AI-Driven De Novo Design and Molecular Modeling for Discovery of Small-Molecule Compounds as Potential Drug Candidates Targeting SARS-CoV-2 Main Protease
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Development of a Deep Generative Neural Network
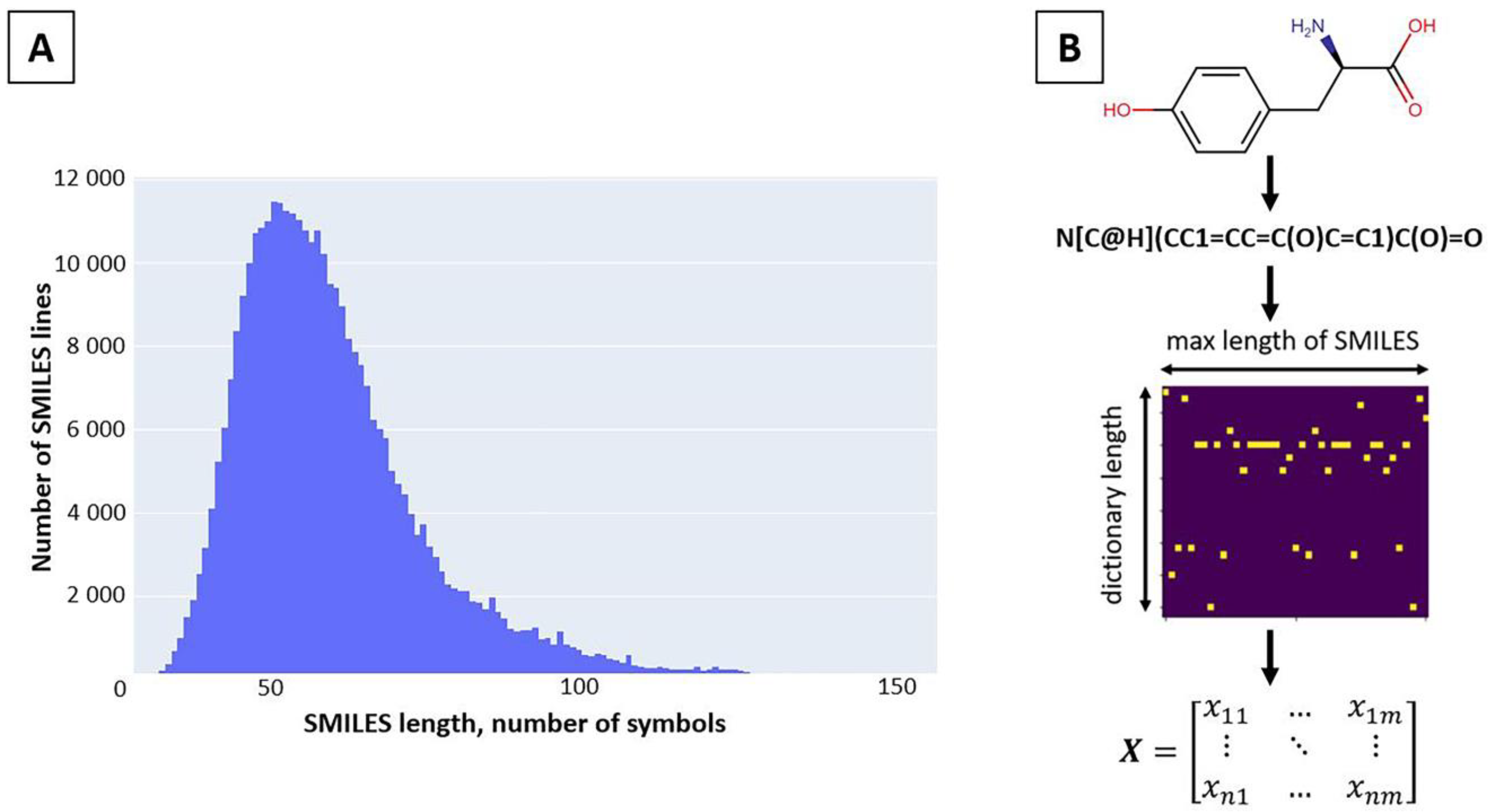
3.1.1. Preparing the Training Dataset
- (A)
- Building pharmacophore models and virtual screening of chemical databases
- (B)
- Molecular docking
3.1.2. SMILES Space Revision and Vectorization
3.1.3. Restoration of Three-Dimensional Structures of Generated Molecules
3.1.4. Preparation of the Mpro Structure
3.1.5. Preparation of Ligand Structures
3.1.6. Computational Protocol of Molecular Docking
3.1.7. Architectures of Deep Generative Models
3.1.8. Training the Models
3.1.9. Deep Learning-Based Compounds’ Generation
3.2. De Novo Design of Potential Inhibitors Targeting SARS-CoV-2 Mpro
3.2.1. Generation of a Wide Set of Potential SARS-CoV-2 Mpro Ligands
3.2.2. Molecular Docking of the Generated Compounds with SARS-CoV-2 Mpro
3.2.3. Molecular Dynamics Simulations
3.2.4. Analysis of Interaction Modes and Binding Affinity Profile
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Katsila, T.; Spyroulias, G.A.; Patrinos, G.P.; Matsoukas, M.-T. Computational approaches in target identification and drug discovery. Comput. Struct. Biotechnol. J. 2016, 14, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Knight-Schrijver, V.R.; Chelliah, V.; Cucurull-Sanchez, L.; Le Novère, N. The promises of quantitative systems pharmacology modelling for drug development. Comput. Struct. Biotechnol. J. 2016, 4, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Vamathevan, J.; Clark, D.; Czodrowski, P.; Dunham, I.; Ferran, E.; Lee, G.; Li, B.; Madabhushi, A.; Shah, P.; Spitzer, M.; et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug. Discov. 2019, 18, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.F.; Maltarollo, V.G.; Oliveira, P.R.; da Silva, A.B.F.; Honorio, K.M. Advances and perspectives in applying deep learning for drug design and discovery. Front. Robot. AI 2019, 6, 108. [Google Scholar] [CrossRef]
- Dobchev, D.A.; Pillai, G.G.; Karelson, M. In silico machine learning methods in drug development. Curr. Top. Med. Chem. 2014, 14, 913–1922. [Google Scholar] [CrossRef]
- Cherkasov, A.; Muratov, E.N.; Fourches, D.; Varnek, A.; Baskin, I.I.; Cronin, M.; Dearden, J.; Gramatica, P.; Martin, Y.C.; Todeschini, R.; et al. QSAR modeling: Where have you been? Where are you going to? J. Med. Chem. 2014, 57, 4977–5010. [Google Scholar] [CrossRef]
- Kinnings, S.L.; Liu, N.; Tonge, P.J.; Jackson, R.M.; Xie, L.; Bourne, P.E. A Machine learning-based method to improve docking scoring functions and its application to drug repurposing. J. Chem. Inf. Model. 2011, 51, 408–419. [Google Scholar] [CrossRef]
- Agastheeswaramoorthy, K.; Sevilimedu, A. Drug REpurposing using AI/ML tools—For Rare Diseases (DREAM-RD): A case study with Fragile X Syndrome (FXS). bioRxiv 2020. [Google Scholar] [CrossRef]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Zıdek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Li, H.; Sze, K.-H.; Lu, G.; Ballester, P.J. Machine-learning scoring functions for structure-based virtual screening. WIREs Comput. Mol. Sci. 2020, 11, e1478. [Google Scholar] [CrossRef]
