Analysis of Circulating Tumor DNA in Synchronous Metastatic Colorectal Cancer at Diagnosis Predicts Overall Patient Survival
Abstract
1. Introduction
2. Results
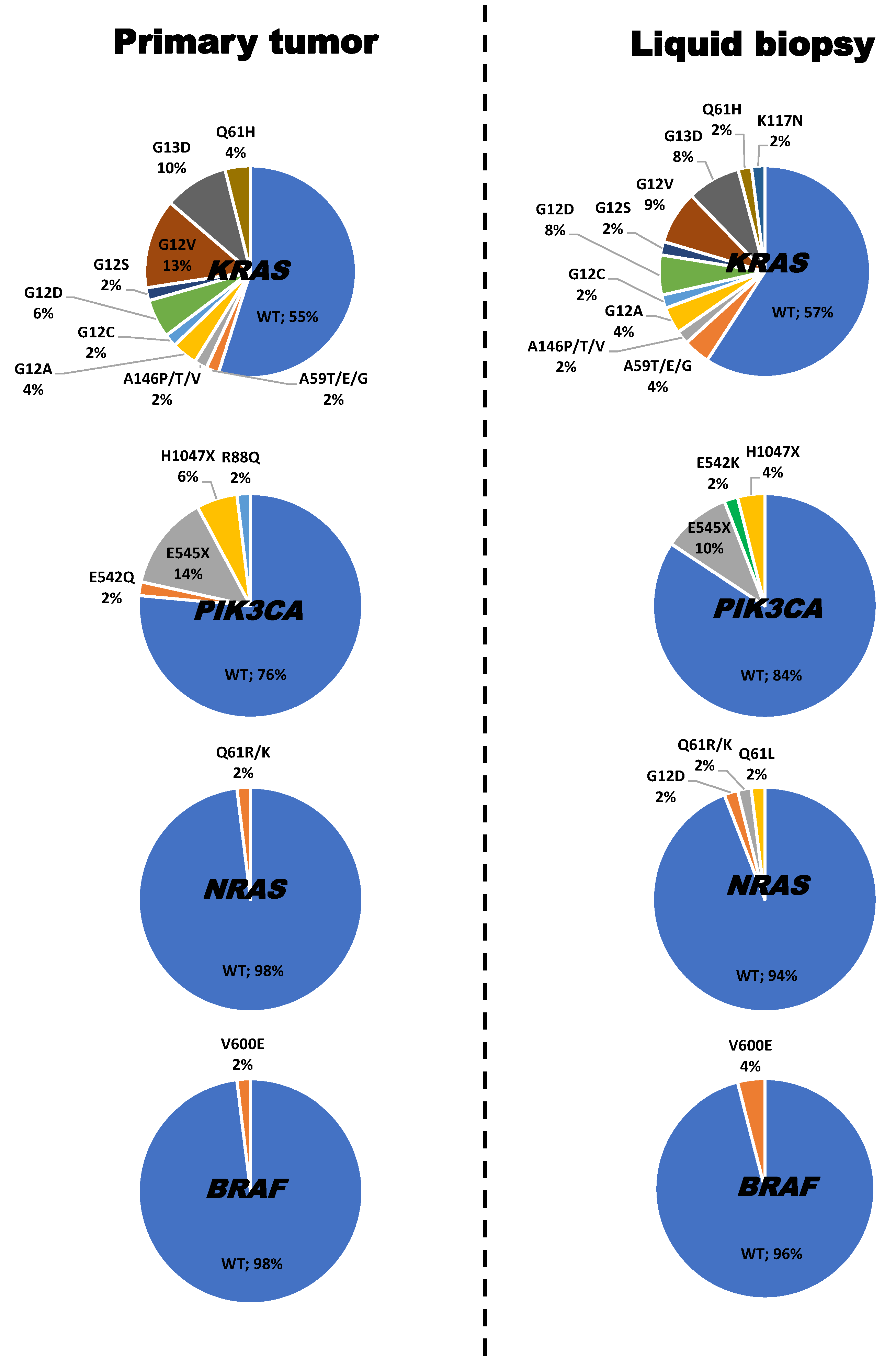
2.1. Frequency, Type and Concordance of KRAS, NRAS, PIK3CA and BRAF Mutations Detected in Tumor Tissue and Plasma of 51 Patients with Synchronous Metastatic Colorectal Cancer (SMCC)
2.2. Association between Mutational Status Detected in Patient Plasma and Other Features of the Disease
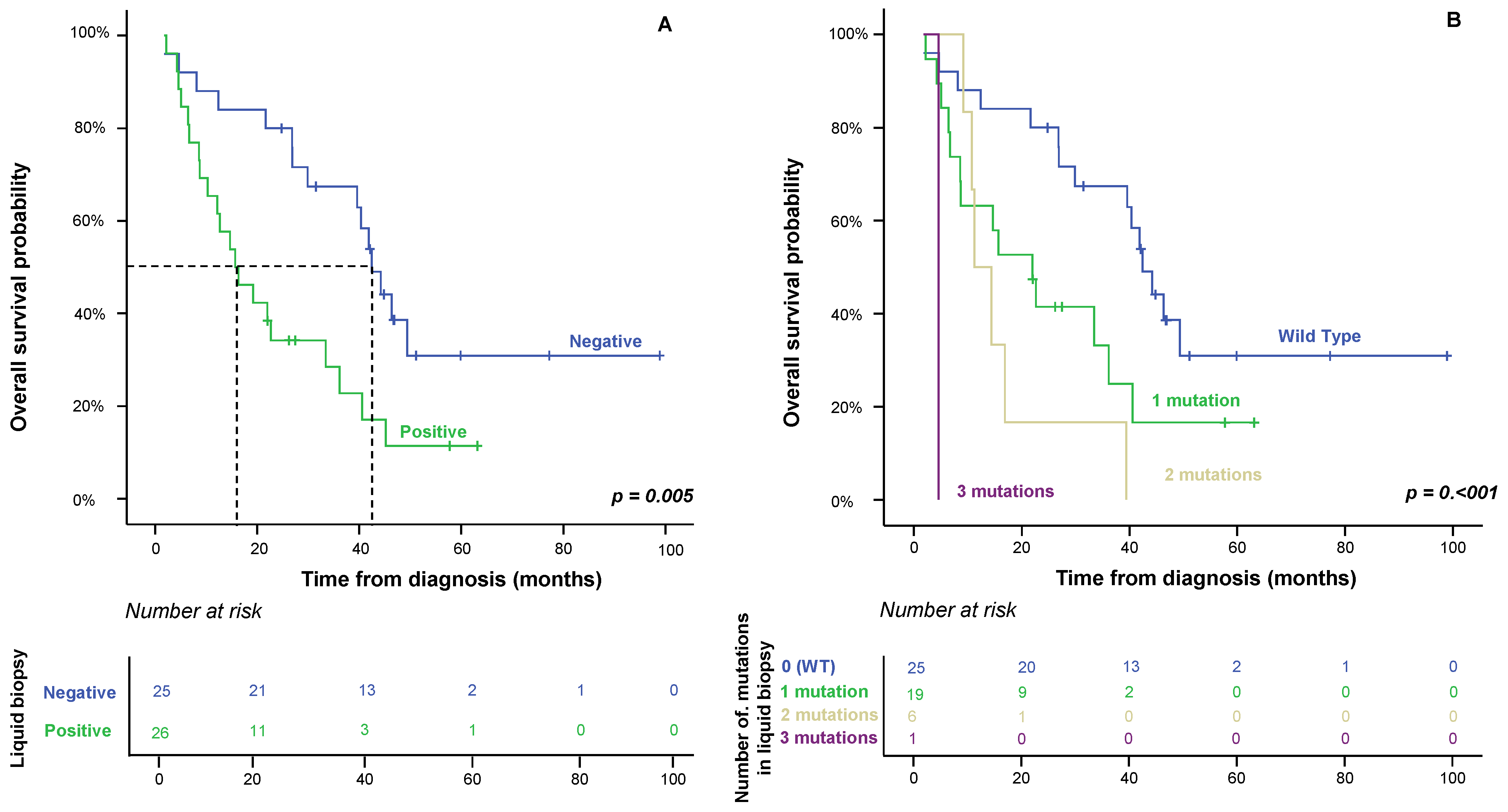
2.3. Impact of Liquid Biopsy on Patient OS
3. Discussion
4. Materials and Methods
4.1. Patients and Samples
4.2. Tissue-Based RAS, BRAF and PIK3CA Mutation Analysis
4.3. Blood-Based RAS, BRAF and PIK3CA Mutation Analysis
4.4. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; van Krieken, J.H.; Aderka, D.; Aguilar, E.A.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- Del Carmen, S.; Sayagués, J.M.; Bengoechea, O.; Anduaga, M.F.; Alcazar, J.A.; Gervas, R.; García, J.; Orfao, A.; Bellvis, L.M.; Sarasquete, M.E.; et al. Spatio-temporal tumor heterogeneity in metastatic CRC tumors: A mutational-based approach. Oncotarget 2018, 9, 34279–34288. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Bellvis, L.; Fontanillo, C.; González-González, M.; Garcia, E.; Iglesias, M.; Esteban, C.; Gutierrez, M.L.; Abad, M.M.; Bengoechea, O.; Rivas, J.D.L.; et al. Unique genetic profile of sporadic colorectal cancer liver metastasis versus primary tumors as defined by high-density single-nucleotide polymorphism arrays. Mod. Pathol. 2012, 25, 590–601. [Google Scholar] [CrossRef] [PubMed]
- González-González, M.; Muñoz-Bellvís, L.; Mackintosh, C.; Fontanillo, C.; Gutiérrez, M.L.; Abad, M.M.; Bengoechea, O.; Teodosio, C.; Fonseca, E.; Fuentes, M.; et al. Prognostic Impact of del(17p) and del(22q) as Assessed by Interphase FISH in Sporadic Colorectal Carcinomas. PLoS ONE 2012, 7, e42683. [Google Scholar] [CrossRef]
