The miRNAs let-7b and miR-141 Coordinately Regulate Vitellogenesis by Modulating Methyl Farnesoate Degradation in the Swimming Crab Portunus trituberculatus
Abstract
:1. Introduction
2. Results
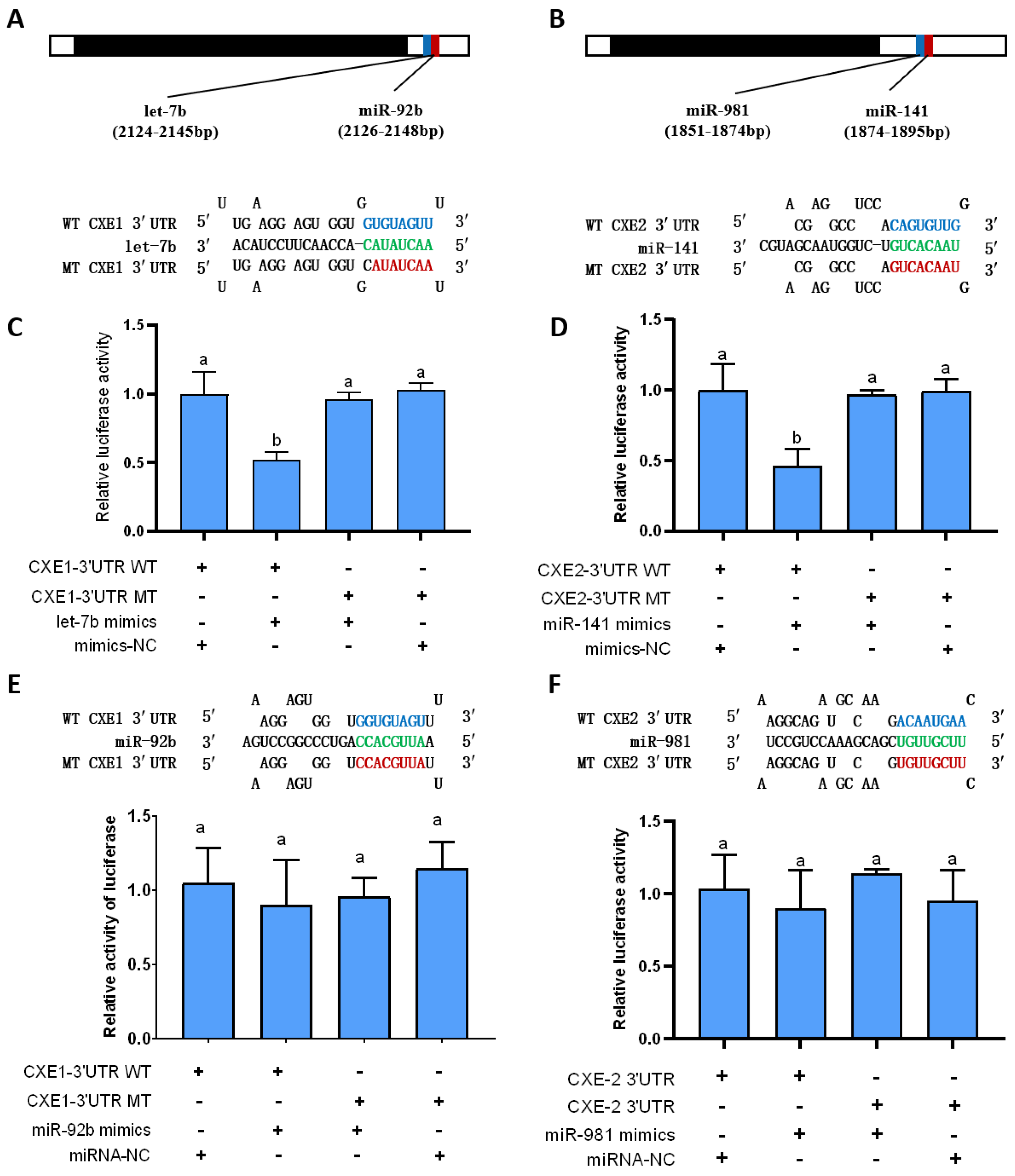
2.1. In Silico Prediction of the miRNAs Targeting CXEs
2.2. In Vitro Validation of the Interaction between the miRNAs and Their Target Genes
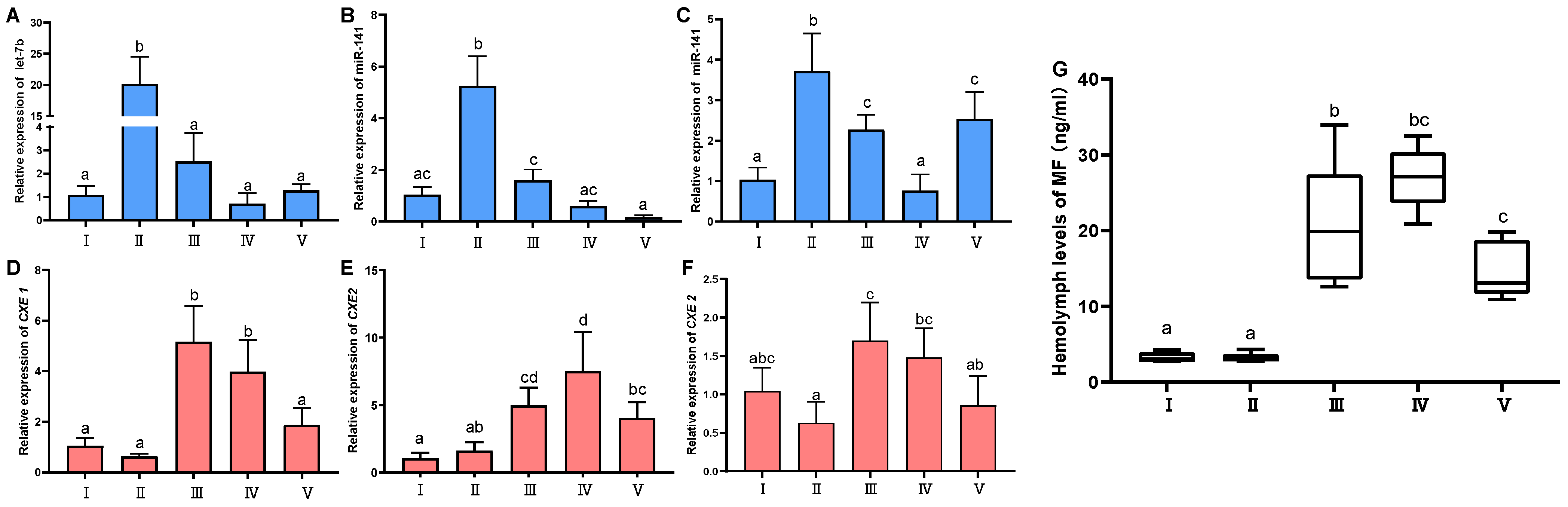
2.3. Expression Profile Analyses of the miRNAs and CXEs, and Variation of Hemolymph MF during Ovarian Development
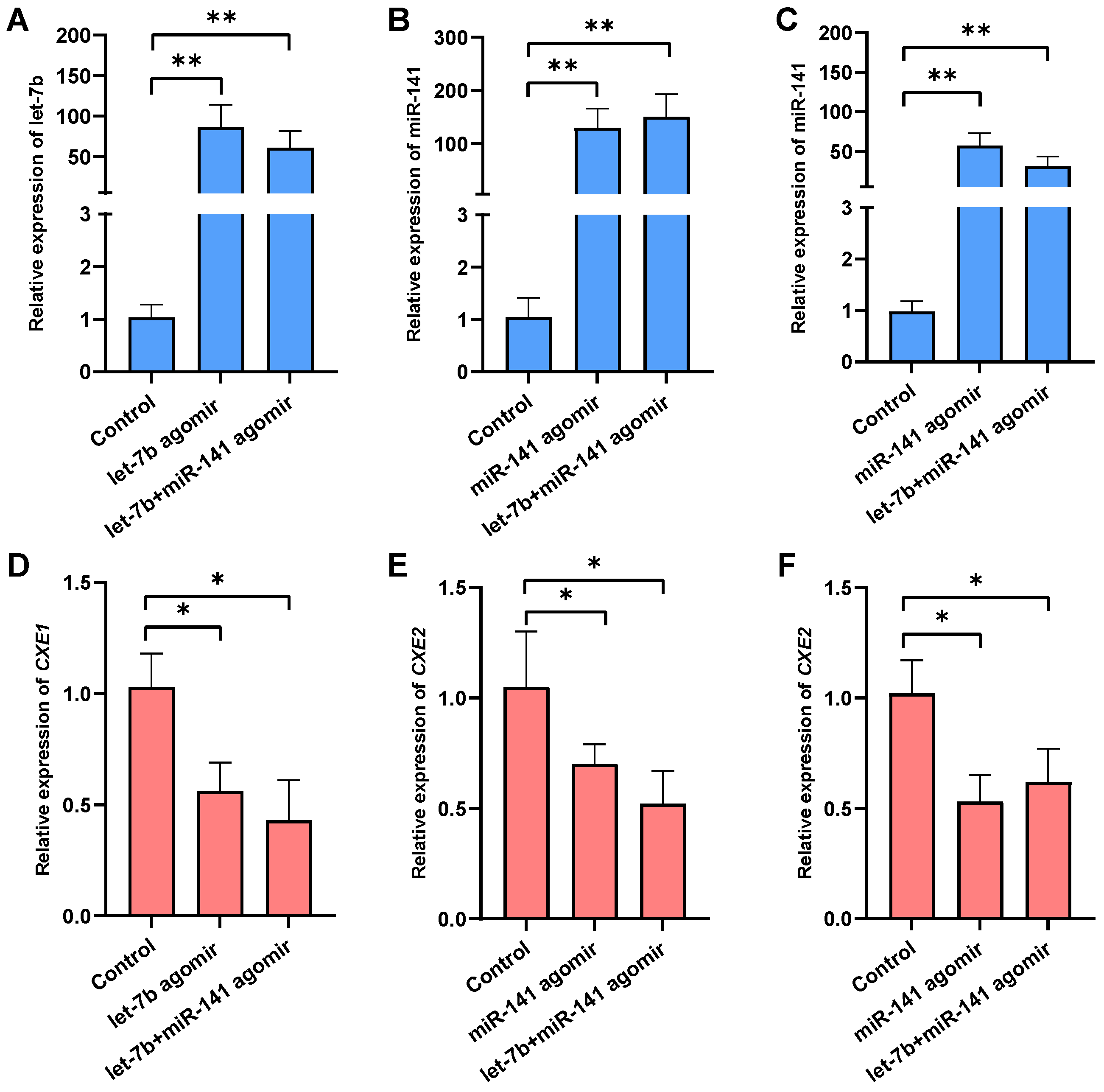
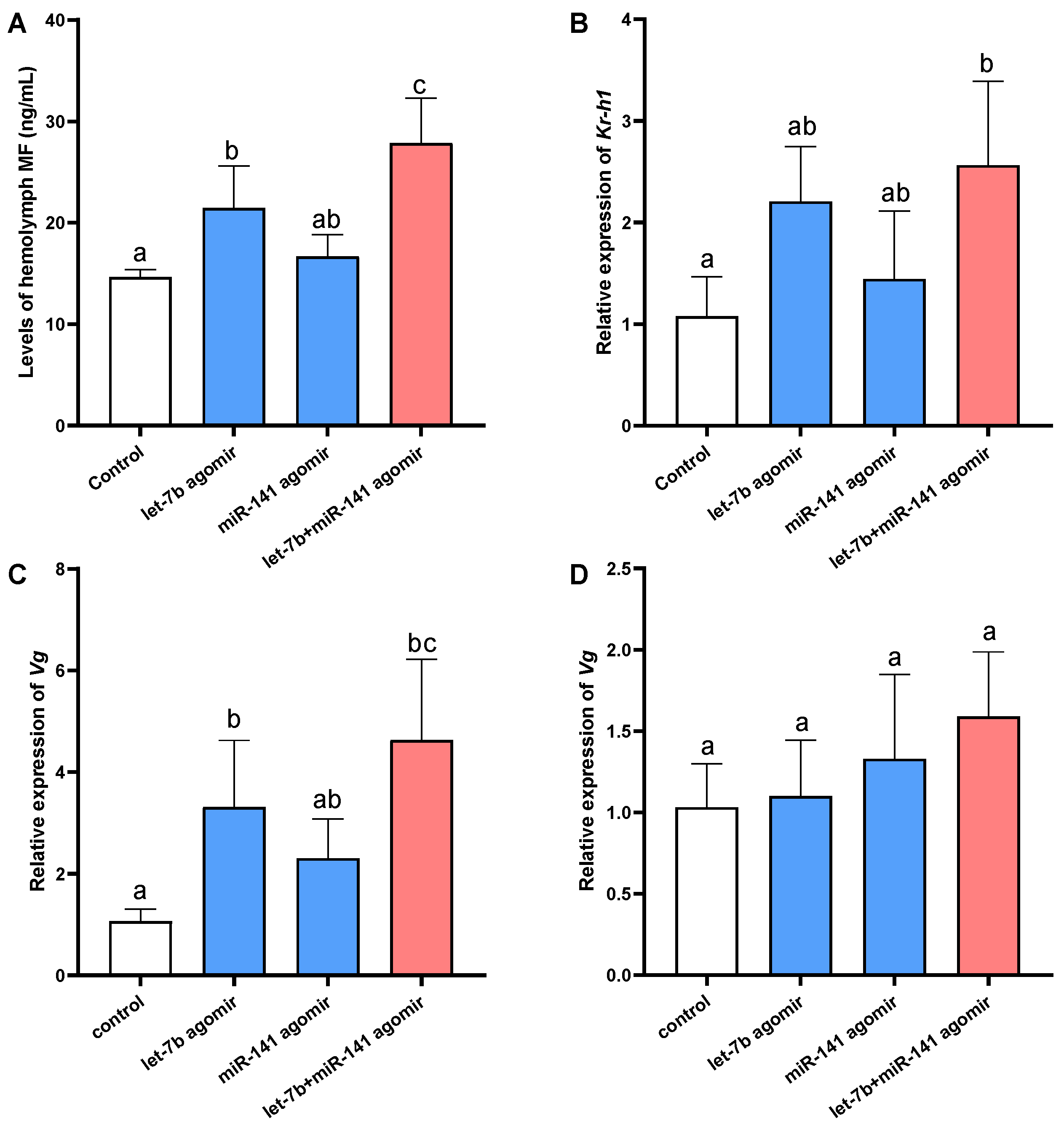
2.4. Effects of the miRNAs Overexpression on Target Gene Expression, MF Level, and Responsive Genes
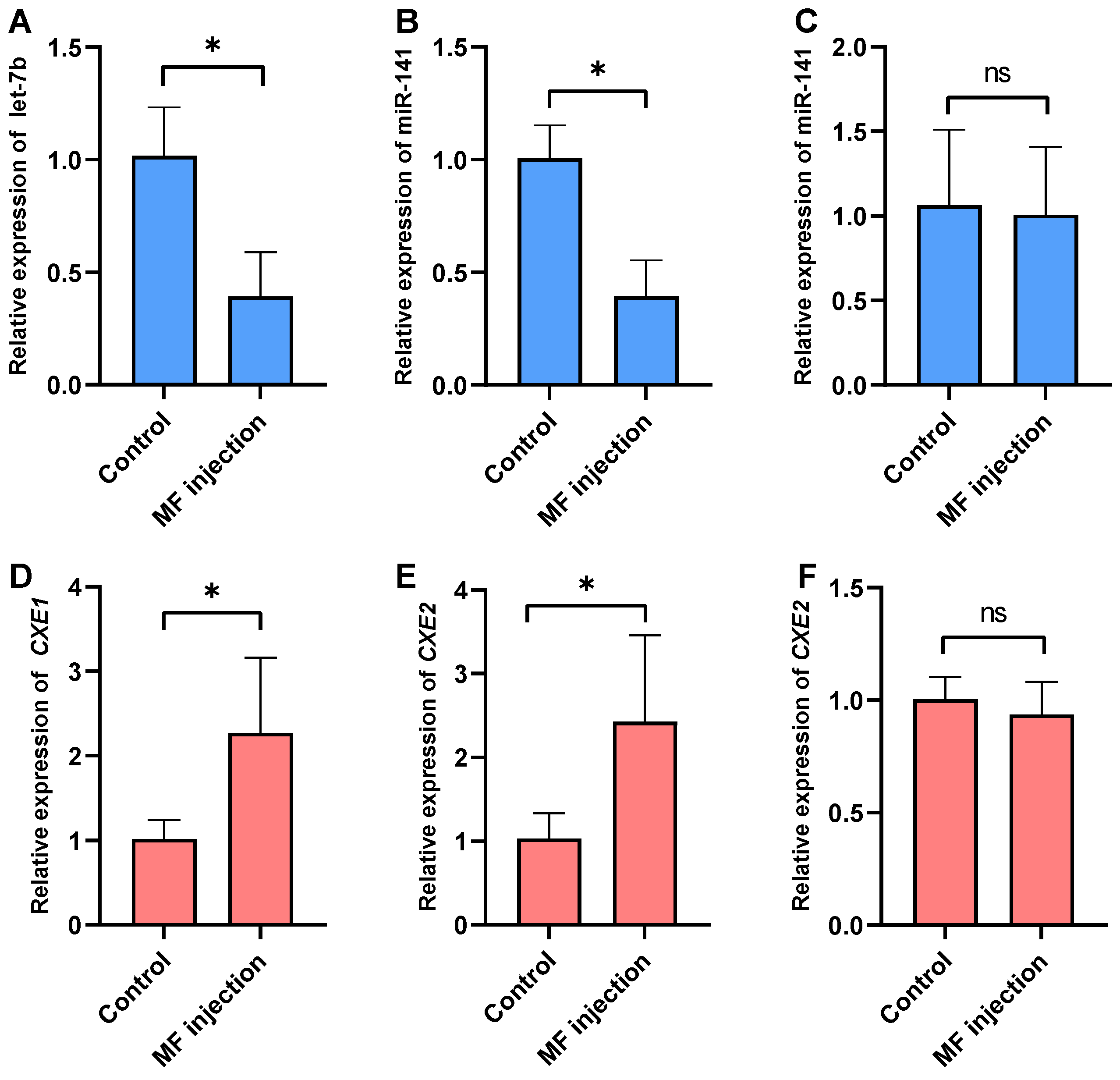
2.5. Effects of MF Injection on the Expression of the miRNAs and Their Targets
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Bioinformatic Prediction of the miRNAs Targeting CXEs
4.3. Dual Luciferase Reporter Assay
4.4. miRNA Agomir
4.5. Hemolymph MF Quantification
4.6. MF Treatment
4.7. RNA Extraction and qRT-PCR
4.8. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wahli, W. Evolution and Expression of Vitellogenin Genes. Trends Genet. 1988, 4, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Subramoniam, T. Mechanisms and Control of Vitellogenesis in Crustaceans. Fish. Sci. 2011, 77, 1–21. [Google Scholar] [CrossRef]
- Laufer, H.; Sagi, A.; Ahl, J.; Homola, E. Methyl Farnesoate Appears to Be a Crustacean Reproductive Hormone. Invertebr. Reprod. Dev. 1992, 22, 17–19. [Google Scholar] [CrossRef]
- Nagaraju, G.P.C. Is Methyl Farnesoate a Crustacean Hormone? Aquaculture 2007, 272, 39–54. [Google Scholar] [CrossRef]