- Xiong, G.-L.; Ye, W.-L.; Shen, C.; Lu, A.-P.; Hou, T.-J.; Cao, D.-S. Improving structure-based virtual screening performance via learning from scoring function components. Brief. Bioinform. 2020, 22, bbaa094. [Google Scholar] [CrossRef] [PubMed]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A deep learning approach to antibiotic discovery. Cell 2020, 180, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Timmons, P.B.; Hewage, C.M. ENNAVIA is a novel method which employs neural networks for antiviral and anti-coronavirus activity prediction for therapeutic peptides. Brief. Bioinform. 2021, 22, bbab258. [Google Scholar] [CrossRef] [PubMed]
- Andrianov, A.M.; Nikolaev, G.I.; Shuldov, N.A.; Bosko, I.P.; Anischenko, A.I.; Tuzikov, A.V. Application of deep learning and molecular modeling to identify small drug-like compounds as potential HIV-1 entry inhibitors. J. Biomol. Struct. Dyn. 2022, 40, 7555–7573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ye, T.; Xi, H.; Juhas, M.; Li, J. Deep learning driven drug discovery: Tackling Severe Acute Respiratory Syndrome Coronavirus 2. Front. Microbiol. 2021, 12, 739684. [Google Scholar] [CrossRef]
- Mercado, R.; Rastemo, T.; Lindelöf, E.; Klambauer, G.; Engkvist, O.; Chen, H.; Bjerrum, E.J. Practical notes on building molecular graph generative models. Appl. AI Lett. 2020, 1. [Google Scholar] [CrossRef]
- Arús-Pous, J.; Blaschke, T.; Ulander, S.; Reymond, J.L.; Chen, H.; Engkvist, O. Exploring the GDB-13 chemical space using deep generative models. J. Cheminform. 2019, 11, 20. [Google Scholar] [CrossRef]
- Prykhodko, O.; Johansson, S.V.; Kotsias, P.C.; Arús-Pous, J.; Bjerrum, E.J.; Engkvist, O.; Chen, H. A de novo molecular generation method using latent vector based generative adversarial network. J. Cheminform. 2019, 11, 74. [Google Scholar] [CrossRef]
- Polykovskiy, D.; Zhebrak, A.; Vetrov, D.; Ivanenkov, Y.; Aladinskiy, V.; Mamoshina, P.; Bozdaganyan, M.; Aliper, A.; Zhavoronkov, A.; Kadurin, A. Entangled conditional adversarial autoencoder for de novo drug discovery. Mol. Pharm. 2018, 15, 4398–4405. [Google Scholar] [CrossRef]
- Zhang, J.; Mercado, R.; Engkvist, O.; Chen, H. Comparative study of deep generative models on chemical space coverage. J. Chem. Inf. Model. 2021, 61, 2572–2581. [Google Scholar] [CrossRef]
- Zhavoronkov, A.; Ivanenkov, Y.A.; Aliper, A.; Veselov, M.S.; Aladinskiy, V.A.; Aladinskaya, A.V.; Terentiev, V.A.; Polykovskiy, D.A.; Kuznetsov, M.D.; Asadulaev, A.; et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019, 37, 1038–1040. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New data content and improved web interfaces. Nucl. Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V.; Hayashi, Y.; Jung, S.H. An overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL protease inhibitors: Peptidomimetics and small molecule chemotherapy. J. Med.Chem. 2016, 59, 6595–6628. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Gao, F. An overview of potential inhibitors targeting non-structural proteins 3 (PLpro and Mac1) and 5 (3CLpro/Mpro) of SARS-CoV-2. Comput. Struct. Biotechnol. J. 2021, 19, 4868–4883. [Google Scholar] [CrossRef]
- Ullrich, S.; Nitsche, C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020, 30, 127377. [Google Scholar] [CrossRef]
- Katre, S.G.; Asnani, A.J.; Pratyush, K.; Sakharkar, N.G.; Bhope, A.G.; Sawarkar, K.T.; Nimbekar, V.S. Review on development of potential inhibitors of SARS-CoV-2 main protease (MPro). Future J. Pharm. Sci. 2022, 8, 36. [Google Scholar] [CrossRef]
- Owen, D.R.; Allerton, C.M.; Anderson, A.S.; Aschenbrenner, L.; Avery, M.; Berritt, S.; Boras, B.; Cardin, R.D.; Carlo, A.; Coffman, K.J.; et al. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 2021, 374, 1586–1593. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Available online: https://www.fda.gov/ (accessed on 27 December 2022).
