Ontogeny of the Cytochrome P450 Superfamily in the Ornate Spiny Lobster (Panulirus ornatus)
Abstract
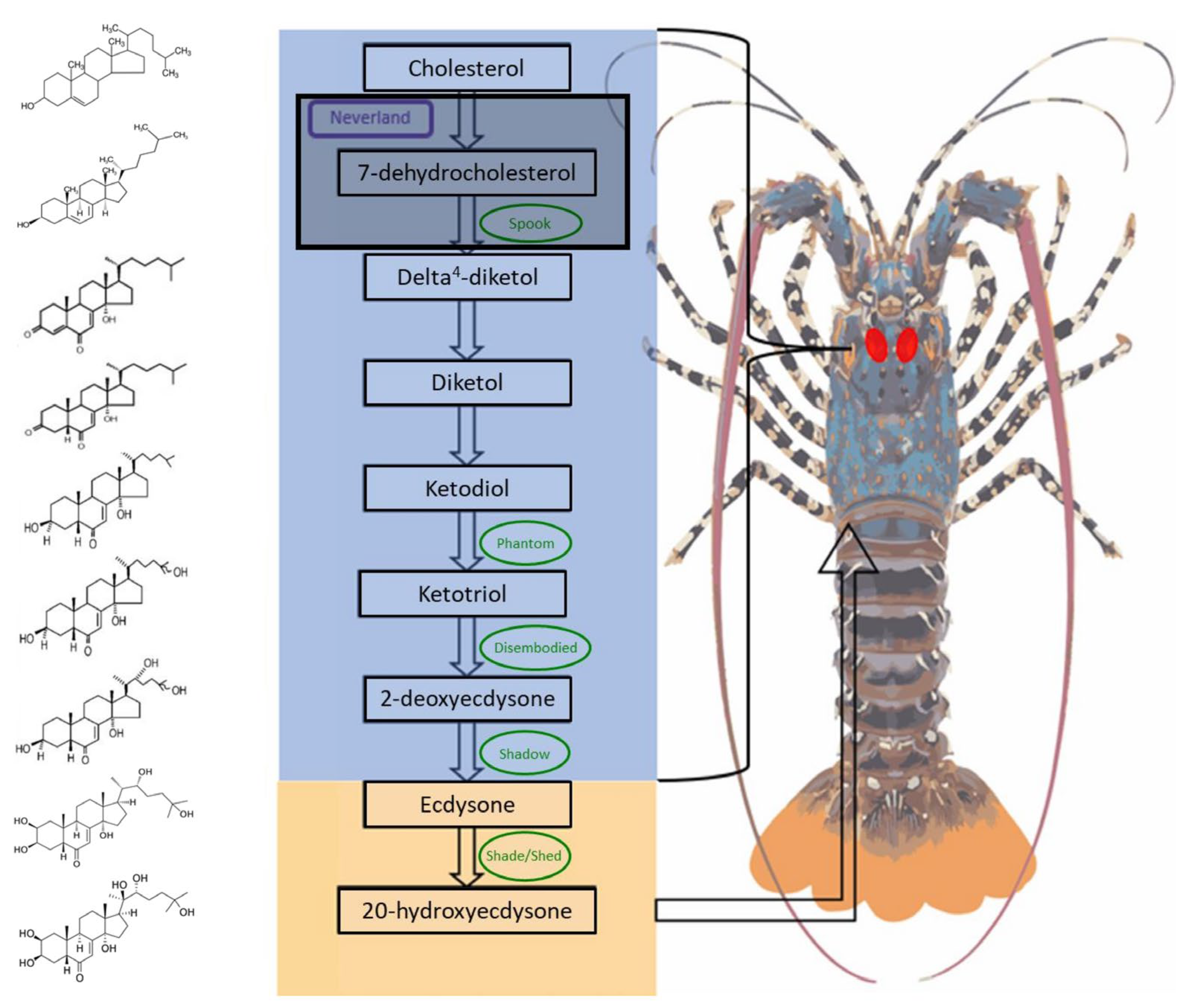
1. Introduction
2. Results
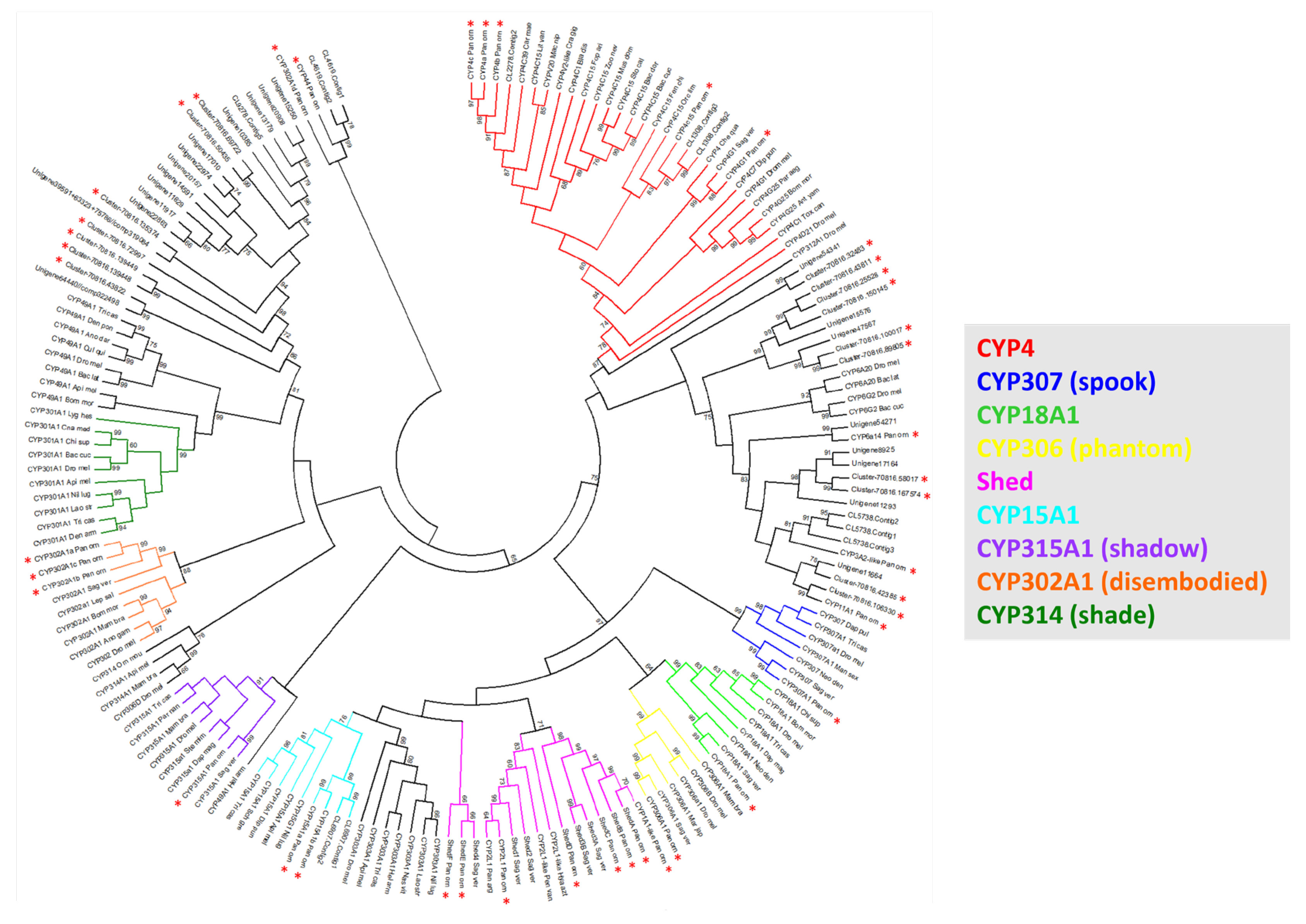
2.1. Phylogenetic Analysis
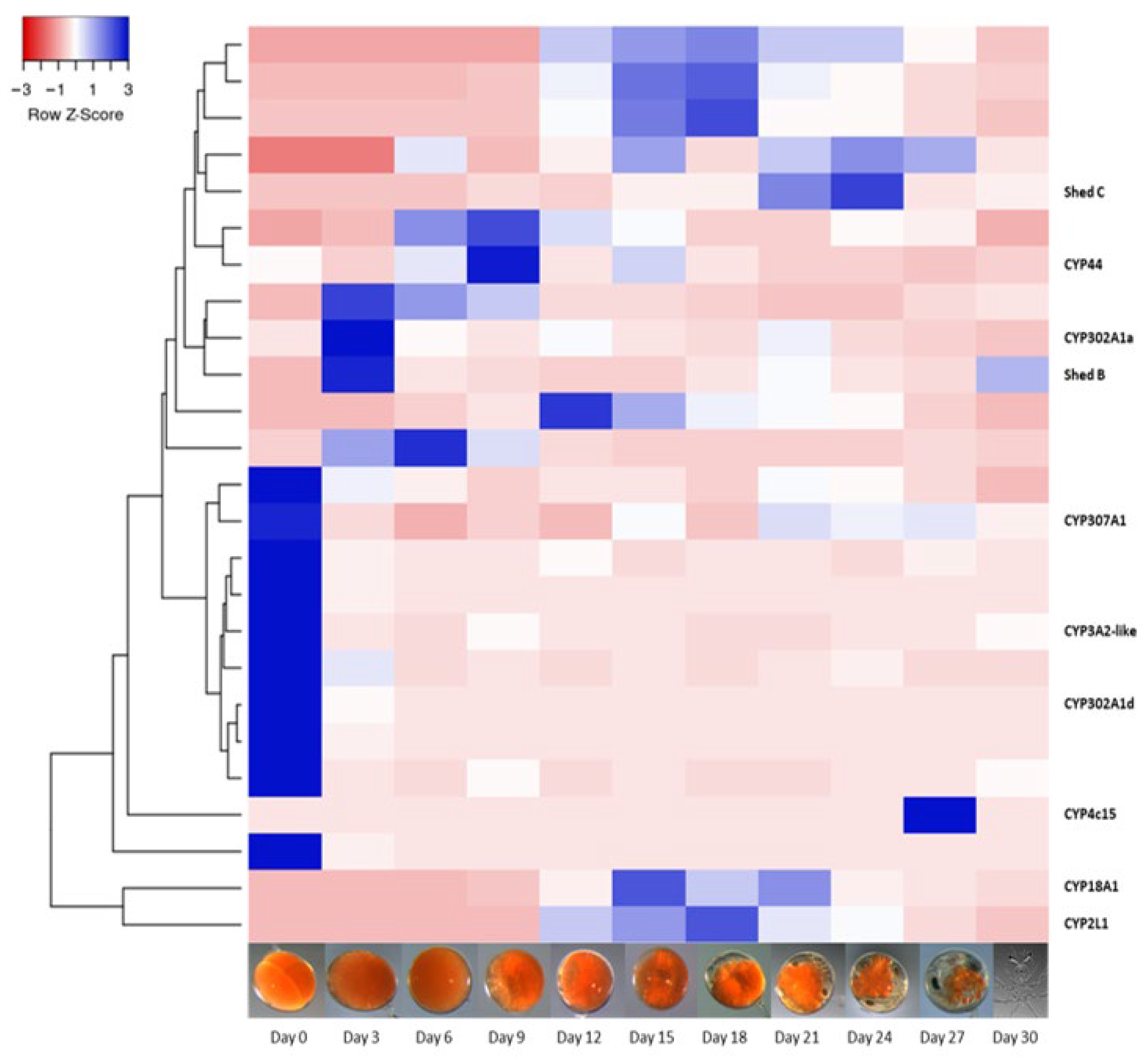
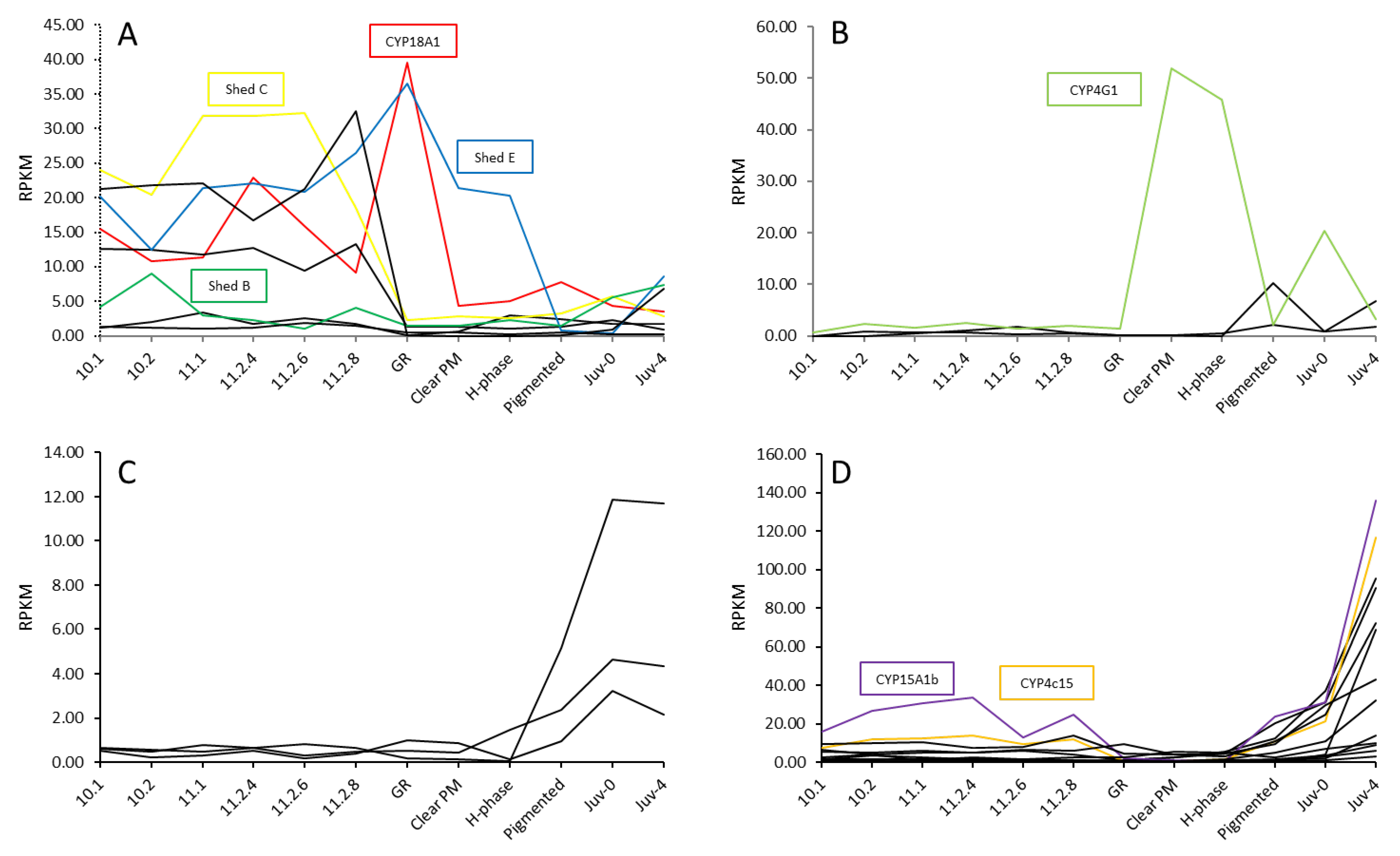
2.2. Differential Gene Expression across Panulirus ornatus Life Stages
2.3. Uncharacterised CYP450s across Life Stages and Tissues of P. ornatus
3. Discussion
4. Materials and Methods
4.1. Transcriptomic Databases
4.2. Annotation of CYP450 Orthologs
4.3. Gene Expression Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nelson, D.R.; Goldstone, J.V.; Stegeman, J.J. The cytochrome P450 genesis locus: The origin and evolution of animal cytochrome P450s. Philos. T R. Soc. B 2013, 368, 20120474. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Bose, U.; Fitzgibbon, Q.P.; Smith, G.G.; Shaw, P.N.; Cummins, S.F.; Elizur, A. CYP450s analysis across spiny lobster metamorphosis identifies a long sought missing link in crustacean development. J. Steroid Biochem. Mol. Biol. 2017, 171, 262–269. [Google Scholar] [CrossRef] [PubMed]
- Dermauw, W.; Van Leeuwen, T.; Feyereisen, R. Diversity and evolution of the P450 family in arthropods. Insect Biochem. Mol. Biol. 2020, 127, 103490. [Google Scholar] [CrossRef]
- Lynch, T.; Price, A. The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician 2007, 76, 391–396. [Google Scholar]
- Meunier, B.; de Visser, S.P.; Shaik, S. Mechanism of oxidation reactions catalyzed by cytochrome P450 enzymes. Chem. Rev. 2004, 104, 3947–3980. [Google Scholar] [CrossRef]
- James, M.O.; Boyle, S.M. Cytochromes P450 in crustacea. Comp. Biochem. Phys. C 1998, 121, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Sugumar, V.; Vijayalakshmi, G.; Saranya, K. Molt cycle related changes and effect of short term starvation on the biochemical constituents of the blue swimmer crab Portunus pelagicus. Saudi J. Biol. Sci. 2013, 20, 93–103. [Google Scholar] [CrossRef]
- Cheong, S.P.S.; Huang, J.; Bendena, W.G.; Tobe, S.S.; Hui, J.H.L. Evolution of ecdysis and metamorphosis in arthropods: The rise of regulation of juvenile hormone. Integr. Comp. Biol. 2015, 55, 878–890. [Google Scholar] [CrossRef]
- Schumannu, I.; Kenny, N.; Hui, J.; Hering, L.; Mayer, G. Halloween genes in panarthropods and the evolution of the early moulting pathway in Ecdysozoa. Roy. Soc. Open Sci. 2018, 5, 180888. [Google Scholar] [CrossRef]