- Wainwright, G.; Prescott, M.; Rees, H.; Webster, S. Mass Spectrometric Determination of Methyl Farnesoate Profiles and Correlation with Ovarian Development in the Edible Crab, Cancer pagurus. J. Mass Spectrom. 1996, 31, 1338–1344. [Google Scholar] [CrossRef]
- Xie, X.; Zhu, D.; Li, Y.; Qiu, X.; Cui, X.; Tang, J. Hemolymph Levels of Methyl Farnesoate during Ovarian Development of the Swimming Crab Portunus trituberculatus, and Its Relation to Transcript Levels of HMG-CoA Reductase and Farnesoic Acid O-Methyltransferase. Biol. Bull. 2015, 228, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Laufer, H.; Biggers, W.J.; Ahl, J.S. Stimulation of Ovarian Maturation in the Crayfishprocambarus Clarkiiby Methyl Farnesoate. Gen. Comp. Endocrinol. 1998, 111, 113–118. [Google Scholar] [CrossRef]
- Jo, Q.-T.; Laufer, H.; Biggers, W.J.; Kang, H.S. Methyl Farnesoate Induced Ovarian Maturation in the Spider Crab, Libinia emarginata. Invertebr. Reprod. Dev. 1999, 36, 79–85. [Google Scholar] [CrossRef]
- Reddy, P.S.; Ramamurthi, R. Methyl Farnesoate Stimulates Ovarian Maturation in the Freshwater Crab Oziotelphusa Senex Senex Fabricius. Curr. Sci. 1998, 74, 68–70. [Google Scholar]
- Nagaraju, G.P.C. Reproductive Regulators in Decapod Crustaceans: An Overview. J. Exp. Biol. 2011, 214, 3–16. [Google Scholar] [CrossRef]
- Lee, S.O.; Jeon, J.M.; Oh, C.W.; Kim, Y.M.; Kang, C.K.; Lee, D.S.; Mykles, D.L.; Kim, H.W. Two Juvenile Hormone Esterase-like Carboxylesterase cDNAs from a Pandalus Shrimp (Pandalopsis japonica): Cloning, Tissue Expression, and Effects of Eyestalk Ablation. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2011, 159, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhao, M.; Deng, Y.; Yang, Y.; Li, X.; Lu, Q.; Ge, J.; Pan, J.; Xu, Z. Molecular Cloning, Characterization and Expression Analysis of Two Juvenile Hormone Esterase-like Carboxylesterase cDNAs in Chinese Mitten Crab, Eriocheir Sinensis. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2017, 205, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, T.; Xu, R.; Huang, M.; Huang, J.; Xie, Q.; Liu, F.; Su, S.; Ma, K. Identification, characterization and mRNA transcript abundance profiles of the carboxylesterase (CXE5) gene in Eriocheir sinensis suggest that it may play a role in methyl farnesoate degradation. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2021, 256, 110630. [Google Scholar] [CrossRef] [PubMed]
- Siomi, H.; Siomi, M.C. Posttranscriptional Regulation of microRNA Biogenesis in Animals. Molecular cell 2010, 38, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Coller, J. What Comes First: Translational Repression or mRNA Degradation? The Deepening Mystery of microRNA Function. Cell Res. 2012, 22, 1322–1324. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Kokoza, V.A.; Zhang, C.; Aksoy, E.; Raikhel, A.S. MicroRNA-277 Targets Insulin-like Peptides 7 and 8 to Control Lipid Metabolism and Reproduction in Aedes Aegypti Mosquitoes. Proc. Natl. Acad. Sci. USA 2017, 114, E8017–E8024. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Li, W.; Zhao, H.; Gao, L.; Fan, Y.; Zhou, S. The microRNAs Let-7 and miR-278 Regulate Insect Metamorphosis and Oogenesis by Targeting the Juvenile Hormone Early-Response Gene Kruppel-Homolog 1. Development 2018, 145, dev170670. [Google Scholar] [CrossRef]
- Song, J.; Li, W.; Zhao, H.; Zhou, S.J. Clustered miR-2, miR-13a, miR-13b and miR-71 Coordinately Target Notch Gene to Regulate Oogenesis of the Migratory Locust Locusta Migratoria. Insect Biochem. Mol. Biol. 2019, 106, 39–46. [Google Scholar] [CrossRef]