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1998, 28, 31–36. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Delaney, J.S. ESOL: Estimating aqueous solubility directly from molecular structure. J. Chem. Inf. Model. 2004, 44, 1000–1005. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Qamar, M.T.; Alqahtani, S.M.; Alamri, M.A.; Chen, L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020, 10, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinity. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; First, E.A. Thermodynamic analysis reveals a temperature-dependent change in the catalytic mechanism of bacillus stearothermophilus tyrosyl-tRNA synthetase. J. Biol. Chem. 2009, 284, 4179–4190. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Hu, Y.; Wang, Z.; Zhang, X.; Zhong, H.; Wang, G.; Yao, X.; Xu, L.; Cao, D.; Hou, T. Can machine learning consistently improve the scoring power of classical scoring functions? Insights into the role of machine learning in scoring functions. Brief. Bioinf. 2021, 22, 497–514. [Google Scholar] [CrossRef] [PubMed]
- Kuseva, C.; Schultz, T.W.; Yordanova, D.; Tankova, K.; Kutsarova, S.; Pavlov, T.; Chapkanov, A.; Georgiev, M.; Gissi, A.; Sobanski, T.; et al. The implementation of RAAF in the OECD QSAR Toolbox. Regul. Toxicol. Pharmacol. 2019, 105, 51–61. [Google Scholar] [CrossRef]
- Sunseri, J.; Koes, D.R. Pharmit: Interactive exploration of chemical space. Nucleic Acids Res. 2016, 44, W442–W448. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Dror, O.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. Deterministic pharmacophore detection via multiple flexible alignment of drug-like molecules. J. Comput. Biol. 2008, 15, 737–754. [Google Scholar] [CrossRef]
- MedChemExpress. Available online: https://www.medchemexpress.com/x77.html (accessed on 21 November 2022).
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Filimonov, D.; Druzhilovskiy, D.; Lagunin, A.; Lagunin, A.A.; Gloriozova, T.A.; Rudik, A.V.; Dmitriev, A.V.; Pogodin, P.V.; Poroikov, V.V. Computer-aided prediction of biological activity spectra for chemical compounds: Opportunities and limitations. Biomed. Chem Res. Methods 2018, 1, e00004. [Google Scholar] [CrossRef]
- Alhossary, A.; Handoko, S.D.; Mu, Y.; Kwoh, C.-K. Fast, accurate, and reliable molecular docking with QuickVina 2. Bioinformatics 2015, 31, 2214–2216. [Google Scholar] [CrossRef] [PubMed]
- Landrum G RDKit: Open-Source Cheminformatics. Available online: http://www.rdkit.org/ (accessed on 21 November 2022).
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Tosco, P.; Stiefl, N.; Landrum, G. Bringing the MMFF force field to the RDKit: Implementation and validation. J. Cheminform. 2014, 6, 37. [Google Scholar] [CrossRef]
- Wang, S.; Witek, J.; Landrum, G.A.; Riniker, S. Improving conformer generation for small rings and macrocycles based on distance geometry and experimental torsional-angle preferences. J. Chem. Inf. Model. 2020, 60, 2044–2058. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, J.; Marsili, M. A new model for calculating atomic charges in molecules. Tetrahedron Lett. 1978, 19, 3181–3184. [Google Scholar] [CrossRef]
- Rappe, A.K.; Casewit, C.J.; Colwell, K.S.; Goddard, W.A.; Skiff, W.M. UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. J. Am. Chem. Soc. 1992, 114, 10024–10035. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Center for Computational Structural Biology. MGL Tools. Available online: https://ccsb.scripps.edu/mgltools/ (accessed on 21 November 2022).
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Epstein, B.; Meir, R. Generalization bounds for unsupervised and semi-supervised learning with autoencoders. arXiv 2019, arXiv:1902.01449. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P.; Brevdo, E.; Chen, Z.; Citro, C.; Corrado, G.S.; Davis, A.; Dean, J.; Devin, M.; et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems. 2015. Available online: https://www.tensorflow.org/ (accessed on 21 November 2022).
