Diverse Virulence Attributes of Pantoea alfalfae sp. nov. CQ10 Responsible for Bacterial Leaf Blight in Alfalfa Revealed by Genomic Analysis
Abstract
1. Introduction
2. Results
2.1. Isolation of a Novel Pantoea Species
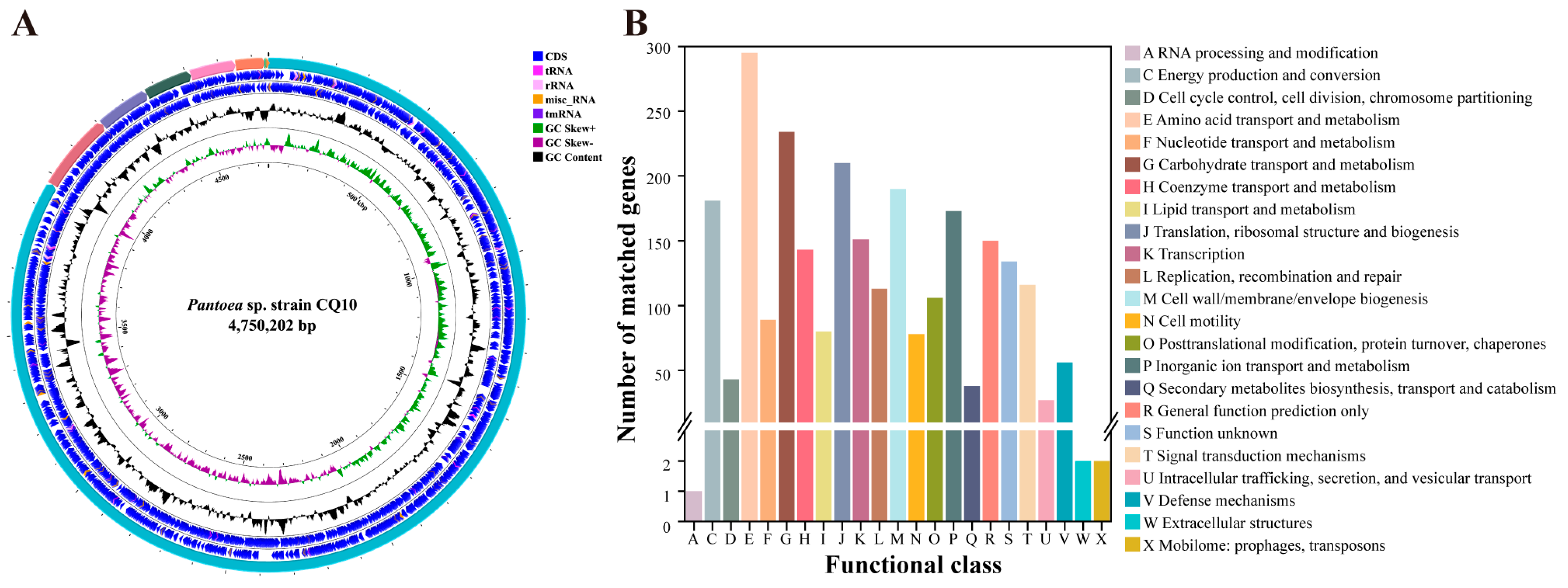
2.2. General Genome Features of Strain CQ10
2.3. Taxonomic Identification of Pantoea sp. Strain CQ10
2.4. Biochemical Characteristics
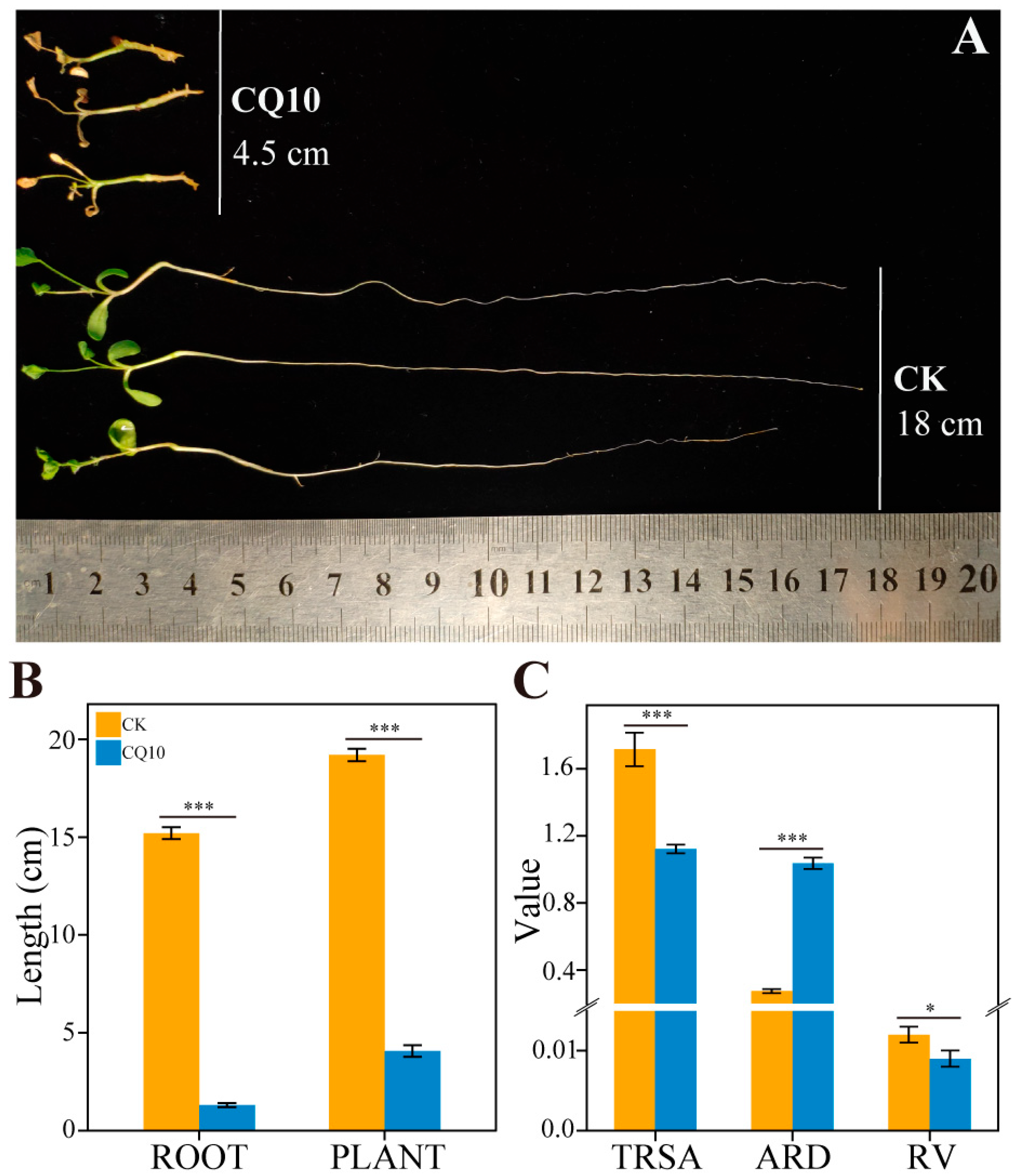
2.5. Alfalfa Bacterial Leaf Blight Caused by Strain CQ10
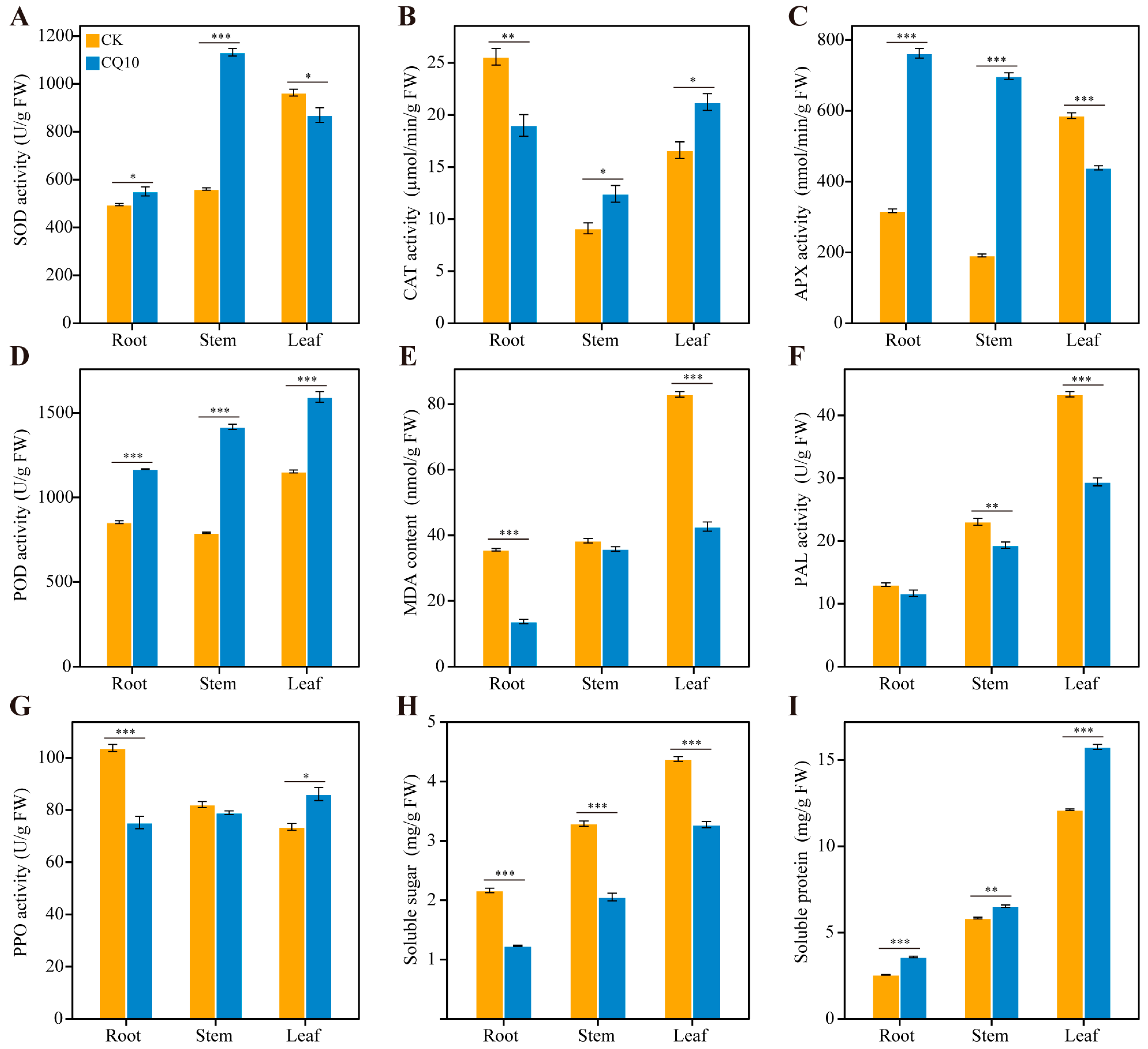
2.6. Changes in the Antioxidative System of Alfalfa Plants
2.7. Analyses of Defense–Related Enzymes in Alfalfa Hosts
2.8. Effect of Strain CQ10 Inoculation on Soluble Sugar and Soluble Protein in Alfalfa Plants
2.9. Correlation between Nine Physiological Parameters
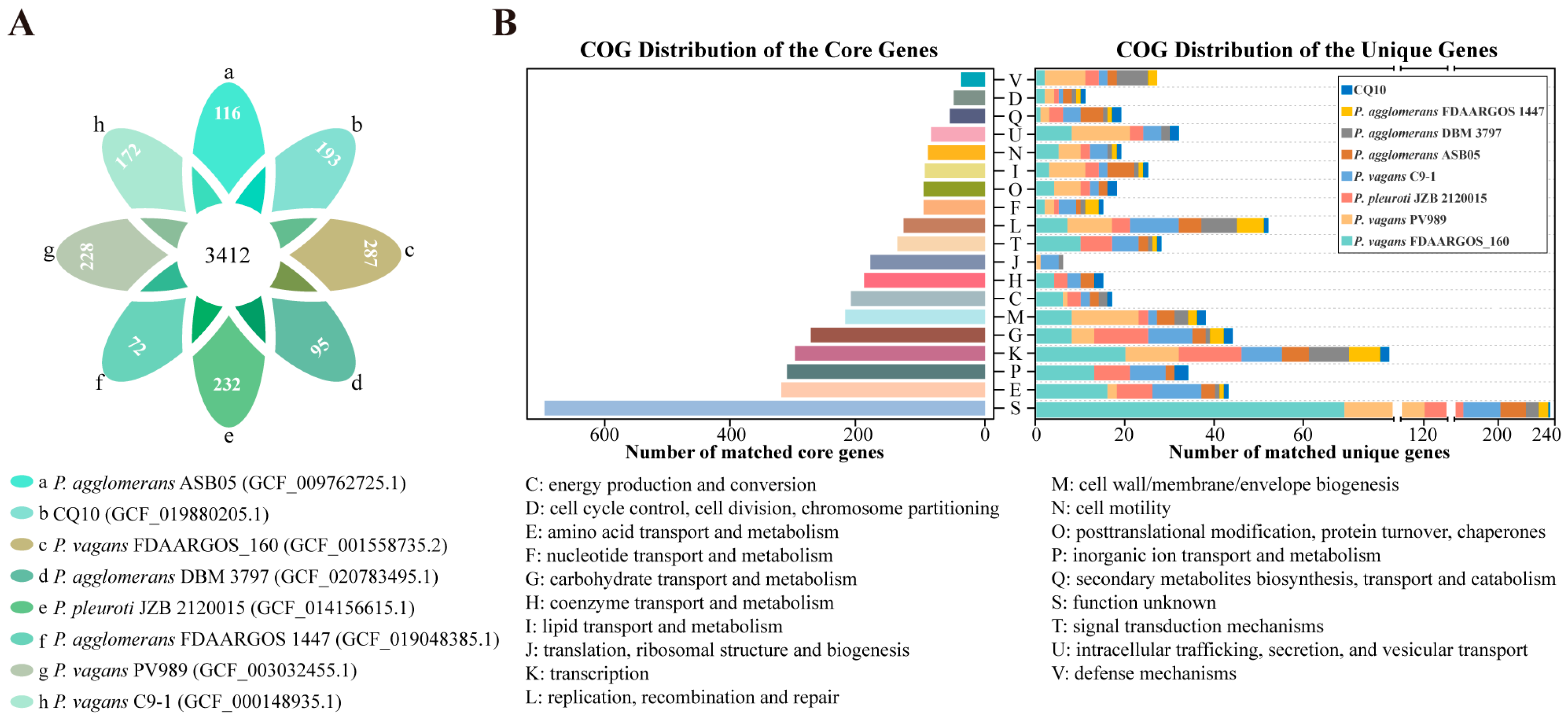
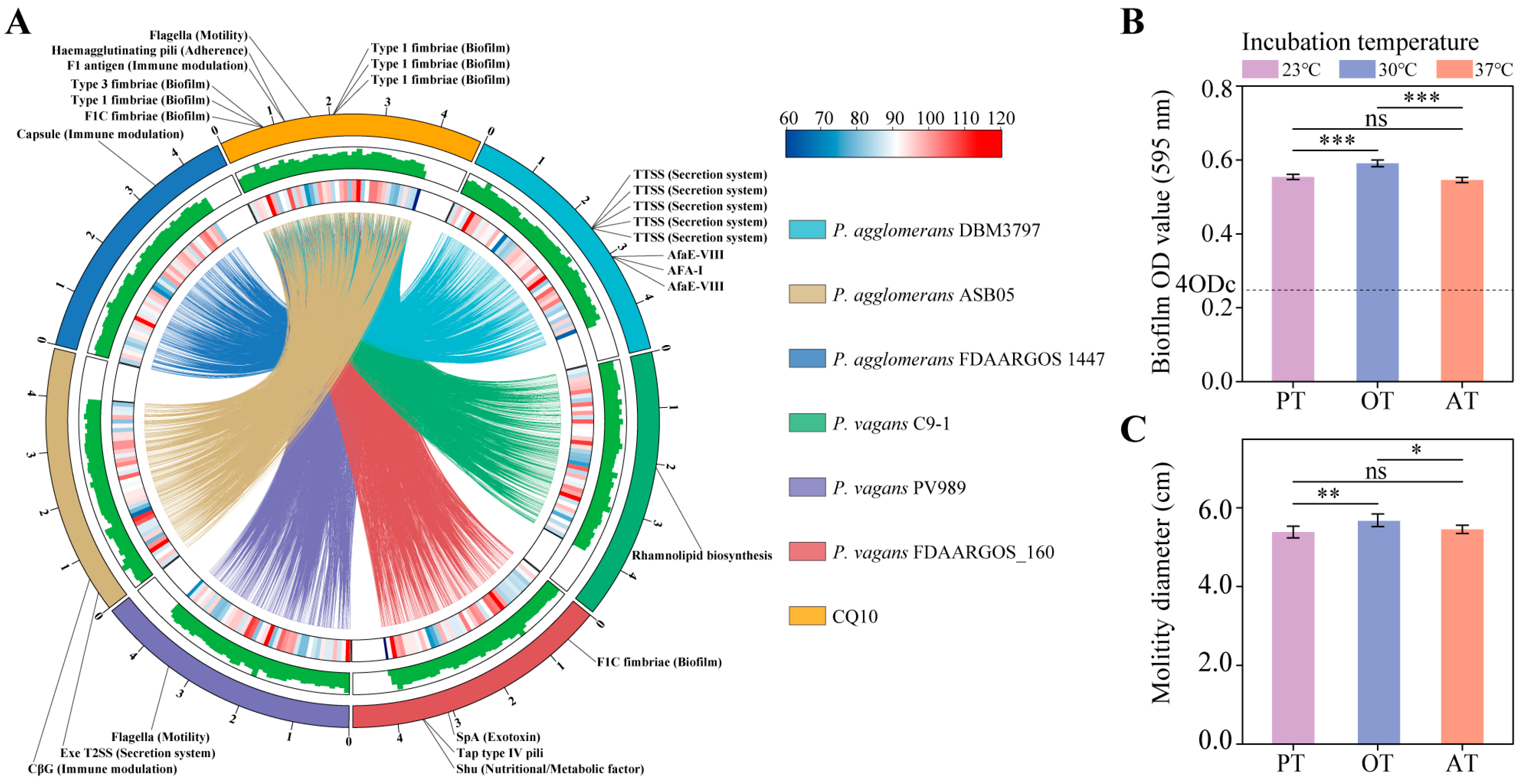
2.10. Pan–Genome Analysis of Pantoea Strains
2.11. Comparative and Functional Genome Analyses
2.12. Biofilm Formation and Motility of Strain CQ10 In Vitro
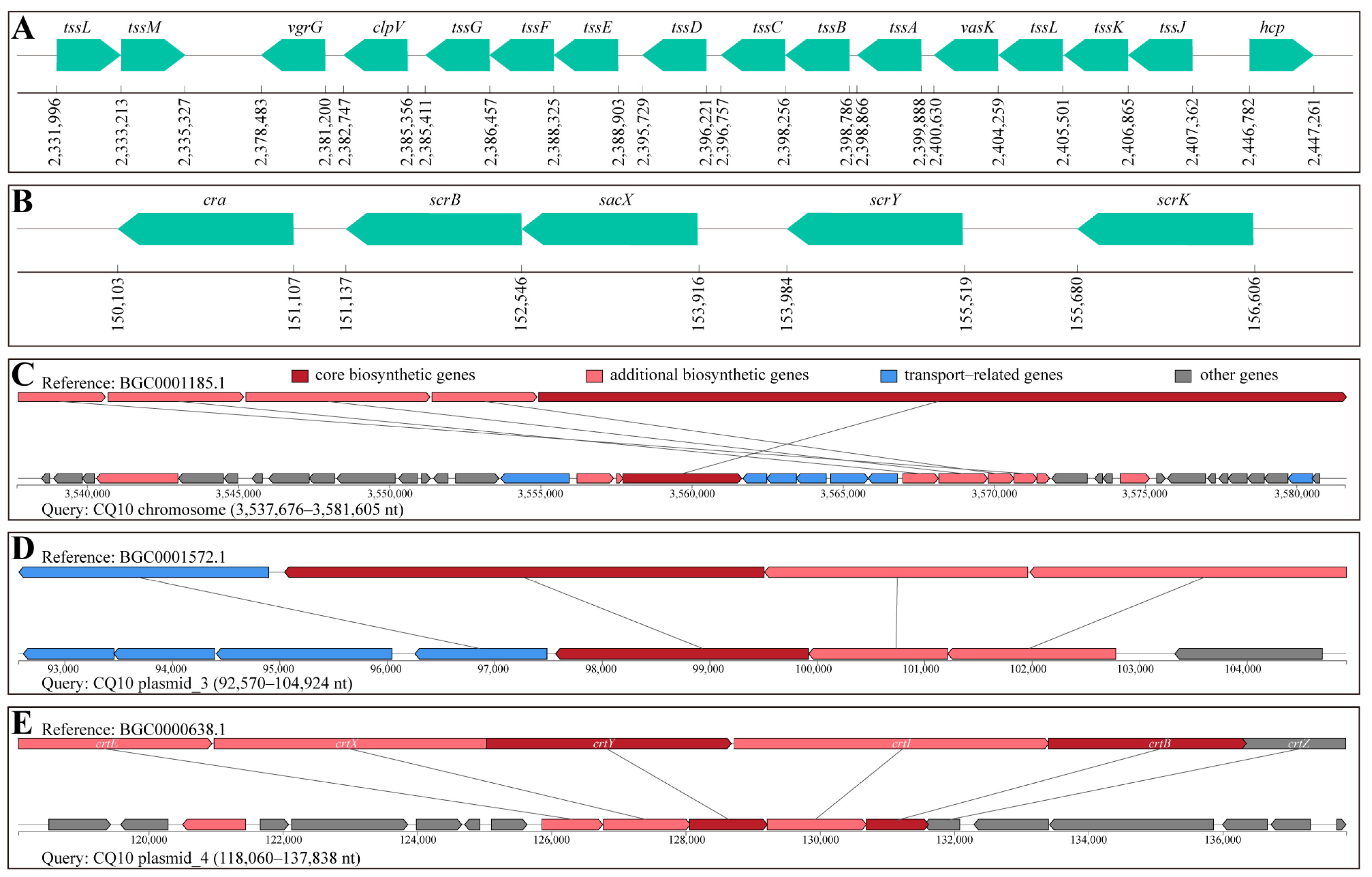
2.13. Analysis of Virulence Attribute of Strain CQ10
3. Discussion
4. Materials and Methods
4.1. Isolation and Characterization of Strain CQ10
4.2. DNA Extraction and 16S rRNA Gene Sequencing
4.3. Whole Genome Sequencing, Assembly, and Annotation
4.4. Phylogenomic Comparison
4.5. Alfalfa Plant Inoculation Assay
4.6. Alfalfa Leaf Gas Exchange Measurements
4.7. Measurement of Various Physiological Indices
4.8. Pan–Genome Analysis
4.9. Genome Comparative Analysis
4.10. Biofilm and Motility Assays
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yu, P.; Christensen, D.A.; McKinnon, J.J.; Markert, J.D. Effect of variety and maturity stage on chemical composition, carbohydrate and protein subfractions, in vitro rumen degradability and energy values of timothy and alfalfa. Can. J. Anim. Sci. 2003, 83, 279–290. [Google Scholar] [CrossRef]