- Martini, G.; Ciardiello, D.; Napolitano, S.; Martinelli, E.; Troiani, T.; Latiano, T.P.; Avallone, A.; Normanno, N.; Di Maio, M.; Maiello, E.; et al. Efficacy and safety of a biomarker-driven cetuximab-based treatment regimen over 3 treatment lines in mCRC patients with RAS/BRAF wild type tumors at start of first line: The CAPRI 2 GOIM trial. Front. Oncol. 2023, 13, 1069370. [Google Scholar] [CrossRef]
- Witt, J.; Haupt, S.; Ahadova, A.; Bohaumilitzky, L.; Fuchs, V.; Ballhausen, A.; Przybilla, M.J.; Jendrusch, M.; Seppälä, T.T.; Fürst, D.; et al. A simple approach for detecting HLA-A *02 alleles in archival formalin-fixed paraffin-embedded tissue samples and an application example for studying cancer immunoediting. Hla 2023, 101, 24–33. [Google Scholar] [CrossRef]
- Yang, Y.C.; Wang, D.; Jin, L.; Yao, H.W.; Zhang, J.H.; Wang, J.; Zhao, X.M.; Shen, C.Y.; Chen, W.; Wang, X.L.; et al. Circulating tumor DNA detectable in early- and late-stage colorectal cancer patients. Biosci. Rep. 2018, 38, BSR20180322. [Google Scholar] [CrossRef]
- Allegretti, M.; Cottone, G.; Carboni, F.; Cotroneo, E.; Casini, B.; Giordani, E.; Amoreo, C.A.; Buglioni, S.; Diodoro, M.; Pescarmona, E.; et al. Cross-sectional analysis of circulating tumor DNA in primary colorectal cancer at surgery and during post-surgery follow-up by liquid biopsy. J. Exp. Clin. Cancer Res. 2020, 39, 69. [Google Scholar] [CrossRef]
- Klein, C.A. Selection and adaptation during metastatic cancer progression. Nature 2013, 501, 365–372. [Google Scholar] [CrossRef]
- Bádon, E.S.; Mokánszki, A.; Mónus, A.; András, C.; Méhes, G. Clonal diversity in KRAS mutant colorectal adenocarcinoma under treatment: Monitoring of cfDNA using reverse hybridization and DNA sequencing platforms. Mol. Cell. Probes 2023, 67, 101891. [Google Scholar] [CrossRef]
- Kawazoe, A.; Shitara, K.; Fukuoka, S.; Kuboki, Y.; Bando, H.; Okamoto, W.; Kojima, T.; Fuse, N.; Yamanaka, T.; Doi, T.; et al. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer 2015, 15, 258. [Google Scholar] [CrossRef]
- Font, N.R.; Garbarino, Y.N.; Castello, O.D.; Amoros, J.M.; Sánchez, P.B.; Lletget, D.C.; Cabello, M.A.L.; Marcet, J.B.; Meca, S.M.; Escape, I.; et al. Concordance analysis between liquid biopsy (ctDNA) and tumor DNA molecular profiles from panel-based next-generation sequencing. Rev. Española De Patol. 2022, 55, 156–162. [Google Scholar] [CrossRef]