- Chang, E.S. Physiological and biochemical changes during the molt cycle in decapod crustaceans: An overview. J. Exp. Mar. Biol. Ecol. 1995, 193, 1–14. [Google Scholar] [CrossRef]
- Mykles, D.L. Ecdysteroid metabolism in crustaceans. J. Steroid Biochem. Mol. Biol. 2011, 127, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Mykles, D.L. Signaling Pathways That Regulate the Crustacean Molting Gland. Front. Endocrinol. 2021, 12, 674711. [Google Scholar] [CrossRef] [PubMed]
- Andersen, Ø.; Johnsen, H.; Wittmann, A.C.; Harms, L.; Thesslund, T.; Berg, R.S.; Siikavuopio, S.; Mykles, D.L. De novo transcriptome assemblies of red king crab (Paralithodes camtschaticus) and snow crab (Chionoecetes opilio) molting gland and eyestalk ganglia-Temperature effects on expression of molting and growth regulatory genes in adult red king crab. Comp. Biochem. Physiol. Part. B Biochem. Mol. Biol. 2022, 257, 110678. [Google Scholar] [CrossRef]
- Benrabaa, S.A.M.; Chang, S.A.; Chang, E.S.; Mykles, D.L. Effects of molting on the expression of ecdysteroid biosynthesis genes in the Y-organ of the blackback land crab, Gecarcinus lateralis. Gen. Comp. Endocr. 2023, 340, 114304. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.; Rewitz, K.F.; Shinoda, T.; Itoyama, K.; Petryk, A.; Rybczynski, R.; Jarcho, M.; Warren, J.T.; Marqués, G.; Shimell, M.J.; et al. Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev. Biol. 2006, 298, 555–570. [Google Scholar] [CrossRef]
- Ventura, T.; Palero, F.; Rotllant, G.; Fitzgibbon, Q.P. Crustacean metamorphosis: An omics perspective. Hydrobiologia 2018, 825, 47–60. [Google Scholar] [CrossRef]
- Mykles, D.L.; Chang, E.S. Hormonal control of the crustacean molting gland: Insights from transcriptomics and proteomics. Gen. Comp. Endocr. 2020, 294, 113493. [Google Scholar] [CrossRef]
- Hyde, C.J.; Elizur, A.; Ventura, T. The crustacean ecdysone cassette: A gatekeeper for molt and metamorphosis. J. Steroid Biochem. Mol. Biol. 2019, 185, 172–183. [Google Scholar] [CrossRef]
- Valadez-Manzano, L.M.; Perez-Gonzalez, R.; Becerra-Arroyo, D.; Borrego, M.I. Settlement of the spiny lobster Panulirus inflatus (Bouvier, 1895) postlarvae in the southeastern Gulf of California, Mexico. Ecosis. Recur. Agropec. 2017, 4, 403–409. [Google Scholar] [CrossRef][Green Version]