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Janocha, K.; Czarnecki, W.M. On loss functions for deep neural networks in classification. Schedae Inform. 2016, 25, 49–59. [Google Scholar] [CrossRef]
- Shuldau, M.A.; Yushkevich, A.M.; Bosko, I.P.; Tuzikov, A.V.; Andrianov, A.M. Generative autoencoders for designing novel small-molecule compounds as potential SARS-CoV-2 main protease Inhibitors. In Pattern Recognition and Information Processing, Proceedings of the15th International Conference, PRIP 2021, Minsk, Belarus, 21–24 September 2021; Tuzikov, A.V., Belotserkovsky, A.M., Lukashevich, M.M., Eds.; Springer: Cham, Switzerland, 2022; Volume 1562. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. NNScore 2.0: A neural-network receptor–ligand scoring function. J. Chem. Inf. Model. 2011, 51, 2897–2903. [Google Scholar] [CrossRef] [PubMed]
- Palacio-Rodriguez, K.; Lans, I.; Cavasotto, C.N.; Cossio, P. Exponential consensus ranking improves the outcome in docking and receptor ensemble docking. Sci. Rep. 2019, 9, 5142. [Google Scholar] [CrossRef]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER 2018; University of California: Oakland, CA, USA, 2018. [Google Scholar]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general Amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Durrant, J.D.; McCammon, J.A. BINANA: A novel algorithm for ligand-binding characterization. J. Mol. Graph. Model. 2011, 29, 888–893. [Google Scholar] [CrossRef]
- Schrödinger, L.; DeLano, W. PyMOL. 2020. Available online: http://www.pymol.org/pymol (accessed on 26 April 2023).