- Güttlein, L.; Luca, M.R.; Esteso, F.; Fresno, C.; Mariani, J.; Otero Pizarro, M.; Brest, E.; Starapoli, S.; Kreimberg, K.; Teves, P.; et al. Liquid biopsy for KRAS, NRAS and BRAF mutation testing in advanced colorectal cancer patients: The Argentinean experience. Future Oncol. Lond. Engl. 2022, 18, 3277–3287. [Google Scholar] [CrossRef]
- Van’t Erve, I.; Greuter, M.J.; Bolhuis, K.; Vessies, D.C.; Leal, A.; Vink, G.R.; van den Broek, D.; Velculescu, V.E.; Punt, C.J.A.; Meijer, G.A.; et al. Diagnostic Strategies toward Clinical Implementation of Liquid Biopsy RAS/BRAF Circulating Tumor DNA Analyses in Patients with Metastatic Colorectal Cancer. J. Mol. Diagn. 2020, 22, 1430–1437. [Google Scholar] [CrossRef]
- Kagawa, Y.; Elez, E.; García-Foncillas, J.; Bando, H.; Taniguchi, H.; Vivancos, A.; Akagi, K.; García, A.; Denda, T.; Ros, J.; et al. Combined Analysis of Concordance between Liquid and Tumor Tissue Biopsies for RAS Mutations in Colorectal Cancer with a Single Metastasis Site: The METABEAM Study. Clin. Cancer Res. 2021, 27, 2515–2522. [Google Scholar] [CrossRef]
- Watson, R.; Liu, T.-C.; Ruzinova, M.B. High frequency of KRAS mutation in early onset colorectal adenocarcinoma: Implications for pathogenesis. Hum. Pathol. 2016, 56, 163–170. [Google Scholar] [CrossRef]
- Taniguchi, F.; Nyuya, A.; Toshima, T.; Yasui, K.; Mori, Y.; Okawaki, M.; Kishimoto, H.; Umeda, Y.; Fujiwara, T.; Tanioka, H.; et al. Concordance of acquired mutations between metastatic lesions and liquid biopsy in metastatic colorectal cancer. Futur. Sci. OA 2021, 7, FSO757. [Google Scholar] [CrossRef]
- Yamada, T.; Matsuda, A.; Koizumi, M.; Shinji, S.; Takahashi, G.; Iwai, T.; Takeda, K.; Ueda, K.; Yokoyama, Y.; Hara, K.; et al. Liquid Biopsy for the Management of Patients with Colorectal Cancer. Digestion 2018, 99, 39–45. [Google Scholar] [CrossRef]