- Fitzgibbon, Q.P.; Jeffs, A.G.; Battaglene, S.C. The Achilles heel for spiny lobsters: The energetics of the non-feeding post-larval stage. Fish. Fish. 2014, 15, 312–326. [Google Scholar] [CrossRef]
- Bascur, M.; Guzman, F.; Mora, S.; Espinoza, P.; Urzua, A. Temporal variation in the fatty acid composition of ovigerous females and embryos of the squat lobster Pleuroncodes monodon (Decapoda, Munididae). J. Mar. Biol. Assoc. 2018, 98, 1977–1990. [Google Scholar] [CrossRef]
- Ventura, T.; Fitzgibbon, Q.P.; Battaglene, S.C.; Elizur, A. Redefining metamorphosis in spiny lobsters: Molecular analysis of the phyllosoma to puerulus transition in Sagmariasus verreauxi. Sci. Rep. 2015, 5, 13537. [Google Scholar] [CrossRef] [PubMed]
- McWilliam, P.S.; Phillips, B.F. Metamorphosis of the final phyllosoma and secondary lecithotrophy in the puerulus of Panulirus cygnus George: A review. Mar. Freshw. Res. 1997, 48, 783–790. [Google Scholar] [CrossRef]
- Martinez-Calderon, R.; Lozano-Alvarez, E.; Briones-Fourzan, P. Morphometric relationships and seasonal variation in size, weight, and a condition index of post-settlement stages of the Caribbean spiny lobster. Peerj 2018, 6, e5297. [Google Scholar] [CrossRef]
- Jeffs, A. Status and challenges of advancing lobster aquaculture globally. J. Mar. Biol. Assoc. India 2010, 52, 320–326. [Google Scholar]
- Boyd, C.E.; D'Abramo, L.R.; Glencross, B.D.; Huyben, D.C.; Juarez, L.M.; Lockwood, G.S.; McNevin, A.A.; Tacon, A.G.J.; Teletchea, F.; Tomasso, J.R.; et al. Achieving sustainable aquaculture: Historical and current perspectives and future needs and challenges. J. World Aquacult. Soc. 2020, 51, 578–633. [Google Scholar] [CrossRef]
- Perera, E.; Simon, C. Digestive physiology of spiny lobsters: Implications for formulated diet development. Rev. Aquacult. 2015, 7, 243–261. [Google Scholar] [CrossRef]
- Chow, S.; Suzuki, N.; Imai, H.; Yoshimura, T. Molecular Species Identification of Spiny Lobster Phyllosoma Larvae of the Genus Panulirus from the Northwestern Pacific. Mar. Biotechnol. 2006, 8, 260–267. [Google Scholar] [CrossRef]
- Kanciruk, P. Ecology of juvenile and adult Palinuridae (spiny lobsters). In The Biology and Management of Lobsters; Academic Press: Cambridge, MA, USA, 1980; Volume 2, pp. 59–96. [Google Scholar]
- Marchese, G.; Fitzgibbon, Q.P.; Trotter, A.J.; Carter, C.G.; Jones, C.M.; Smith, G.G. The influence of flesh ingredients format and krill meal on growth and feeding behaviour of juvenile tropical spiny lobster Panulirus ornatus. Aquaculture 2019, 499, 128–139. [Google Scholar] [CrossRef]
