDL1, a homolog of the autosomal recessive polycystic kidney disease gene, encodes a receptor with inducible T lymphocyte expression. Hum. Mol. Genet. 2003, 12, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Sadelain, M.; Riviere, I.; Riddell, S. Therapeutic T cell engineering. Nature 2017, 545, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [PubMed]
- Sade-Feldman, M.; Yizhak, K.; Bjorgaard, S.L.; Ray, J.P.; de Boer, C.G.; Jenkins, R.W.; Lieb, D.J.; Chen, J.H.; Frederick, D.T.; Barzily-Rokni, M.; et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2018, 175, 998–1013.e20. [Google Scholar] [CrossRef] [PubMed]
- Willsmore, Z.N.; Coumbe, B.G.T.; Crescioli, S.; Reci, S.; Gupta, A.; Harris, R.J.; Chenoweth, A.; Chauhan, J.; Bax, H.J.; McCraw, A.; et al. Combined anti-PD-1 and anti-CTLA-4 checkpoint blockade: Treatment of melanoma and immune mechanisms of action. Eur. J. Immunol. 2021, 51, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Willsmore, Z.N.; Harris, R.J.; Crescioli, S.; Hussein, K.; Kakkassery, H.; Thapa, D.; Cheung, A.; Chauhan, J.; Bax, H.J.; Chenoweth, A.; et al. B Cells in Patients with Melanoma: Implications for Treatment with Checkpoint Inhibitor Antibodies. Front. Immunol. 2020, 11, 622442. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Zakka, L.R.; Mihm, M.C., Jr.; Schatton, T. Tumour-infiltrating lymphocytes in melanoma prognosis and cancer immunotherapy. Pathology 2016, 48, 177–187. [Google Scholar] [CrossRef]
- Salas-Benito, D.; Vercher, E.; Conde, E.; Glez-Vaz, J.; Tamayo, I.; Hervas-Stubbs, S. Inflammation and immunity in ovarian cancer. EJC Suppl. 2020, 15, 56–66. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, M.; Ng, D.M.; Haleem, M.; Yi, T.; Hu, S.; Zhu, H.; Zhao, G.; Liao, Q. Multi-omics Data Analyses Construct TME and Identify the Immune-Related Prognosis Signatures in Human LUAD. Mol. Ther. Nucleic Acids 2020, 21, 860–873. [Google Scholar] [CrossRef]
- Jin, S.; Deng, Y.; Hao, J.W.; Li, Y.; Liu, B.; Yu, Y.; Shi, F.D.; Zhou, Q.H. NK cell phenotypic modulation in lung cancer environment. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.; Kim, K.E. Interleukin-18 Is a Prognostic Biomarker Correlated with CD8(+) T Cell and Natural Killer Cell Infiltration in Skin Cutaneous Melanoma. J. Clin. Med. 2019, 8, 1993. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Komura, D.; Konishi, H.; Suzuki, R.; Yamamoto, A.; Kakiuchi, M.; Sato, R.; Ushiku, T.; Yamamoto, S.; Tatsuno, K.; et al. Immunogenetic Profiling for Gastric Cancers Identifies Sulfated Glycosaminoglycans as Major and Functional B Cell Antigens in Human Malignancies. Cell Rep. 2017, 20, 1073–1087. [Google Scholar] [CrossRef] [PubMed]
- van Herpen, C.M.; van der Voort, R.; van der Laak, J.A.; Klasen, I.S.; de Graaf, A.O.; van Kempen, L.C.; de Vries, I.J.; Boer, T.D.; Dolstra, H.; Torensma, R.; et al. Intratumoral rhIL-12 administration in head and neck squamous cell carcinoma patients induces B cell activation. Int. J. Cancer 2008, 123, 2354–2361. [Google Scholar] [CrossRef]