- Xu, L.; Sun, H.; Li, Y.; Wang, J.; Hou, T.; Xu, L.; Sun, H.; Li, Y.; Wang, J.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 3. The impact of force fields and ligand charge models. J. Phys. Chem. B 2013, 117, 8408–8421. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Li, Y.; Tian, S.; Xu, L.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014, 16, 16719–16729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Stone, E.A.; Deshmukh, M.; Ippolito, J.A.; Ghahremanpour, M.M.; Tirado-Rives, J.; Spasov, K.A.; Zhang, S.; Takeo, Y.; Kudalkar, S.N.; et al. Potent noncovalent inhibitors of the main Protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free energy perturbation calculations. ACS Cent. Sci. 2021, 7, 467–475. [Google Scholar] [CrossRef] [PubMed]
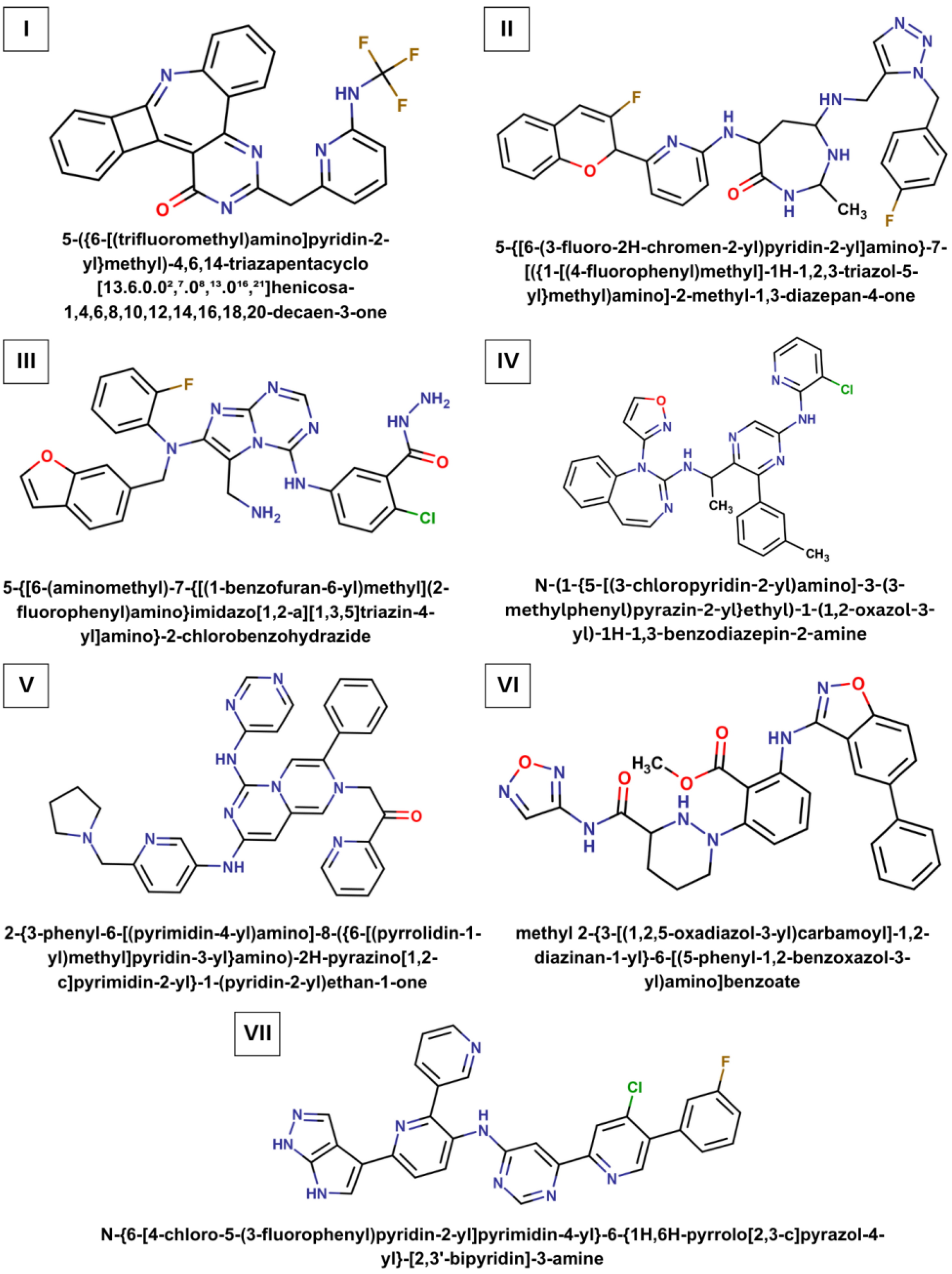
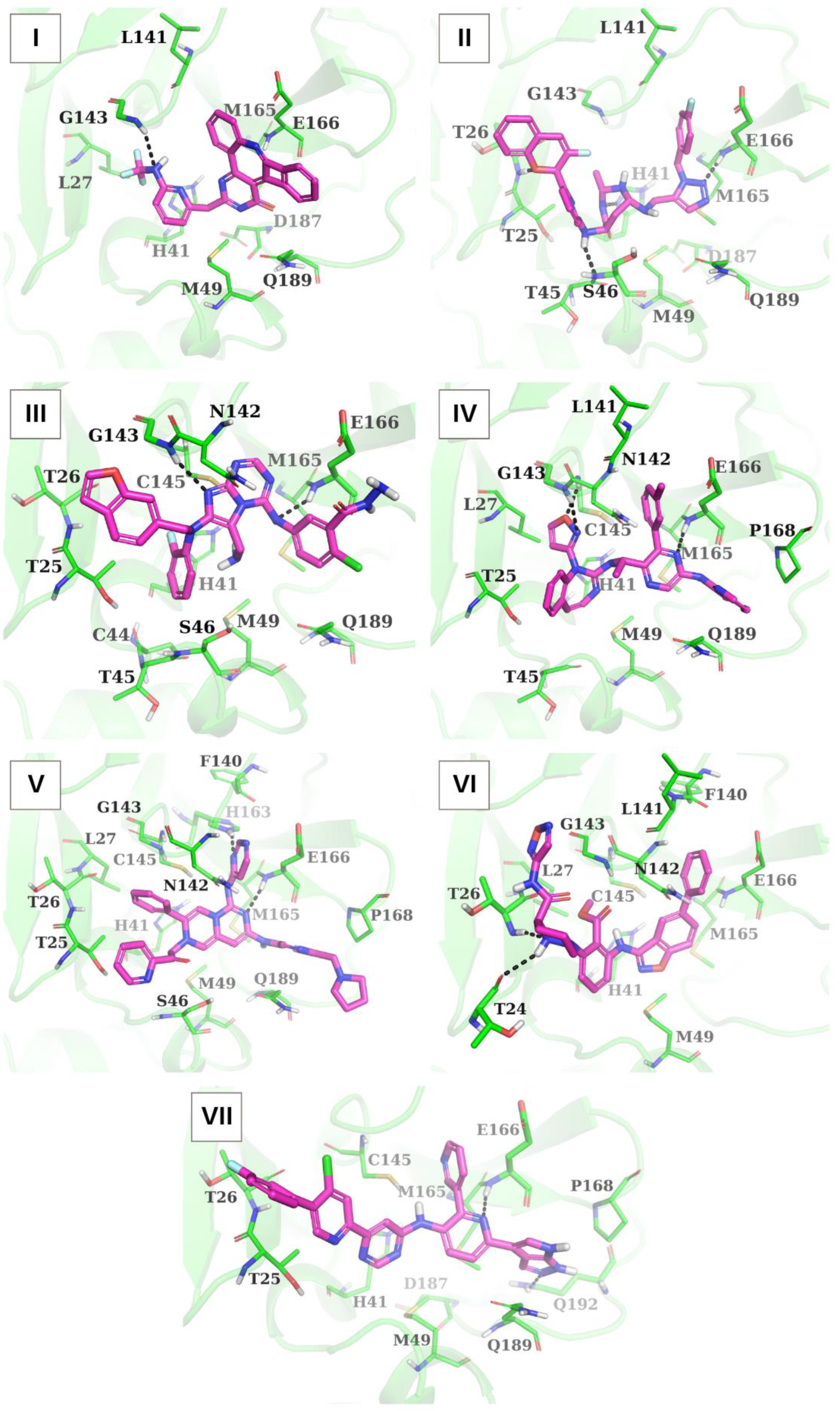
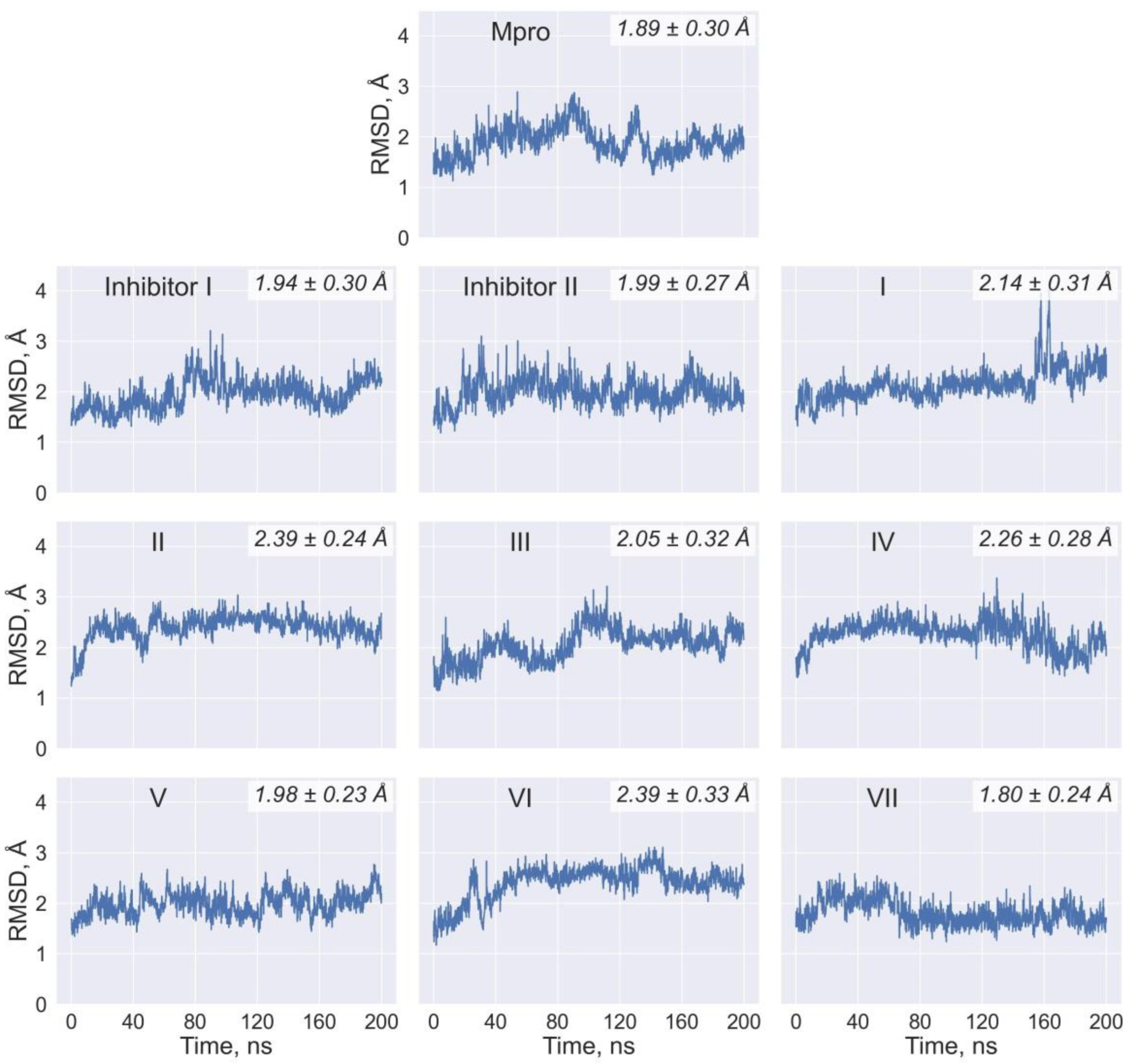
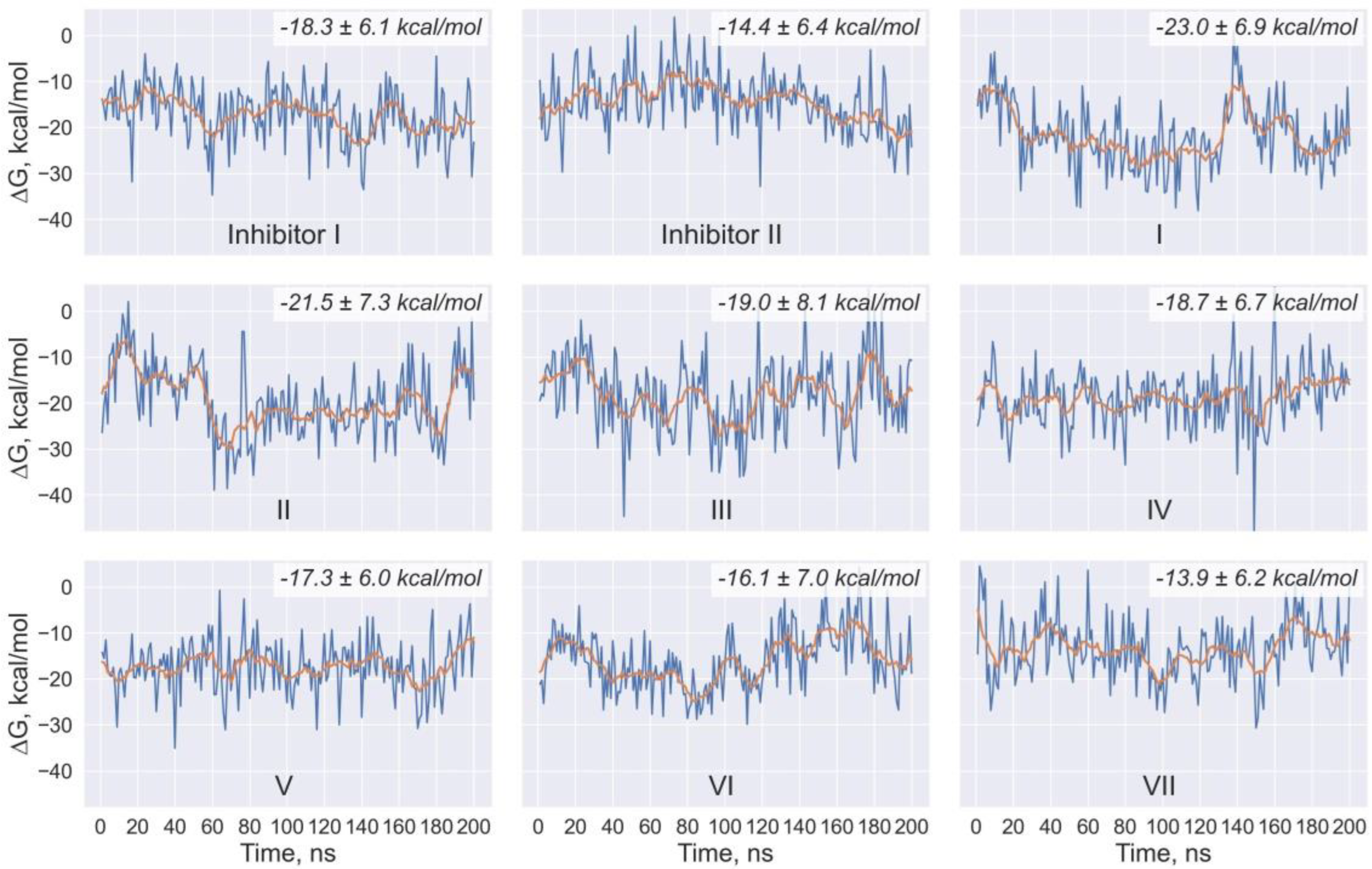

| Ligand | Chemical Formula | Molecular Weight (Da) | LogP | Number of H-Bond Donors | Number of H-Bond Acceptors |
|---|---|---|---|---|---|
| I | C25H14F3N5O | 457.4 | 4.43 | 1 | 8 |
| II | C30H30F2N8O2 | 572.6 | 2.93 | 4 | 9 |
| III | C28H23ClFN9O2 | 572.0 | 3.48 | 5 | 8 |
| IV | C30H25ClN8O | 549.0 | 4.73 | 2 | 6 |
| V | C34H32N10O | 596.7 | 3.62 | 2 | 7 |
| VI | C28H25N7O5 | 539.5 | 3.43 | 3 | 9 |
| VII | C30H19ClFN9 | 560.0 | 4.93 | 3 | 7 |
| Ligand | Decimal Logarithm of the Molar Solubility in Water LogS | Synthetic Accessibility SA |
|---|---|---|
| I | −5.68 | 3.55 |
| II | −5.40 | 5.70 |
| III | −6.37 | 4.07 |
| IV | −6.62 | 5.23 |
| V | −6.79 | 4,52 |
| VI | −6.28 | 4.83 |
| VII | −6.75 | 3.89 |