- Chen, E.; Chen, Z.; Li, S.; Xing, D.; Guo, H.; Liu, J.; Ji, X.; Lin, Y.; Liu, S.; Xia, Q. Bmo-miR-2739 and the Novel microRNA miR-167 Coordinately Regulate the Expression of the Vitellogenin Receptor in Bombyx mori Oogenesis. Development 2020, 147, dev183723. [Google Scholar] [CrossRef]
- Hao, J.; Luo, J.; Chen, Z.; Ren, Q.; Guo, J.; Liu, X.; Chen, Q.; Wu, F.; Wang, Z.; Luo, J.; et al. MicroRNA-275 and Its Target Vitellogenin-2 Are Crucial in Ovary Development and Blood Digestion of Haemaphysalis Longicornis. Parasit Vectors 2017, 10, 253. [Google Scholar] [CrossRef]
- Waiho, K.; Fazhan, H.; Zhang, Y.; Zhang, Y.; Li, S.; Zheng, H.; Liu, W.; Ikhwanuddin, M.; Ma, H. Gonadal microRNA Expression Profiles and Their Potential Role in Sex Differentiation and Gonadal Maturation of Mud Crab Scylla paramamosain. Mar. Biotechnol. 2019, 21, 320–334. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Zhou, M.; Zou, Z.; Lin, P.; Wang, Y.; Zhang, Z. Identification and Comparative Analysis of the Ovary and Testis microRNAome of Mud Crab Scylla paramamosain. Mol. Reprod. Dev. 2018, 85, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, X.; Li, J.; Liu, P. Identification and Comparative Profiling of Ovarian and Testicular microRNAs in the Swimming Crab Portunus trituberculatus. Gene 2018, 640, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-N.; Shi, L.-L.; Liu, Z.-Q.; Qiu, G.-F. Global Analysis of the Ovarian microRNA Transcriptome: Implication for miR-2 and miR-133 Regulation of Oocyte Meiosis in the Chinese Mitten Crab, Eriocheir sinensis (Crustacea: Decapoda). BMC Genom. 2014, 15, 547. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Luo, B.Y.; Feng, J.B.; Zhou, L.X.; Ma, K.Y.; Qiu, G.F. Identification and Profiling of microRNAs during Gonadal Development in the Giant Freshwater Prawn Macrobrachium rosenbergii. Sci. Rep. 2019, 9, 2406. [Google Scholar] [CrossRef] [PubMed]