- Liao, S.F.; Liang, C.H.; Ho, M.Y.; Hsu, T.L.; Tsai, T.I.; Hsieh, Y.S.; Tsai, C.M.; Li, S.T.; Cheng, Y.Y.; Tsao, S.M.; et al. Immunization of fucose-containing polysaccharides from Reishi mushroom induces antibodies to tumor-associated Globo H-series epitopes. Proc. Natl. Acad. Sci. USA 2013, 110, 13809–13814. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.J.; Richter, A.; Drayson, M.T.; Middleton, G.W. The role of B lymphocytes in the immuno-biology of non-small-cell lung cancer. Cancer Immunol. Immunother. CII 2020, 69, 325–342. [Google Scholar] [CrossRef]
- Tokunaga, R.; Naseem, M.; Lo, J.H.; Battaglin, F.; Soni, S.; Puccini, A.; Berger, M.D.; Zhang, W.; Baba, H.; Lenz, H.J. B cell and B cell-related pathways for novel cancer treatments. Cancer Treat. Rev. 2019, 73, 10–19. [Google Scholar] [CrossRef]
- Kinker, G.S.; Vitiello, G.A.F.; Ferreira, W.A.S.; Chaves, A.S.; Cordeiro de Lima, V.C.; Medina, T.D.S. B Cell Orchestration of Anti-tumor Immune Responses: A Matter of Cell Localization and Communication. Front. Cell Dev. Biol. 2021, 9, 678127. [Google Scholar] [CrossRef]
- Tao, H.; Lu, L.; Xia, Y.; Dai, F.; Wang, Y.; Bao, Y.; Lundy, S.K.; Ito, F.; Pan, Q.; Zhang, X.; et al. Antitumor effector B cells directly kill tumor cells via the Fas/FasL pathway and are regulated by IL-10. Eur. J. Immunol. 2015, 45, 999–1009. [Google Scholar] [CrossRef]
- DiLillo, D.J.; Yanaba, K.; Tedder, T.F. B cells are required for optimal CD4+ and CD8+ T cell tumor immunity: Therapeutic B cell depletion enhances B16 melanoma growth in mice. J. Immunol. 2010, 184, 4006–4016. [Google Scholar] [CrossRef]
- Garaud, S.; Buisseret, L.; Solinas, C.; Gu-Trantien, C.; de Wind, A.; Van den Eynden, G.; Naveaux, C.; Lodewyckx, J.N.; Boisson, A.; Duvillier, H.; et al. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight 2019, 5, e129641. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Wang, J.; Fagerberg, E.; Chen, P.M.; Connolly, K.A.; Damo, M.; Cheung, J.F.; Mao, T.; Askari, A.S.; Chen, S.; et al. Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell 2021, 184, 6101–6118. [Google Scholar] [CrossRef] [PubMed]
- Burger, J.A.; Wiestner, A. Targeting B cell receptor signalling in cancer: Preclinical and clinical advances. Nat. Rev. Cancer 2018, 18, 148–167. [Google Scholar] [CrossRef] [PubMed]
- Selitsky, S.R.; Mose, L.E.; Smith, C.C.; Chai, S.; Hoadley, K.A.; Dittmer, D.P.; Moschos, S.J.; Parker, J.S.; Vincent, B.G. Prognostic value of B cells in cutaneous melanoma. Genome Med. 2019, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.N.; Tong, W.L.; Callahan, B.M.; Chobrutskiy, B.I.; Blanck, G. B-cell Receptor Recombinations in Lung Adenocarcinoma Exome Files Correlate with a Higher Overall Survival Rate. Anticancer Res. 2020, 40, 2043–2051. [Google Scholar] [CrossRef] [PubMed]
- Melero, I.; Rouzaut, A.; Motz, G.T.; Coukos, G. T-cell and NK-cell infiltration into solid tumors: A key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014, 4, 522–526. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pages, F.; Sautes-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Seo, A.N.; Lee, H.J.; Kim, E.J.; Kim, H.J.; Jang, M.H.; Lee, H.E.; Kim, Y.J.; Kim, J.H.; Park, S.Y. Tumour-infiltrating CD8+ lymphocytes as an independent predictive factor for pathological complete response to primary systemic therapy in breast cancer. Br. J. Cancer 2013, 109, 2705–2713. [Google Scholar] [CrossRef]
- Skuciova, V.; Drahosova, S.; Vybohova, D.; Cigerova, V.; Adamkov, M. The relationships between PD-L1 expression, CD8+ TILs and clinico-histomorphological parameters in malignant melanomas. Pathol. Res. Pract. 2020, 216, 153071. [Google Scholar] [CrossRef]