- McGranahan, N.; Swanton, C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017, 168, 613–628. [Google Scholar] [CrossRef]
- Liebs, S.; Keilholz, U.; Kehler, I.; Schweiger, C.; Haybäck, J.; Nonnenmacher, A. Detection of mutations in circulating cell-free DNA in relation to disease stage in colorectal cancer. Cancer Med. 2019, 8, 3761–3769. [Google Scholar] [CrossRef]
- Hrebien, S.; Citi, V.; Garcia-Murillas, I.; Cutts, R.; Fenwick, K.; Kozarewa, I.; McEwen, R.; Ratnayake, J.; Maudsley, R.; Carr, T.; et al. Early ctDNA dynamics as a surrogate for progression-free survival in advanced breast cancer in the BEECH trial. Ann. Oncol. 2019, 30, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Sun, X.; Wang, W.; Lu, S. Hypoxia-inducible factor-1alpha modulates the down-regulation of the homeodomain protein CDX2 in colorectal cancer. Oncol. Rep. 2010, 24, 97–104. [Google Scholar] [PubMed]
- Singh, D.; Attri, B.K.; Gill, R.K.; Bariwal, J. Review on EGFR Inhibitors: Critical Updates. Mini. Rev. Med. Chem. 2016, 16, 1134–1166. [Google Scholar] [CrossRef] [PubMed]
- Klein-Scory, S.; Wahner, I.; Maslova, M.; Al-Sewaidi, Y.; Pohl, M.; Mika, T.; Ladigan, S.; Schroers, R.; Baraniskin, A. Evolution of RAS Mutational Status in Liquid Biopsies During First-Line Chemotherapy for Metastatic Colorectal Cancer. Front. Oncol. 2020, 10, 1115. [Google Scholar] [CrossRef] [PubMed]
- Van Helden, E.J.; Angus, L.; Menke-van der Houven van Oordt, C.W.; Heideman, D.A.M.; Boon, E.; van Es, S.C.; Radema, S.A.; van Herpen, C.M.L.; de Jan, J.A.D.; de Vries, E.G.E.; et al. RAS and BRAF mutations in cell-free DNA are predictive for outcome of cetuximab monotherapy in patients with tissue-tested RAS wild-type advanced colorectal cancer. Mol. Oncol. 2019, 13, 2361–2374. [Google Scholar] [CrossRef] [PubMed]
- Gazzaniga, P.; Raimondi, C.; Urbano, F.; Cortesi, E. EGFR Inhibitor as Second-Line Therapy in a Patient with Mutant RAS Metastatic Colorectal Cancer: Circulating Tumor DNA to Personalize Treatment. JCO Precis. Oncol. 2018, 2, 1–6. [Google Scholar] [CrossRef]