- Niwa, R.; Sakudoh, T.; Matsuya, T.; Namiki, T.; Kasai, S.; Tomita, T.; Kataoka, H. Expressions of the cytochrome P450 monooxygenase gene Cyp4g1 and its homolog in the prothoracic glands of the fruit fly Drosophila melanogaster (Diptera: Drosophilidae) and the silkworm Bombyx mori (Lepidoptera: Bombycidae). Appl. Entomol. Zool. 2011, 46, 533–543. [Google Scholar] [CrossRef]
- Lewis, C.L.; Fitzgibbon, Q.P.; Smith, G.G.; Elizur, A.; Ventura, T. Transcriptomic analysis and time to hatch visual prediction of embryo development in the ornate spiny lobster (Panulirus ornatus). Front. Mar. Sci. 2022, 9, 889317. [Google Scholar] [CrossRef]
- Hyde, C.J.; Fitzgibbon, Q.P.; Elizur, A.; Smith, G.G.; Ventura, T. Transcriptional profiling of spiny lobster metamorphosis reveals three new additions to the nuclear receptor superfamily. Bmc Genom. 2019, 20, 531. [Google Scholar] [CrossRef]
- Ventura, T.; Chandler, J.C.; Nguyen, T.V.; Hyde, C.J.; Elizur, A.; Fitzgibbon, Q.P.; Smith, G.G. Multi-tissue transcriptome analysis identifies key sexual development-related genes of the ornate spiny lobster (Panulirus ornatus). Genes 2020, 11, 1150. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Vraspir, L.; Zhou, W.; Durica, D.S.; Mykles, D.L. Transcriptomic analysis of differentially expressed genes in the molting gland (Y-organ) of the blackback land crab, Gecarcinus lateralis, during molt-cycle stage transitions. Comp. Biochem. Physiol. Part. D Genom. Proteom. 2018, 28, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Marchal, E.; Zhang, J.R.; Badisco, L.; Verlinden, H.; Hult, E.F.; Van Wielendaele, P.; Yagi, K.J.; Tobe, S.S.; Vanden Broeck, J. Final steps in juvenile hormone biosynthesis in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2011, 41, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, J.P. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochem. Pharmacol. 2008, 75, 2263–2275. [Google Scholar] [CrossRef] [PubMed]
- Wojcikowski, J.; Haduch, A.; Daniel, W.A. Effect of classic and atypical neuroleptics on cytochrome P450 3A (CYP3A) in rat liver. Pharmacol. Rep. 2012, 64, 1411–1418. [Google Scholar] [CrossRef]