- Paul, S.; Lal, G. The Molecular Mechanism of Natural Killer Cells Function and Its Importance in Cancer Immunotherapy. Front. Immunol. 2017, 8, 1124. [Google Scholar] [CrossRef]
- Jassar, A.S.; Suzuki, E.; Kapoor, V.; Sun, J.; Silverberg, M.B.; Cheung, L.; Burdick, M.D.; Strieter, R.M.; Ching, L.M.; Kaiser, L.R.; et al. Activation of tumor-associated macrophages by the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid induces an effective CD8+ T-cell-mediated antitumor immune response in murine models of lung cancer and mesothelioma. Cancer Res. 2005, 65, 11752–11761. [Google Scholar] [CrossRef] [PubMed]
- Russick, J.; Joubert, P.E.; Gillard-Bocquet, M.; Torset, C.; Meylan, M.; Petitprez, F.; Dragon-Durey, M.A.; Marmier, S.; Varthaman, A.; Josseaume, N.; et al. Natural killer cells in the human lung tumor microenvironment display immune inhibitory functions. J. Immunother. Cancer 2020, 8, e001054. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Lostao, L.; Anel, A.; Pardo, J. How Do Cytotoxic Lymphocytes Kill Cancer Cells? Clin. Cancer Res. 2015, 21, 5047–5056. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Kim, K.E. Prognostic Value of Interleukin-32 Expression and Its Correlation with the Infiltration of Natural Killer Cells in Cutaneous Melanoma. J. Clin. Med. 2021, 10, 4691. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.; Kim, K.E. Systematic Multiomic Analysis of Ly75 Gene Expression and Its Prognostic Value Through the Infiltration of Natural Killer (NK) Cells in Skin Cutaneous Melanoma. J. Clin. Med. 2020, 9, 1383. [Google Scholar] [CrossRef] [PubMed]
- Lanzavecchia, A.; Sallusto, F. Dynamics of T lymphocyte responses: Intermediates, effectors, and memory cells. Science 2000, 290, 92–97. [Google Scholar] [CrossRef] [PubMed]
- von Andrian, U.H.; Mackay, C.R. T-cell function and migration. Two sides of the same coin. N. Engl. J. Med. 2000, 343, 1020–1034. [Google Scholar] [CrossRef]
- Janeway, C.A., Jr. How the immune system works to protect the host from infection: A personal view. Proc. Natl. Acad. Sci. USA 2001, 98, 7461–7468. [Google Scholar] [CrossRef]
- Robertson, M.J. Role of chemokines in the biology of natural killer cells. J. Leukoc. Biol. 2002, 71, 173–183. [Google Scholar] [CrossRef]
- Harlin, H.; Meng, Y.; Peterson, A.C.; Zha, Y.; Tretiakova, M.; Slingluff, C.; McKee, M.; Gajewski, T.F. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009, 69, 3077–3085. [Google Scholar] [CrossRef]
- Huang, B.; Han, W.; Sheng, Z.F.; Shen, G.L. Identification of immune-related biomarkers associated with tumorigenesis and prognosis in cutaneous melanoma patients. Cancer Cell Int. 2020, 20, 195. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, L.; Cheng, S.; Fan, Z.; Shen, Z.; Xue, W.; Zheng, Y.; Li, F.; Wang, D.; Zhang, K.; et al. Lung adenocarcinoma-intrinsic GBE1 signaling inhibits anti-tumor immunity. Mol. Cancer 2019, 18, 108. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Matosevic, S. Chemokine networks modulating natural killer cell trafficking to solid tumors. Cytokine Growth Factor Rev. 2021, 59, 36–45. [Google Scholar] [CrossRef] [PubMed]