- Saetan, U.; Chotigeat, W. Identification of Ovarian miRNAs in Banana Shrimp (Fenneropenaeus merguiensis) during Ovarian Development between Non-Vitellogenic and Vitellogenic Shrimp. Aquaculture 2019, 511, 734240. [Google Scholar] [CrossRef]
- Zhao, C.; Fan, S.; Qiu, L. Identification of microRNAs and Their Target Genes Associated with Ovarian Development in Black Tiger Shrimp (Penaeus monodon) Using High-Throughput Sequencing. Sci. Rep. 2018, 8, 11602. [Google Scholar] [CrossRef]
- Jia, X.; Fang, Z.; Zeng, X.; Zhang, X.; Zhang, Z.; Wang, Y. Regulation of VIH by miR-277 in the Eyestalk of Mud Crab Scylla paramamosain. Aquaculture 2021, 534, 736254. [Google Scholar] [CrossRef]
- Zhou, M.; Jia, X.; Wan, H.; Wang, S.; Zhang, X.; Zhang, Z.; Wang, Y.B. miR-9 and miR-263 Regulate the Key Genes of the ERK Pathway in the Ovary of Mud Crab Scylla paramamosain. Mar. Biotechnol. 2020, 22, 594–606. [Google Scholar] [CrossRef]
- Mengqian, Z.; Jingyan, Z.; Hongxing, G.; Ping, L.; Jian, L.; Xianliang, M. Preliminary Functional Study of Foxl2 in Portunus trituberculatus and Analysis of Its Related miRNA. Acta Oceanol. Sin. 2022, 44, 85–94. (In Chinese) [Google Scholar]
- Fishery Department of Ministry of Agriculture and Rural Affairs of China. China Fishery Statistical Yearbook 2022; China Agriculture Press: Beijing, China, 2022. [Google Scholar]
- Kruger, J.; Rehmsmeier, M. RNAhybrid: microRNA Target Prediction Easy, Fast and Flexible. Nucleic Acids Res. 2006, 34, W451–W454. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D. MicroRNA Targets in Drosophila. Genome Biol. 2003, 4, P8. [Google Scholar] [CrossRef]