- Slominski, A.T.; Li, W.; Kim, T.K.; Semak, I.; Wang, J.; Zjawiony, J.K.; Tuckey, R.C. Novel activities of CYP11A1 and their potential physiological significance. J. Steroid. Biochem. Mol. Biol. 2015, 151, 25–37. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Guittard, E.; Blais, C.; Maria, A.; Parvy, J.P.; Pasricha, S.; Lumb, C.; Lafont, R.; Daborn, P.J.; Dauphin-Villemant, C. CYP18A1, a key enzyme of Drosophila steroid hormone inactivation, is essential for metamorphosis. Dev. Biol. 2011, 349, 35–45. [Google Scholar] [CrossRef]
- Helvig, C.; Koener, J.F.; Unnithan, G.C.; Feyereisen, R. CYP15A1, the cytochrome P450 that catalyzes epoxidation of methyl farnesoate to juvenile hormone III in cockroach Corpora allata. Proc. Natl. Acad. Sci. USA 2004, 101, 4024–4029. [Google Scholar] [CrossRef] [PubMed]
- Kubota, A.; Stegeman, J.J.; Goldstone, J.V.; Nelson, D.R.; Kim, E.Y.; Tanabe, S.; Iwata, H. Cytochrome P450 CYP2 genes in the common cormorant: Evolutionary relationships with 130 diapsid CYP2 clan sequences and chemical effects on their expression. Comp. Biochem. Phys. C 2011, 153, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Gurusamy, U.; Shewade, D.G. Pharmacogenomics in India. In Handbook of Pharmacogenomics and Stratified Medicine; Padmanabhan, S., Ed.; Academic Press: Cambridge, MA, USA, 2014; pp. 1037–1059. [Google Scholar]
- Baldwin, W.S.; Marko, P.B.; Nelson, D.R. The cytochrome P450 (CYP) gene superfamily in Daphnia pulex. BMC Genom. 2009, 10, 169. [Google Scholar] [CrossRef]
- Pan, F.; Fu, Y.; Zhang, W.; Jiang, S.; Xiong, Y.; Yan, Y.; Gong, Y.; Qiao, H.; Fu, H. Characterization, expression and functional analysis of CYP306a1 in the oriental river prawn, Macrobrachium nipponense. Aquacult. Rep. 2022, 22. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, M.; Liu, Y.; Zhou, Z.; Guo, J. Identification of cytochrome P450 monooxygenase genes and their expression in response to high temperature in the alligatorweed flea beetle Agasicles hygrophila (Coleoptera: Chrysomelidae). Sci. Rep. 2018, 8, 17847. [Google Scholar] [CrossRef] [PubMed]
- Hyde, C.J.; Fitzgibbon, Q.P.; Elizur, A.; Smith, G.G.; Ventura, T. CrustyBase: An interactive online database for crustacean transcriptomes. BMC Genom. 2020, 21, 637. [Google Scholar] [CrossRef]
- Campos-Ortega, J.A.; Hartenstein, V. Stages of Drosophila Embryogenesis. In The Embryonic Development of Drosophila melanogaster; Campos-Ortega, J.A., Hartenstein, V., Eds.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 9–102. [Google Scholar]