- Baekkevold, E.S.; Yamanaka, T.; Palframan, R.T.; Carlsen, H.S.; Reinholt, F.P.; von Andrian, U.H.; Brandtzaeg, P.; Haraldsen, G. The CCR7 ligand elc (CCL19) is transcytosed in high endothelial venules and mediates T cell recruitment. J. Exp. Med. 2001, 193, 1105–1112. [Google Scholar] [CrossRef] [PubMed]
- Karin, N. Chemokines and cancer: New immune checkpoints for cancer therapy. Curr. Opin. Immunol. 2018, 51, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Verma, A.K.; Dev, K.; Goyal, Y.; Bhatt, D.; Alsahli, M.A.; Rahmani, A.H.; Almatroudi, A.; Almatroodi, S.A.; Alrumaihi, F.; et al. Role of Cytokines and Chemokines in NSCLC Immune Navigation and Proliferation. Oxid. Med. Cell. Longev. 2021, 2021, 5563746. [Google Scholar] [CrossRef]
- Johnston, H.E.; Carter, M.J.; Larrayoz, M.; Clarke, J.; Garbis, S.D.; Oscier, D.; Strefford, J.C.; Steele, A.J.; Walewska, R.; Cragg, M.S. Proteomics Profiling of CLL Versus Healthy B-cells Identifies Putative Therapeutic Targets and a Subtype-independent Signature of Spliceosome Dysregulation. Mol. Cell. Proteom. 2018, 17, 776–791. [Google Scholar] [CrossRef]
- Gao, X.; Jiang, C.; Yao, S.; Ma, L.; Wang, X.; Cao, Z. Identification of hub genes related to immune cell infiltration in periodontitis using integrated bioinformatic analysis. J. Periodontal. Res. 2022, 57, 392–401. [Google Scholar] [CrossRef]
- Zhu, M.; Rhee, I.; Liu, Y.; Zhang, W. Negative regulation of Fc epsilonRI-mediated signaling and mast cell function by the adaptor protein LAX. J. Biol. Chem. 2006, 281, 18408–18413. [Google Scholar] [CrossRef]
- Alari-Pahissa, E.; Grandclement, C.; Jeevan-Raj, B.; Leclercq, G.; Veillette, A.; Held, W. Activation by SLAM Family Receptors Contributes to NK Cell Mediated “Missing-Self” Recognition. PLoS ONE 2016, 11, e0153236. [Google Scholar] [CrossRef][Green Version]
- Margraf, S.; Garner, L.I.; Wilson, T.J.; Brown, M.H. A polymorphism in a phosphotyrosine signalling motif of CD229 (Ly9, SLAMF3) alters SH2 domain binding and T-cell activation. Immunology 2015, 146, 392–400. [Google Scholar] [CrossRef]
- Chen, S.; Yang, M.; Du, J.; Li, D.; Li, Z.; Cai, C.; Ma, Y.; Zhang, L.; Tian, Z.; Dong, Z. The Self-Specific Activation Receptor SLAM Family Is Critical for NK Cell Education. Immunity 2016, 45, 292–304. [Google Scholar] [CrossRef] [PubMed]
- de Lima, D.S.; Cardozo, L.E.; Maracaja-Coutinho, V.; Suhrbier, A.; Mane, K.; Jeffries, D.; Silveira, E.L.V.; Amaral, P.P.; Rappuoli, R.; de Silva, T.I.; et al. Long noncoding RNAs are involved in multiple immunological pathways in response to vaccination. Proc. Natl. Acad. Sci. USA 2019, 116, 17121–17126. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Luo, H.; Cao, J.; Zheng, X.; Zhang, J.; Zhang, Y.; Fu, Z. Identification of a Novel Tumor Microenvironment-Associated Eight-Gene Signature for Prognosis Prediction in Lung Adenocarcinoma. Front. Mol. Biosci. 2020, 7, 571641. [Google Scholar] [CrossRef] [PubMed]
- Lenk, L.; Carlet, M.; Vogiatzi, F.; Spory, L.; Winterberg, D.; Cousins, A.; Vossen-Gajcy, M.; Ibruli, O.; Vokuhl, C.; Cario, G.; et al. CD79a promotes CNS-infiltration and leukemia engraftment in pediatric B-cell precursor acute lymphoblastic leukemia. Commun. Biol. 2021, 4, 73. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Schmiedel, B.J.; Singh, D.; Madrigal, A.; Valdovino-Gonzalez, A.G.; White, B.M.; Zapardiel-Gonzalo, J.; Ha, B.; Altay, G.; Greenbaum, J.A.; McVicker, G.; et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018, 175, 1701–1715.e16. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Franz, M.; Rodriguez, H.; Lopes, C.; Zuberi, K.; Montojo, J.; Bader, G.D.; Morris, Q. GeneMANIA update 2018. Nucleic Acids Res. 2018, 46, W60–W64. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]