- Heydari, A.; Khodakaramian, G.; Zafari, D. Occurrence, genetic diversity and pathogenicity characteristics of Pseudomonas viridiflava inducing alfalfa bacterial wilt and crown root rot disease in Iran. Eur. J. Plant Pathol. 2014, 139, 299–307. [Google Scholar] [CrossRef]
- Yaripour, Z.; Mohsen Taghavi, S.; Osdaghi, E.; Lamichhane, J.R. Host range and phylogenetic analysis of Xanthomonas alfalfae causing bacterial leaf spot of alfalfa in Iran. Eur. J. Plant Pathol. 2018, 150, 267–274. [Google Scholar] [CrossRef]
- Kataria, R.; Duhan, N.; Kaundal, R. Computational systems biology of alfalfa–bacterial blight host–pathogen interactions: Uncovering the complex molecular networks for developing durable disease resistant crop. Front. Plant Sci. 2022, 12, 807354. [Google Scholar] [CrossRef]
- Kimball, A.M. Risky Trade: Infectious Disease in the Era of Global Trade; Routledge: London, UK, 2016. [Google Scholar]
- Yao, B.; Huang, R.; Zhang, Z.F.; Shi, S.L. Seed–borne Erwinia persicina affects the growth and physiology of alfalfa (Medicago sativa L.). Front. Microbiol. 2022, 13, 891188. [Google Scholar] [CrossRef]
- Zhang, Z.F. Seed–Borne Bacteria of Lucerne (Medicago sativa) and Their Pathogenicity. Ph.D Thesis, Lanzhou University, Lanzhou, China, 2013. [Google Scholar]
- López, J.L.; Alvarez, F.; Príncipe, A.; Lozano, M.J.; Draghi, W.O.; Jofré, E.; Lagares, A. Isolation, taxonomic analysis, and phenotypic characterization of bacterial endophytes present in alfalfa (Medicago sativa) seeds. J. Biotechnol. 2018, 267, 55–62. [Google Scholar] [CrossRef]