- Xie, X.; Liu, M.; Jiang, Q.; Zheng, H.; Zheng, L.; Zhu, D. Role of Kruppel Homolog 1 (Kr-H1) in Methyl Farnesoate-Mediated Vitellogenesis in the Swimming Crab Portunus trituberculatus. Gene 2018, 679, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Saha, T.T.; Zou, Z.; Raikhel, A.S. Regulatory Pathways Controlling Female Insect Reproduction. Annu. Rev. Entomol. 2018, 63, 489–511. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, H.D.; Hussain, M.; Parry, R.; Etebari, K.; Hedges, L.M.; Zhang, G.; Schulz, B.L.; Asgari, S. Human Blood microRNA Hsa-miR-21-5p Induces Vitellogenin in the Mosquito Aedes aegypti. Commun. Biol. 2021, 4, 856. [Google Scholar] [CrossRef] [PubMed]
- Duan, T.; Li, L.; Wang, H.; Pang, B. MicroRNA miR-2765-3p Regulates Reproductive Diapause by Targeting FoxO in Galeruca Daurica. Insect Sci. 2023, 30, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Liao, J.; Zhang, Z.; Li, Y.; Jia, X.; Zeng, X.; Wang, Y. Regulation of Vtg and VtgR in Mud Crab Scylla Paramamosain by miR-34. Mol. Biol. Rep. 2022, 49, 7367–7376. [Google Scholar] [CrossRef]
- Kuballa, A.V.; Guyatt, K.; Dixon, B.; Thaggard, H.; Ashton, A.R.; Paterson, B.; Merritt, D.; Elizur, A. Isolation and expression analysis of multiple isoforms of putative farnesoic acid O-methyltransferase in several crustacean species. Gen. Comp. Endocrinol. 2007, 150, 48–58. [Google Scholar] [CrossRef]
- Qu, Z.; Bendena, W.G.; Nong, W.; Siggens, K.W.; Noriega, F.G.; Kai, Z.; Zang, Y.; Koon, A.C.; Chan, H.Y.E.; Chan, T.F.; et al. MicroRNAs Regulate the Sesquiterpenoid Hormonal Pathway in Drosophila and Other Arthropods. Proc. R. Soc. B Biol. Sci. 2017, 284, 20171827. [Google Scholar]