| Ligand | Hydrogen Bonds 1 | Van Der Waals Contacts 2 | Cation-π Interactions, Salt Bridges, and π-π Stacking 3 |
|---|---|---|---|
| I | N...*HN[G143] | E166(5), M49(3), L141(2), H41(2), M165(1), Q189(1), L27(1) | − |
| II | NH...**N[H41] NH...*N[S46] O...*HN[T26] N...* HN[E166] | E166(7), M165(3), T25(2), T26(2), L141(2), T45(1), H41(1), M49(1), S46(1), G143(1) | − |
| III | N...*HN[G143] N...*HN[E166] | E166(4), N142(3), T25(3), T45(2), M49(2), T26(1), S46(1), C44(1), H41(1), M165(1), Q189(1) | H41(2) (cation-π interaction); E166 (salt bridge) |
| IV | N...*HN[E166] O...*HN[C145] N...*HN[G143] | E166(8), L27(6), Q189(6), T25(3), L141(3), P168(3), H41(1), M49(1), T45(1), G143(1), C145(1), M165(1), N142(1) | − |
| V | N...*HN[E166] N...**HN[H163] | Q189(7), P168(6), T25(5), E166(4), F140(2), S46(2), L27(2), G143(2), M49(1), M165(1), H41(1), T26(1) | H41 (cation-π interaction) |
| VI | NH...*O[T24] N...*HN[T26] | E166(8), F140(3), L141(3), G143(3), M165(2), H41(1), L27(1), C145(1) | H41 (cation-π interaction); H41 (π-π stacking) |
| VII | N...*HN[E166] N...**HN[Q192] | Q189(8), E166(5), T25(4), M165(3), H41(2), P168(2), M49(1), T26(1) | H41 (π-π stacking) |
| Ligand | ΔGVINA 1 kcal/mol | KdVINA 1 μM | ΔGRFScore4 2 kcal/mol | KdRFScore4 2 μM | ΔGNNScore2.0 2 kcal/mol | KdNNScore2.0 2 μM |
|---|---|---|---|---|---|---|
| I | −9.1 | 0.384 | −10.9 | 0.022 | −11.9 | 0.0041 |
| II | −10.3 | 0.055 | −11.0 | 0.016 | −11.6 | 0.0069 |
| III | −8.7 | 0.735 | −11.1 | 0.015 | −12.7 | 0.0012 |
| IV | −10.0 | 0.089 | −11.1 | 0.014 | −13.0 | 0.0007 |
| V | −9.2 | 0.326 | −10.9 | 0.022 | −13.4 | 0.0004 |