- Charkowski, A.O.; Barak, J.D.; Sarreal, C.Z.; Mandrell, R.E. Differences in growth of Salmonella enterica and Escherichia coli O157:H7 on alfalfa sprouts. Appl. Environ. Microbiol. 2002, 68, 3114–3120. [Google Scholar] [CrossRef]
- Brady, C.L.; Cleenwerck, I.; Venter, S.; Vancanneyt, M.; Swings, J.; Coutinho, T. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 2008, 31, 447–460. [Google Scholar] [CrossRef]
- Coutinho, T.A.; Venter, S.N. Pantoea ananatis: An unconventional plant pathogen. Mol. Plant Pathol. 2009, 10, 325–335. [Google Scholar] [CrossRef]
- Weller-Stuart, T.; De Maayer, P.; Coutinho, T. Pantoea ananatis: Genomic insights into a versatile pathogen. Mol. Plant Pathol. 2017, 18, 1191–1198. [Google Scholar] [CrossRef]
- Asai, N.; Koizumi, Y.; Yamada, A.; Sakanashi, D.; Watanabe, H.; Kato, H.; Shiota, A.; Hagihara, M.; Suematsu, H.; Yamagishi, Y.; et al. Pantoea dispersa bacteremia in an immunocompetent patient: A case report and review of the literature. J. Med. Case Rep. 2019, 13, 33. [Google Scholar] [CrossRef]
- Lotte, L.; Sindt, A.; Ruimy, R.; Neri, D.; Lotte, R.; Weiss, N.; Vassallo, M. Description of the first case of catheter–related bloodstream infection due to Pantoea eucrina in a cancer patient. SN Compr. Clin. Med. 2019, 1, 142–145. [Google Scholar] [CrossRef]
- Toh, W.K.; Loh, P.C.; Wong, H.L. First report of leaf blight of rice caused by Pantoea ananatis and Pantoea dispersa in Malaysia. Plant Dis. 2019, 103, 1764. [Google Scholar] [CrossRef]
- Tufail, M.R.; Fayyaz, A.; Sarfraz, S.; Waqas, M.; Ahmad, M.Z.; Hameed, A.; Masood, A.; Abdin, Z.U.; Amrao, L.; Sahi, S.T.; et al. First report of Pantoea leaf blight of cotton caused by Pantoea spp. in Punjab, Pakistan. Plant Dis. 2020, 104, 1534. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Mackiewicz, B.; Kinga Lemieszek, M.; Golec, M.; Milanowski, J. Pantoea agglomerans: A mysterious bacterium of evil and good. Part III. deleterious effects: Infections of humans, animals and plants. Ann. Agric. Environ. Med. 2016, 23, 197–205. [Google Scholar] [CrossRef]
- Josenhans, C.; Suerbaum, S. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 2002, 291, 605–614. [Google Scholar] [CrossRef]
- Yaron, S.; Römling, U. Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechnol. 2014, 7, 496–516. [Google Scholar] [CrossRef]
- Shyntum, D.Y.; Theron, J.; Venter, S.N.; Moleleki, L.N.; Toth, I.K.; Coutinho, T.A. Pantoea ananatis utilizes a type VI secretion system for pathogenesis and bacterial competition. Mol. Plant–Microbe Interact. 2015, 28, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.L.; Venter, S.N.; Cleenwerck, I.; Vandemeulebroecke, K.; De Vos, P.; Coutinho, T.A. Transfer of Pantoea citrea, Pantoea punctata and Pantoea terrea to the genus Tatumella emend. as Tatumella citrea comb. nov., Tatumella punctata comb. nov. and Tatumella terrea comb. nov. and description of Tatumella morbirosei sp. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 484–494. [Google Scholar] [CrossRef]
- Palmer, M.; Steenkamp, E.T.; Coetzee, M.P.A.; Avontuur, J.R.; Chan, W.Y.; Van Zyl, E.; Blom, J.; Venter, S.N. Mixta gen. nov., a new genus in the Erwiniaceae. Int. J. Syst. Evol. Microbiol. 2018, 68, 1396–1407. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Tambong, J.T. Taxogenomics and systematics of the genus Pantoea. Front. Microbiol. 2019, 10, 2463. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA–DNA hybridization for microbial species delineation by means of genome–to–genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Walterson, A.M.; Stavrinides, J. Pantoea: Insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015, 39, 968–984. [Google Scholar] [CrossRef]