- Song, J.; Zhou, S. Post-Transcriptional Regulation of Insect Metamorphosis and Oogenesis. Cell. Mol. Life Sci. 2020, 77, 1893–1909. [Google Scholar] [CrossRef]
- Jagadeeswaran, G.; Zheng, Y.; Sumathipala, N.; Jiang, H.; Arrese, E.L.; Soulages, J.L.; Zhang, W.; Sunkar, R. Deep Sequencing of Small RNA Libraries Reveals Dynamic Regulation of Conserved and Novel microRNAs and microRNA-Stars during Silkworm Development. BMC Genom. 2010, 11, 52. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Jia, X.; Wan, H.; Wang, S.; Zhang, X.; Zhang, Z.; Wang, Y. miR-34 Regulates Reproduction by Inhibiting the Expression of MIH, CHH, EcR, and FAMeT Genes in Mud Crab Scylla paramamosain. Mol. Reprod. Dev. 2019, 86, 122–131. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Xiao, H.; Sun, Y.; Situ, G.; Xi, Y.; Li, F. microRNA-14 as an Efficient Suppressor to Switch off Ecdysone Production after Ecdysis in Insects. RNA Biol. 2019, 16, 1313–1325. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ge, X.; Li, Z.; Wang, Y.; Song, Q.; Stanley, D.W.; Tan, A.; Huang, Y. MicroRNA-281 Regulates the Expression of Ecdysone Receptor (EcR) Isoform B in the Silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2013, 43, 692–700. [Google Scholar] [CrossRef]
- Tao, T.; Xie, X.; Liu, M.; Jiang, Q.; Zhu, D. Cloning of Two Carboxylesterase cDNAs from the Swimming Crab Portunus trituberculatus: Molecular Evidences for Their Putative Roles in Methyl Farnesotae Degradation. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 203, 100–107. [Google Scholar] [CrossRef]
| Primer | Sequence (5′-3′) |
|---|---|
| β-actin-F | CGAAACCTTCAACACTCCCG |
| β-actin-R | GGGACAGTGTGTGAAACGCC |
| CXE1-F [46] | CACGGGAAAGGTGTCTGGTAT |