- Dauphin-Villemant, C.; Böcking, D.; Tom, M.; Maïbèche, M.; Lafont, R. Cloning of a novel cytochrome P450 (CYP4C15) differentially expressed in the steroidogenic glands of an arthropod. Biochem. Biophys. Res. Commun. 1999, 264, 413–418. [Google Scholar] [CrossRef]
- Helluy, S.M.; Beltz, B.S. Embryonic Development of the American Lobster (Homarus americanus): Quantitative Staging and Characterization of an Embryonic Molt Cycle. Biol. Bull. 1991, 180, 355–371. [Google Scholar] [CrossRef]
- Duggan, S.; McKinnon, A.D. The early larval developmental stages of the spiny lobster Panulirus ornatus (Fabricius, 1798) cultured under laboratory conditions. Crustaceana 2003, 76, 313–332. [Google Scholar]
- Saunders, M.I.; Thompson, P.A.; Jeffs, A.G.; Säwström, C.; Sachlikidis, N.; Beckley, L.E.; Waite, A.M. Fussy feeders: Phyllosoma larvae of the western rocklobster (Panulirus cygnus) demonstrate prey preference. PLoS ONE 2012, 7, e36580. [Google Scholar] [CrossRef][Green Version]
- Aragon, S.; Claudinot, S.; Blais, C.; Maibeche, M.; Dauphin-Villemant, C. Molting cycle-dependent expression of CYP4C15, a cytochrome P450 enzyme putatively involved in ecdysteroidogenesis in the crayfish, Orconectes limosus. Insect Biochem. Mol. 2002, 32, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Hu, K.; Hou, Y.; Wang, Y.; Yao, Y.; Lei, X.; Yan, B.; Jiang, Q.; Xiong, C.; Xu, L.; et al. Transcriptome analysis of hepatopancreas of Eriocheir sinensis with hepatopancreatic necrosis disease (HPND). PLoS ONE 2020, 15, e0228623. [Google Scholar] [CrossRef] [PubMed]
- Xichao, X.; Wenfeng, W.; Xin, L.; Xueshen, W.; Li, L.; Ning, Q. A novel CYP450 gene (CYP4V18) of the shrimp Penaeus schmitti Burkenroad, 1936 (Decapoda, Dendrobranchiata): Cloning and down-regulation of CYP4V18 expression by the imidazole derivative KK-42. Crustaceana 2013, 86, 1179–1190. [Google Scholar] [CrossRef]
- Swall, M.E.; Benrabaa, S.A.M.; Tran, N.M.; Tran, T.D.; Ventura, T.; Mykles, D.L. Characterization of Shed genes encoding ecdysone 20-monooxygenase (CYP314A1) in the Y-organ of the blackback land crab, Gecarcinus lateralis. Gen. Comp. Endocrinol. 2021, 301, 113658. [Google Scholar] [CrossRef]
- de Oliveira, A.P.L.; Calcino, A.; Wanninger, A. Ancient origins of arthropod moulting pathway components. Elife 2019, 8, e46113. [Google Scholar] [CrossRef]
- Dam, E.; Rewitz, K.F.; Styrishave, B.; Andersen, O. Cytochrome P450 expression is moult stage specific and regulated by ecdysteroids and xenobiotics in the crab Carcinus maenas. Biochem. Biophys. Res. Commun. 2008, 377, 1135–1140. [Google Scholar] [CrossRef]