- Rossez, Y.; Wolfson, E.B.; Holmes, A.; Gally, D.L.; Holden, N.J. Bacterial flagella: Twist and stick, or dodge across the kingdoms. PLoS Pathog. 2015, 11, e1004483. [Google Scholar] [CrossRef] [PubMed]
- Chaban, B.; Hughes, H.V.; Beeby, M. The flagellum in bacterial pathogens: For motility and a whole lot more. Semin. Cell Dev. Biol. 2015, 46, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Sauer, K.; Stoodley, P.; Goeres, D.M.; Hall-Stoodley, L.; Burmølle, M.; Stewart, P.S.; Bjarnsholt, T. The biofilm life cycle: Expanding the conceptual model of biofilm formation. Nat. Rev. Microbiol. 2022, 20, 608–620. [Google Scholar] [CrossRef]
- Castiblanco, L.F.; Sundin, G.W. New insights on molecular regulation of biofilm formation in plant–associated bacteria. J. Integr. Plant Biol. 2016, 58, 362–372. [Google Scholar] [CrossRef]
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Kapitein, N.; Mogk, A. Deadly syringes: Type VI secretion system activities in pathogenicity and interbacterial competition. Curr. Opin. Microbiol. 2013, 16, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Bernal, P.; Llamas, M.A.; Filloux, A. Type VI secretion systems in plant–associated bacteria. Environ. Microbiol. 2018, 20, 1–15. [Google Scholar] [CrossRef]
- De Maayer, P.; Venter, S.N.; Kamber, T.; Duffy, B.; Coutinho, T.A.; Smits, T.H. Comparative genomics of the type VI secretion systems of Pantoea and Erwinia species reveals the presence of putative effector islands that may be translocated by the VgrG and Hcp proteins. BMC Genom. 2011, 12, 576. [Google Scholar] [CrossRef]