| Cancer | SKCM | LUAD | ||
|---|---|---|---|---|
| Correlation | p | Correlation | p | |
| Purity | −0.40445 | 1.88 × 10−19 | −0.35726 | 2.56 × 10−16 |
| B cells | 0.370997 | 4.58 × 10−16 | 0.419973 | 4.2 × 10−22 |
| CD8+ T cells | 0.390797 | 1.97 × 10−17 | 0.335172 | 3 × 10−14 |
| CD4+ T cells | 0.354657 | 1.15 × 10−14 | 0.199543 | 9.73 × 10−6 |
| Macrophages | 0.21392 | 4.35 × 10−6 | 0.196683 | 1.28 × 10−5 |
| Neutrophils | 0.409228 | 1.12 × 10−19 | 0.252514 | 1.84 × 10−8 |
| Dendritic Cells | 0.34351 | 8.49 × 10−14 | 0.208905 | 3.25 × 10−6 |
| Immune Cells | Statistical Test Method | SKCM (n = 471) | LUAD (n = 515) | ||
|---|---|---|---|---|---|
| Rho | p | Rho | p | ||
| B cells | EPIC | 0.512 | 5.83 × 10−32 | 0.519 | 2.32 × 10−35 |
| QUANTISEQ | 0.531 | 1.30 × 10−34 | 0.497 | 4.44 × 10−32 | |
| MCP_COUNTER | 0.542 | 2.56 × 10−36 | 0.551 | 2.08 × 10−40 | |
| CIBERSORT_ABS | 0.403 | 2.53 × 10−12 | 0.403 | 1.01 × 10−20 | |
| CD8+ T cells | MCP_COUNTER | 0.433 | 7.25 × 10−72 | 0.279 | 2.73 × 10−10 |
| CIBERSORT_ABS | 0.436 | 3.49 × 10−50 | 0.365 | 3.49 × 10−50 | |
| QUANTISEQ | 0.36 | 6.91 × 10−14 | 0.287 | 6.91 × 10−14 | |
| XCELL | 0.347 | 6.91 × 10−14 | 0.259 | 6.91 × 10−14 | |
| NK cells | MCP_COUNTER | 0.433 | 2.40 × 10−22 | 0.263 | 1.47 × 10−10 |
| Activated CIBERSORT_ABS | 0.326 | 9.09 × 10−13 | 0.22 | 8.399 × 10−7 | |
| Resting CIBERSORT | −0.196 | 2.50 × 10−5 | −0.196 | 2.16 × 10−1 | |
| Resting CIBERSORT_ABS | −0.164 | 4.45 × 10−4 | −0.05 | 2.68 × 10−1 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, J.Y.; Yang, J.; Lee, H.; Park, S.; Gil, M.; Kim, K.E. Systematic Multiomic Analysis of PKHD1L1 Gene Expression and Its Role as a Predicting Biomarker for Immune Cell Infiltration in Skin Cutaneous Melanoma and Lung Adenocarcinoma. Int. J. Mol. Sci. 2024, 25, 359. https://doi.org/10.3390/ijms25010359
Kang JY, Yang J, Lee H, Park S, Gil M, Kim KE. Systematic Multiomic Analysis of PKHD1L1 Gene Expression and Its Role as a Predicting Biomarker for Immune Cell Infiltration in Skin Cutaneous Melanoma and Lung Adenocarcinoma. International Journal of Molecular Sciences. 2024; 25(1):359. https://doi.org/10.3390/ijms25010359
Chicago/Turabian StyleKang, Ji Young, Jisun Yang, Haeryung Lee, Soochul Park, Minchan Gil, and Kyung Eun Kim. 2024. "Systematic Multiomic Analysis of PKHD1L1 Gene Expression and Its Role as a Predicting Biomarker for Immune Cell Infiltration in Skin Cutaneous Melanoma and Lung Adenocarcinoma" International Journal of Molecular Sciences 25, no. 1: 359. https://doi.org/10.3390/ijms25010359
APA StyleKang, J. Y., Yang, J., Lee, H., Park, S., Gil, M., & Kim, K. E. (2024). Systematic Multiomic Analysis of PKHD1L1 Gene Expression and Its Role as a Predicting Biomarker for Immune Cell Infiltration in Skin Cutaneous Melanoma and Lung Adenocarcinoma. International Journal of Molecular Sciences, 25(1), 359. https://doi.org/10.3390/ijms25010359