| VI | −9.6 | 0.171 | −11.2 | 0.012 | −11.9 | 0.0043 |
| VII | −9.9 | 0.105 | −11.2 | 0.012 | −12.9 | 0.0007 |
| Inhibitor I | −8.3 | 1.407 | −11.0 | 0.018 | −8.1 | 1.9 |
| Inhibitor II | −8.5 | 1.017 | −11.1 | 0.015 | −7.9 | 2.9 |
| Ligand | <ΔH> kcal/mol | ΔHSTD kcal/mol | <TΔS> kcal/mol | (TΔS)STD kcal/mol | <ΔG> kcal/mol | ΔGSTD kcal/mol |
|---|---|---|---|---|---|---|
| I | −45.3 | 5.2 | −22.3 | 4.5 | −23.0 | 6.9 |
| II | −46.0 | 6.2 | −24.4 | 4.7 | −21.5 | 7.3 |
| III | −46.6 | 6.0 | −27.6 | 6.0 | −19.0 | 8.1 |
| IV | −44.6 | 3.9 | −25.9 | 5.3 | −18.7 | 6.7 |
| V | −45.5 | 4.3 | −28.2 | 4.3 | −17.3 | 6.0 |
| VI | −40.9 | 6.2 | −24.8 | 4.4 | −16.1 | 7.0 |
| VII | −37.4 | 4.8 | −23.4 | 3.9 | −13.9 | 6.2 |
| Inhibitor I | −42.8 | 4.1 | −24.5 | 4.9 | −18.3 | 6.1 |
| Inhibitor II | −39.0 | 4.1 | −24.5 | 4.5 | −14.4 | 6.4 |
| Residue Contribution to the Binding Energy (kcal/mol) 1,2,3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue of Mpro | Compounds | ||||||||
| Inhibitor I | Inhibitor II | I | II | III | IV | V | VI | VII | |
| Thr-25 | - | −0.6 ± 0.4 | - | - | −1.9 ± 0.9 | −0.8 ± 0.4 | - | −0.7 ± 0.6 | −1.7 ± 0.4 |
| Leu-27 | −0.5 ± 0.3 | −1.2 ± 0.4 | - | - | −1.1 ± 0.3 | −0.5 ± 0.2 | −0.6 ± 0.2 | −2.1 ± 0.7 | −1.0 ± 0.2 |
| His-41 | −2.2 ± 0.6 | - | −2.1 ± 0.4 | −0.9 ± 0.6 | −0.7 ± 0.3 | −0.9 ± 0.5 | −1.4 ± 0.4 | - | −1.2 ± 0.3 |
| Ser-46 | - | - | - | - | - | −1.5 ± 0.9 | −1.2 ± 0.4 | - | −0.6 ± 0.4 |
| Met-49 | −1.1 ± 0.6 | −0.9 ± 0.6 | −2.7 ± 1.5 | −1.8 ± 0.9 | −1.8 ± 0.5 | −0.8 ± 0.3 | −1.2 ± 0.3 | −0.8 ± 0.6 | −1.2 ± 0.3 |
| Leu-141 | - | −0.5 ± 0.3 | - | - | - | - | −1.1 ± 0.3 | −0.6 ± 0.3 | - |