- De Santiago, B.G.; López-Gómez, M.; Delgado-López, P.D.; Gordo, A.J.; Neria, F.; Thuissard-Vasallo, I.J.; Gómez-Raposo, C.; Tevar, F.Z.; Moreno-Rubio, J.; Hernández, A.M. RAS Mutational Status in Advanced Colorectal Adenocarcinoma Treated with Anti-angiogenics: Preliminary Experience with Liquid Biopsy. In Vivo 2021, 35, 2841–2844. [Google Scholar] [CrossRef]
- Vassy, J.L.; Christensen, K.D.; Schonman, E.F.; Blout, C.L.; Robinson, J.O.; Krier, J.B.; Diamond, P.M.; Lebo, M.; Machini, K.; Azzariti, D.R.; et al. The Impact of Whole-Genome Sequencing on the Primary Care and Outcomes of Healthy Adult Patients: A Pilot Randomized Trial. Ann. Intern. Med. 2017, 167, 159–169. [Google Scholar] [CrossRef]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.-L.; Christie, M.; et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra92. [Google Scholar] [CrossRef]
- Sclafani, F.; Chau, I.; Cunningham, D.; Hahne, J.C.; Vlachogiannis, G.; Eltahir, Z.; Lampis, A.; Braconi, C.; Kalaitzaki, E.; De Castro, D.G.; et al. KRAS and BRAF mutations in circulating tumour DNA from locally advanced rectal cancer. Sci. Rep. 2018, 8, 1445. [Google Scholar] [CrossRef]
- Weiser, M.R. AJCC 8th Edition: Colorectal Cancer. Ann. Surg. Oncol. 2018, 25, 1454–1455. [Google Scholar] [CrossRef]
| Variable | Negative Liquid Biopsy Patients (n = 25) | Positive Liquid Biopsy Patients (n = 26) | p | Total (n = 51) |
|---|---|---|---|---|
| Age (years) * | 65 (43–83) | 67 (50–81) | 0.89 | 66 (43–83) |
| Gender | ||||
| Female | 5 (20%) | 9 (35%) | 0.51 | 14 (27%) |
| Male | 20 (80%) | 17 (55%) | 37 (63%) | |
| Site of PT | ||||
| Right colon | 7 (28%) | 11 (42%) | 18 (35%) | |
| Left colon | 1 (4%) | 3 (12%) | 0.11 | 4 (8%) |
| Rectum | 17 (68%) | 12 (46%) | 29 (57%) | |
| Treatment type | ||||
| Chemotherapy + anti-EGFR | 19 (76%) | 5 (19%) | 24 (47%) | |
| Chemotherapy + anti-VEGF | 2 (8%) | 16 (62%) | <0.001 | 18 (35%) |
| Chemotherapy | 4 (16%) | 5 (19%) | 9 (18%) | |
| CEA serum levels * | ||||
| ≤7.5 ng/mL | 10 (40%) | 1 (4%) | 0.002 | 11 (22%) |
| >7.5 ng/mL | 15 (60%) | 25 (96%) | 40 (78%) | |
| Mutational status of PT | ||||
| Mutated | 16 (64%) | 3 (12%) | <0.001 | 19 (27%) |