- Gottardi, M.; Kretschmann, A.; Cedergreen, N. Measuring cytochrome P450 activity in aquatic invertebrates: A critical evaluation of in vitro and in vivo methods. Ecotoxicology 2016, 25, 419–430. [Google Scholar] [CrossRef]
- Dejong, C.A.; Wilson, J.Y. The cytochrome P450 superfamily complement (CYPome) in the annelid Capitella teleta. PLoS ONE 2014, 9, e107728. [Google Scholar] [CrossRef]
- Legrand, E.; Bachvaroff, T.; Schock, T.B.; Chung, J.S. Understanding molt control switches: Transcriptomic and expression analysis of the genes involved in ecdysteroidogenesis and cholesterol uptake pathways in the Y-organ of the blue crab, Callinectes sapidus. PLoS ONE 2021, 16, e0256735. [Google Scholar] [CrossRef]
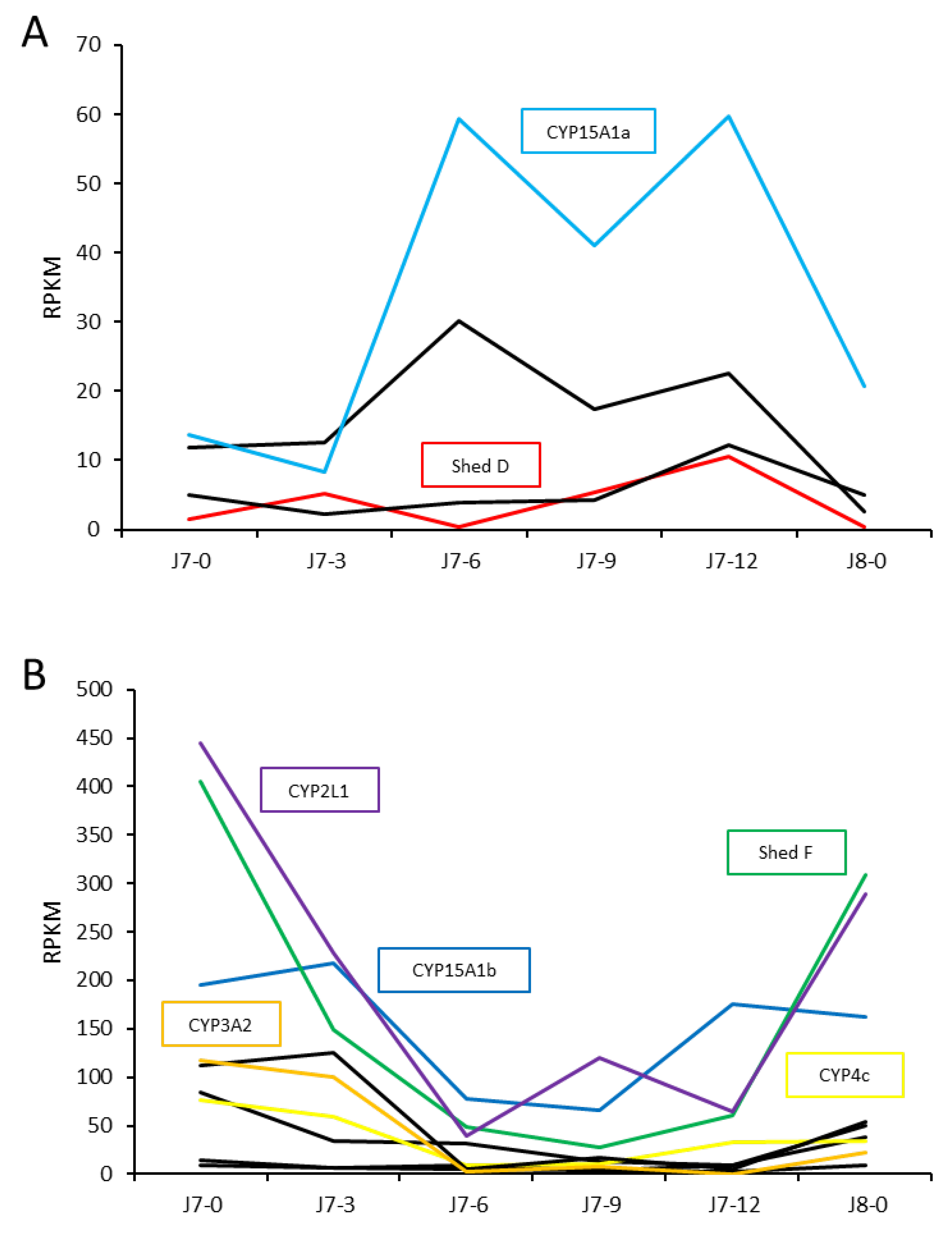
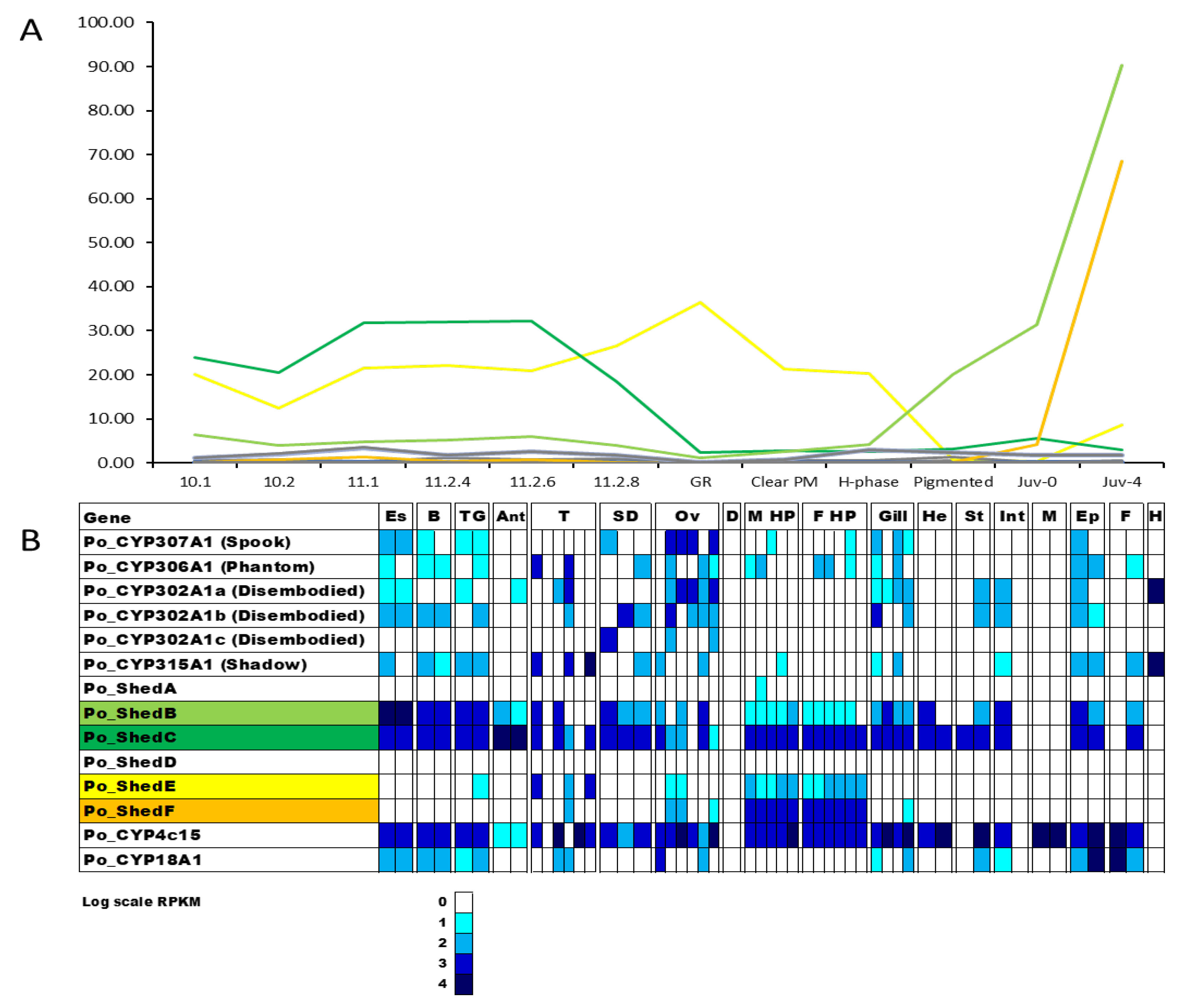
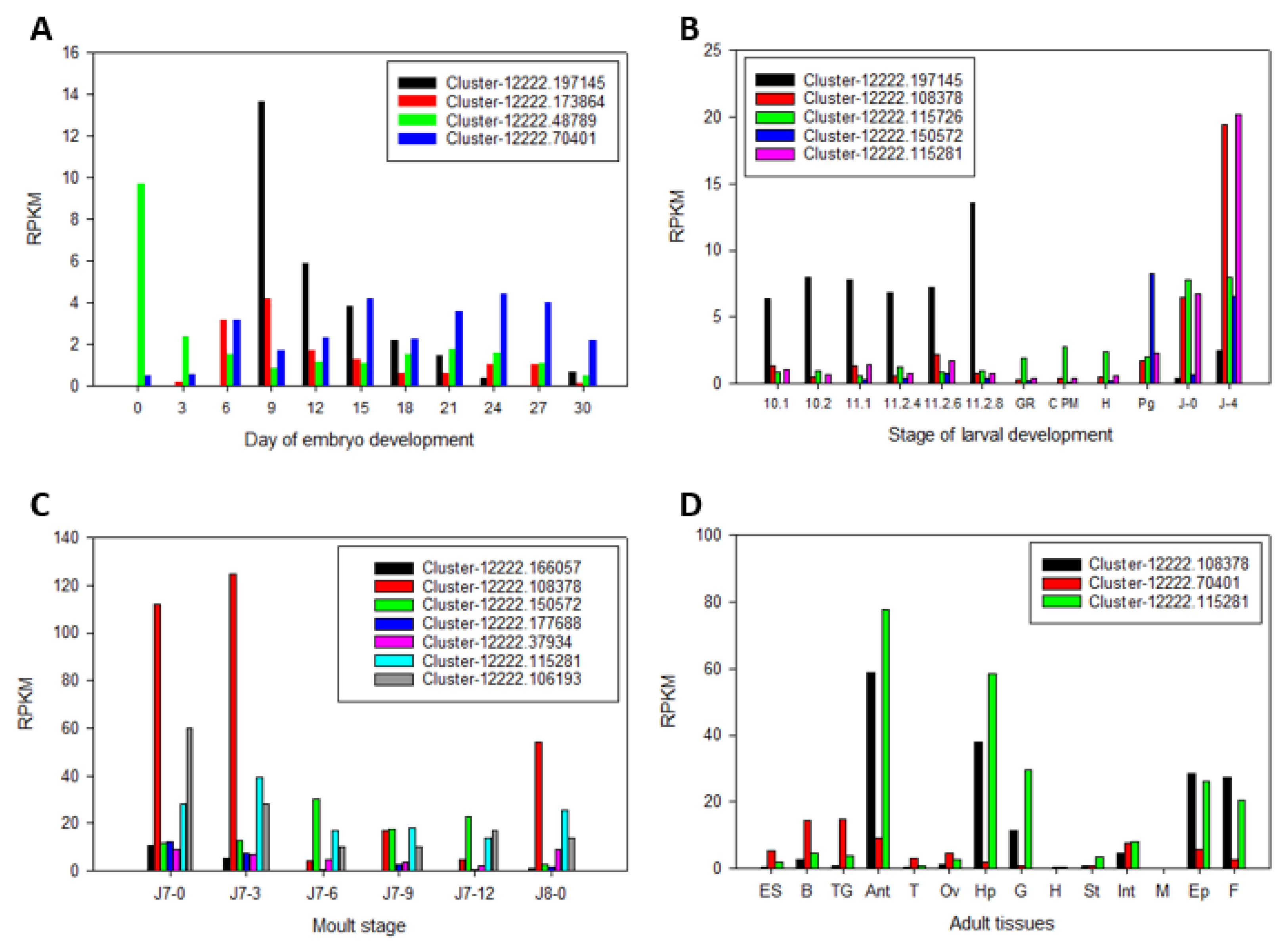
| Transcripts | Annotation | Associated Pathway | Clan |
|---|---|---|---|
| Cluster-12222.190192 | CYP307A1_Pan_orn (Spook) | Ecdysteroidogenesis | CYP2 |
| Cluster-12222.47152 | Shed A_Pan_orn | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.105348 | Shed B_Pan_orn | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.144497 | Shed C_Pan_orn | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.19622 | Shed D_Pan_orn | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.37028 | Shed E_Pan_orn | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.37028 | Shed F_Pan_orn | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.26280 | CYP315A1_Pan_orn (shadow) | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.74444 | CYP302A1a_Pan_orn (disembodied) | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.32150 | CYP302A1b_Pan_orn (disembodied) | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.175902 | CYP302A1c_Pan_orn (disembodied) | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.184161 | CYP302A1d_Pan_orn (disembodied) | Ecdysteroidogenesis | Mitochondrial CYP |
| Cluster-12222.93853 | CYP4c15_Pan_orn | Ecdysteroidogenesis | CYP4 |
| Cluster-12222.171626 | CYP306A1_Pan_orn (phantom) | Ecdysteroidogenesis | CYP2 |
| Cluster-12222.113231 | CYP18A1_Pan_orn | Ecdysteroidogenesis | CYP2 |
| Cluster-12222.52575 | CYP15A1a_Pan_orn | Juvenile hormone biosynthesis [36] | CYP2 |
| Cluster-12222.175641 | CYP15A1b_Pan_orn | Juvenile hormone biosynthesis | CYP2 |