- Horbowicz, M.; Obendorf, R.L.; McKersie, B.D.; Viands, D.R. Soluble saccharides and cyclitols in alfalfa (Medicago sativa L.) somatic embryos, leaflets, and mature seeds. Plant Sci. 1995, 109, 191–198. [Google Scholar] [CrossRef]
- Jung, M.Y.; Jung, H.M.; Lee, J.; Oh, M.K. Alleviation of carbon catabolite repression in Enterobacter aerogenes for efficient utilization of sugarcane molasses for 2, 3-butanediol production. Biotechnol. Biofuels 2015, 8, 106. [Google Scholar] [CrossRef]
- Reid, S.J.; Abratt, V.R. Sucrose utilisation in bacteria: Genetic organisation and regulation. Appl. Microbiol. biotechnol. 2005, 67, 312–321. [Google Scholar] [CrossRef]
- Expert, D.; Enard, C.; Masclaux, C. The role of iron in plant host–pathogen interactions. Trends Microbiol. 1996, 4, 232–237. [Google Scholar] [CrossRef]
- Khun, H.H.; Kirby, S.D.; Lee, B.C. A Neisseria meningitidis fbpABC mutant is incapable of using nonheme iron for growth. Infect. Immun. 1998, 66, 2330–2336. [Google Scholar] [CrossRef]
- Anderson, D.S.; Adhikari, P.; Nowalk, A.J.; Chen, C.Y.; Mietzner, T.A. The hFbpABC transporter from Haemophilus influenzae functions as a binding–protein–dependent ABC transporter with high specificity and affinity for ferric iron. J. Bacteriol. 2004, 186, 6220–6229. [Google Scholar] [CrossRef]
- Liu, Y.; Kong, D.; Wu, H.L.; Ling, H.Q. Iron in plant–pathogen interactions. J. Exp. Bot. 2021, 72, 2114–2124. [Google Scholar] [CrossRef] [PubMed]
- Abergel, R.J.; Zawadzka, A.M.; Hoette, T.M.; Raymond, K.N. Enzymatic hydrolysis of trilactone siderophores: Where chiral recognition occurs in enterobactin and bacillibactin iron transport. J. Am. Chem. Soc. 2009, 131, 12682–12692. [Google Scholar] [CrossRef]