| CXE1-R [46] | TTGGCGTAGGGAATGGAGTAG |
| CXE2-F [46] | GCACCTTCCGCTATGAGTTT |
| CXE2-R [46] | CCGCCAGTGAATAGATAGAACA |
| Kr-h1-F [34] | TACATCCCACATCCCCAACA |
| Kr-h1-R [34] | CGTCACATCAGGGACCATTT |
| Vitellogenin-F [34] | ATTGCTGGTCGCCTGTTGT |
| Vitellogenin-R [34] | ATCCTGCCTCAGTCCCCTTA |
| U6-F U6-R | CTTGCTTCGGCAGAACATATACT AACGCTTCACGATTTTGCGT |
| let-7b | GCAACTATACAACCTACTACCTCA |
| miR-141 | CAACACTGTCTGGTAAAGATGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Zhang, M.; Liu, P.; Li, J.; Gao, B.; Meng, X. The miRNAs let-7b and miR-141 Coordinately Regulate Vitellogenesis by Modulating Methyl Farnesoate Degradation in the Swimming Crab Portunus trituberculatus. Int. J. Mol. Sci. 2024, 25, 279. https://doi.org/10.3390/ijms25010279
Yu X, Zhang M, Liu P, Li J, Gao B, Meng X. The miRNAs let-7b and miR-141 Coordinately Regulate Vitellogenesis by Modulating Methyl Farnesoate Degradation in the Swimming Crab Portunus trituberculatus. International Journal of Molecular Sciences. 2024; 25(1):279. https://doi.org/10.3390/ijms25010279
Chicago/Turabian StyleYu, Xuee, Mengqian Zhang, Ping Liu, Jitao Li, Baoquan Gao, and Xianliang Meng. 2024. "The miRNAs let-7b and miR-141 Coordinately Regulate Vitellogenesis by Modulating Methyl Farnesoate Degradation in the Swimming Crab Portunus trituberculatus" International Journal of Molecular Sciences 25, no. 1: 279. https://doi.org/10.3390/ijms25010279
APA StyleYu, X., Zhang, M., Liu, P., Li, J., Gao, B., & Meng, X. (2024). The miRNAs let-7b and miR-141 Coordinately Regulate Vitellogenesis by Modulating Methyl Farnesoate Degradation in the Swimming Crab Portunus trituberculatus. International Journal of Molecular Sciences, 25(1), 279. https://doi.org/10.3390/ijms25010279