| Cluster-12222.93855 | CYP4G1_Pan_orn | Lipid metabolism | CYP4 |
| Cluster-12222.55035 | CYP4a_Pan_orn | Omega hydroxylation [37] | CYP4 |
| Cluster-12222.52849 | CYP4b_Pan_orn | Omega hydroxylation | CYP4 |
| Cluster-12222.55185 | CYP4c_Pan_orn | Omega hydroxylation | CYP4 |
| Cluster-12222.102364 | CYP2L1-like_Pan_orn | Estrogen synthesis | CYP2 |
| Cluster-12222.82549 | CYP3A2-like_Pan_orn | Oxidative metabolism [38] | CYP3 |
| Cluster-12222.159369 | CYP6a14_Pan_orn | Hormone biosynthesis | CYP3 |
| Cluster-12222.134345 | CYP44_Pan_orn | Oxidative metabolism | Mitochondrial CYP |
| Cluster-12222.139601 | CYP1A1-like_Pan_orn | Xenobiotic metabolism | CYP2 |
| Cluster-12222.56205 | CYP11A1a_Pan_orn | Steroidogenesis [39] | Mitochondrial CYP |
| Cluster-12222.166057 | CYP11A1b_Pan_orn | Steroidogenesis | Mitochondrial CYP |
| Embryo | Larval | Juvenile Hp | Adult | |
|---|---|---|---|---|
| Cluster-12222.166057 | ||||
| Cluster-12222.108378 | ||||
| Cluster-12222.150572 | ||||
| Cluster-12222.177688 | ||||
| Cluster-12222.37934 | ||||
| Cluster-12222.115281 | ||||
| Cluster-12222.106193 | ||||
| Cluster-12222.70401 | ||||
| Cluster-12222.197145 | ||||
| Cluster-12222.173864 | ||||
| Cluster-12222.48789 | ||||
| Cluster-12222.115726 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lewis, C.L.; Fitzgibbon, Q.P.; Smith, G.G.; Elizur, A.; Ventura, T. Ontogeny of the Cytochrome P450 Superfamily in the Ornate Spiny Lobster (Panulirus ornatus). Int. J. Mol. Sci. 2024, 25, 1070. https://doi.org/10.3390/ijms25021070
Lewis CL, Fitzgibbon QP, Smith GG, Elizur A, Ventura T. Ontogeny of the Cytochrome P450 Superfamily in the Ornate Spiny Lobster (Panulirus ornatus). International Journal of Molecular Sciences. 2024; 25(2):1070. https://doi.org/10.3390/ijms25021070
Chicago/Turabian StyleLewis, Courtney L., Quinn P. Fitzgibbon, Gregory G. Smith, Abigail Elizur, and Tomer Ventura. 2024. "Ontogeny of the Cytochrome P450 Superfamily in the Ornate Spiny Lobster (Panulirus ornatus)" International Journal of Molecular Sciences 25, no. 2: 1070. https://doi.org/10.3390/ijms25021070
APA StyleLewis, C. L., Fitzgibbon, Q. P., Smith, G. G., Elizur, A., & Ventura, T. (2024). Ontogeny of the Cytochrome P450 Superfamily in the Ornate Spiny Lobster (Panulirus ornatus). International Journal of Molecular Sciences, 25(2), 1070. https://doi.org/10.3390/ijms25021070