| Wild type | 9 (36%) | 23 (89%) | 32 (63%) | |
| Number of deaths | 15 (60%) | 21 (81%) | 0.009 | 36 (71%) |
| Overall survival (months) | 52 (38–67) | 24 (16–31) | <0.001 | 40 (30–51) |
| Variable | N * | Univariate Analysis | Multivariate Analysis | HR (95% CI) |
|---|---|---|---|---|
| Age | ||||
| <66 years | 26 (51%) | 0.889 | ||
| ≥66 years | 25 (49%) | |||
| Gender | ||||
| Male | 37 (76%) | 0.879 | ||
| Female | 14 (24%) | |||
| Site of PT | ||||
| Right colon | 18 (35%) | |||
| Left colon | 4 (8%) | 0.114 | ||
| Rectum | 29 (57%) | |||
| CEA serum levels | ||||
| <7.5 ng | 11 (22%) | 0.073 | NS | |
| ≥7.5 ng | 40 (78%) | |||
| Treatment type | ||||
| Chemotherapy + anti-EGFR | 24 (47%) | |||
| Chemotherapy + anti-VEGF | 18 (35%) | 0.053 | NS | |
| Chemotherapy | 9 (18%) | |||
| Microsatellite instability | ||||
| No | 50 (98%) | 0.879 | ||
| Yes | 1 (2%) | |||
| KRAS mutation in PT | ||||
| Yes | 23 (45%) | 0.050 | NS | |
| No | 28 (55%) | |||
| NRAS mutation in PT | ||||
| Yes | 1 (2%) | 0.487 | ||
| No | 50 (98%) | |||
| BRAF mutation in PT | ||||
| Yes | 1 (2%) | 0.937 | ||
| No | 50 (98%) | |||
| PIK3CA mutation in PT | ||||
| Yes | 12 (24%) | 0.728 | ||
| No | 39 (76%) | |||
| KRAS mutation in plasma | ||||
| Yes | 23 (45%) | 0.004 | NS | |
| No | 28 (55%) | |||
| NRAS mutation in plasma | ||||
| Yes | 3 (6%) | 0.420 | ||
| No | 48 (94%) | |||
| PIK3CA mutation in plasma | ||||
| Yes | 8 (16%) | 0.185 | ||
| No | 43 (84%) | |||
| BRAF mutation in plasma | ||||
| Yes | 2 (4%) | 0.472 | ||
| No | 49 (96%) | |||
| Liquid biopsy at diagnosis | ||||
| Positive | 26 (53%) | 0.005 | 0.007 | 0.388 (0.196–0.768) |
| Negative | 25 (47%) | |||
| Mutations in liquid biopsy | ||||
| Wild type | 25 (49%) | <0.001 | ||
| 1 mutation | 19 (37%) | 0.001 | 2.018 (0.316–3.095) | |
| 2 mutations | 6 (12%) | |||
| 3 mutations | 1 (2%) |
| Plasma | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| KRAS | R/p | NRAS | R/p | BRAF | R/p | PIK3CA | R/p | |||||
| Primary Tumor | WT (n = 29) | Mutated (n = 22) | WT (n = 48) | Mutated (n = 3) | WT (n = 49) | Mutated (n = 2) | WT (n = 43) | Mutated (n = 8) | ||||