- Choi, O.; Cho, J.; Kang, B.; Lee, Y.; Kim, J. Negatively regulated aerobactin and desferrioxamine E by Fur in Pantoea ananatis are required for full siderophore production and antibacterial activity, but not for virulence. Appl. Environ. Microbiol. 2022, 88, e02405-21. [Google Scholar] [CrossRef] [PubMed]
- Howard, M.B.; Hutcheson, S.W. Growth dynamics of Salmonella enterica strains on alfalfa sprouts and in waste seed irrigation water. Appl. Environ. Microbiol. 2003, 69, 548–553. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow Chichester, M., Eds.; John Wiley & Sons: Hoboken, NJ, USA, 1991; pp. 115–175. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Chen, Y.X.; Chen, Y.S.; Shi, C.M.; Huang, Z.B.; Zhang, Y.; Li, S.K.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration–supported software for integrated quality control and preprocessing of high–throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; Sahu, J.; Vijaylakshmi lyer, S.; Khamari, L.; De Silva, N.; et al. PHI–base in 2022: A multi–species phenotype database for Pathogen–Host Interactions. Nucleic Acids Res. 2022, 50, D837–D847. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate–active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence–based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Von Caemmerer, S.; Farquhar, G.D. Some relations between the biochemistry of photosynthesis and the gas exchange of leaves. Planta 1981, 153, 376–387. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, J.; Yang, J.; Sun, S.; Xiao, J.; Yu, J. PGAP: Pan–genomes analysis pipeline. Bioinformatics 2012, 28, 416–418. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter–plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Meth. 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Mahmud, S.A.; Qureshi, M.A.; Sapkota, M.; Pellegrino, M.W. A pathogen branched–chain amino acid catabolic pathway subverts host survival by impairing energy metabolism and the mitochondrial UPR. PLoS Pathog. 2020, 16, e1008918. [Google Scholar] [CrossRef]


| Characteristic | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Utilization of | ||||
| Amygdalina | + | − | − | + |
| L–fucose | + | − | − | + |
| L–pyroglutamic acid | + | − | − | − |
| Tween 40 | − | − | + | + |
| D–fucose | + | − | + | + |
| D–arabitol | + | + | − | + |
| Raffinose | − | − | − | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, B.; Huang, R.; Zhang, Z.; Shi, S. Diverse Virulence Attributes of Pantoea alfalfae sp. nov. CQ10 Responsible for Bacterial Leaf Blight in Alfalfa Revealed by Genomic Analysis. Int. J. Mol. Sci. 2023, 24, 8138. https://doi.org/10.3390/ijms24098138
Yao B, Huang R, Zhang Z, Shi S. Diverse Virulence Attributes of Pantoea alfalfae sp. nov. CQ10 Responsible for Bacterial Leaf Blight in Alfalfa Revealed by Genomic Analysis. International Journal of Molecular Sciences. 2023; 24(9):8138. https://doi.org/10.3390/ijms24098138
Chicago/Turabian StyleYao, Bo, Rong Huang, Zhenfen Zhang, and Shangli Shi. 2023. "Diverse Virulence Attributes of Pantoea alfalfae sp. nov. CQ10 Responsible for Bacterial Leaf Blight in Alfalfa Revealed by Genomic Analysis" International Journal of Molecular Sciences 24, no. 9: 8138. https://doi.org/10.3390/ijms24098138
APA StyleYao, B., Huang, R., Zhang, Z., & Shi, S. (2023). Diverse Virulence Attributes of Pantoea alfalfae sp. nov. CQ10 Responsible for Bacterial Leaf Blight in Alfalfa Revealed by Genomic Analysis. International Journal of Molecular Sciences, 24(9), 8138. https://doi.org/10.3390/ijms24098138