| KRAS | ||||||||||||
| WT (n = 28) | 24 (86) | 4 (14) | 0.65/<0.001 | |||||||||
| Mutated (n = 23) | 5 (22) | 18 (78) | ||||||||||
| NRAS | ||||||||||||
| WT (n = 50) | 48 (96) | 2 (4) | 0.57/0.05 | |||||||||
| Mutated (n = 1) | 0 (0) | 1 (100) | ||||||||||
| BRAF | ||||||||||||
| WT (n = 50) | 49 (98) | 1 (2) | 0.77/<0.001 | |||||||||
| Mutated (n = 1) | 0 (0) | 1 (100) | ||||||||||
| PIK3CA | ||||||||||||
| WT (n = 39) | 37 (95) | 2 (5) | 0.53/0.001 | |||||||||
| Mutated (n = 12) | 6 (50) | 6 (50) | ||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sayagués, J.M.; Montero, J.C.; Jiménez-Pérez, A.; del Carmen, S.; Rodríguez, M.; Vidal Tocino, R.; Montero, E.; Sanz, J.; Abad, M. Analysis of Circulating Tumor DNA in Synchronous Metastatic Colorectal Cancer at Diagnosis Predicts Overall Patient Survival. Int. J. Mol. Sci. 2023, 24, 8438. https://doi.org/10.3390/ijms24098438
Sayagués JM, Montero JC, Jiménez-Pérez A, del Carmen S, Rodríguez M, Vidal Tocino R, Montero E, Sanz J, Abad M. Analysis of Circulating Tumor DNA in Synchronous Metastatic Colorectal Cancer at Diagnosis Predicts Overall Patient Survival. International Journal of Molecular Sciences. 2023; 24(9):8438. https://doi.org/10.3390/ijms24098438
Chicago/Turabian StyleSayagués, José María, Juan Carlos Montero, Andrea Jiménez-Pérez, Sofía del Carmen, Marta Rodríguez, Rosario Vidal Tocino, Enrique Montero, Julia Sanz, and Mar Abad. 2023. "Analysis of Circulating Tumor DNA in Synchronous Metastatic Colorectal Cancer at Diagnosis Predicts Overall Patient Survival" International Journal of Molecular Sciences 24, no. 9: 8438. https://doi.org/10.3390/ijms24098438
APA StyleSayagués, J. M., Montero, J. C., Jiménez-Pérez, A., del Carmen, S., Rodríguez, M., Vidal Tocino, R., Montero, E., Sanz, J., & Abad, M. (2023). Analysis of Circulating Tumor DNA in Synchronous Metastatic Colorectal Cancer at Diagnosis Predicts Overall Patient Survival. International Journal of Molecular Sciences, 24(9), 8438. https://doi.org/10.3390/ijms24098438