| Asn-142 | −2.5 ± 0.6 | −2.5 ± 0.6 | - | - | −1.4 ± 0.7 | −0.7 ± 0.8 | −0.7 ± 0.5 | −3.3 ± 1.2 | - |
| Gly-143 | −1.8 ± 0.3 | −2.3 ± 0.5 | - | - | −1.9 ± 0.4 | - | −0.5 ± 0.2 | −2.2 ± 0.6 | - |
| Ser-144 | −0.7 ± 0.4 | −1.0 ± 0.4 | - | - | - | - | - | −1.7 ± 0.6 | - |
| Cys-145 | −1.4 ± 0.3 | −1.6 ± 0.5 | - | - | −1.1 ± 0.3 | −0.9 ± 0.3 | −1.3 ± 0.3 | −2.2 ± 0.7 | −1.4 ± 0.3 |
| His-163 | −1.7 ± 0.3 | −1.7 ± 0.4 | - | - | - | - | −1.2 ± 0.5 | −0.6 ± 0.2 | −1.5 ± 0.4 |
| His-164 | - | - | −0.5 ± 0.3 | - | - | - | −2.9 ± 0.8 | - | −0.9 ± 0.2 |
| Met-165 | −2.6 ± 0.4 | −2.6 ± 0.7 | −2.9 ± 0.4 | −2.5 ± 0.7 | −3.0 ± 0.4 | −2.9 ± 0.4 | −3.5 ± 0.5 | - | −3.7 ± 0.5 |
| Glu-166 | −1.4 ± 0.6 | −1.2 ± 0.7 | −1.0 ± 0.5 | - | −2.6 ± 0.6 | −1.6 ± 0.7 | −2.0 ± 0.8 | −0.6 ± 0.8 | −1.1 ± 0.6 |
| Leu-167 | - | - | −1.4 ± 0.5 | −2.1 ± 0.6 | −1.0 ± 0.4 | −0.8 ± 0.2 | −0.7 ± 0.4 | - | - |
| Pro-168 | - | - | −0.6 ± 0.3 | −2.0 ± 0.5 | −0.9 ± 0.4 | −1.3 ± 0.3 | −1.1 ± 0.4 | - | - |
| Phe-185 | - | - | - | −1.4 ± 0.5 | - | - | - | - | - |
| Asp-187 | −1.9 ± 0.7 | −0.7 ± 0.9 | −2.4 ± 0.4 | −1.2 ± 0.7 | - | - | - | - | −1.4 ± 0.3 |
| Arg-188 | - | - | −1.2 ± 0.6 | −0.5 ± 0.5 | - | - | - | - | −1.0 ± 0.4 |
| Gln-189 | −1.0 ± 0.8 | −1.3 ± 0.6 | −2.5 ± 0.8 | −2.9 ± 1.1 | −2.9 ± 1.6 | −3.3 ± 0.7 | −0.7 ± 0.8 | - | −1.1 ± 0.6 |
| Thr-190 | - | - | −0.6 ± 0.4 | −1.2 ± 0.8 | - | −0.6 ± 0.2 | - | - | - |
| Gln-192 | - | - | - | −1.3 ± 0.6 | - | −0.5 ± 0.2 | - | - | - |
| Compounds | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue of Mpro | I | II | III | IV | V | VI | VII | Inhibitor I | Inhibitor II |
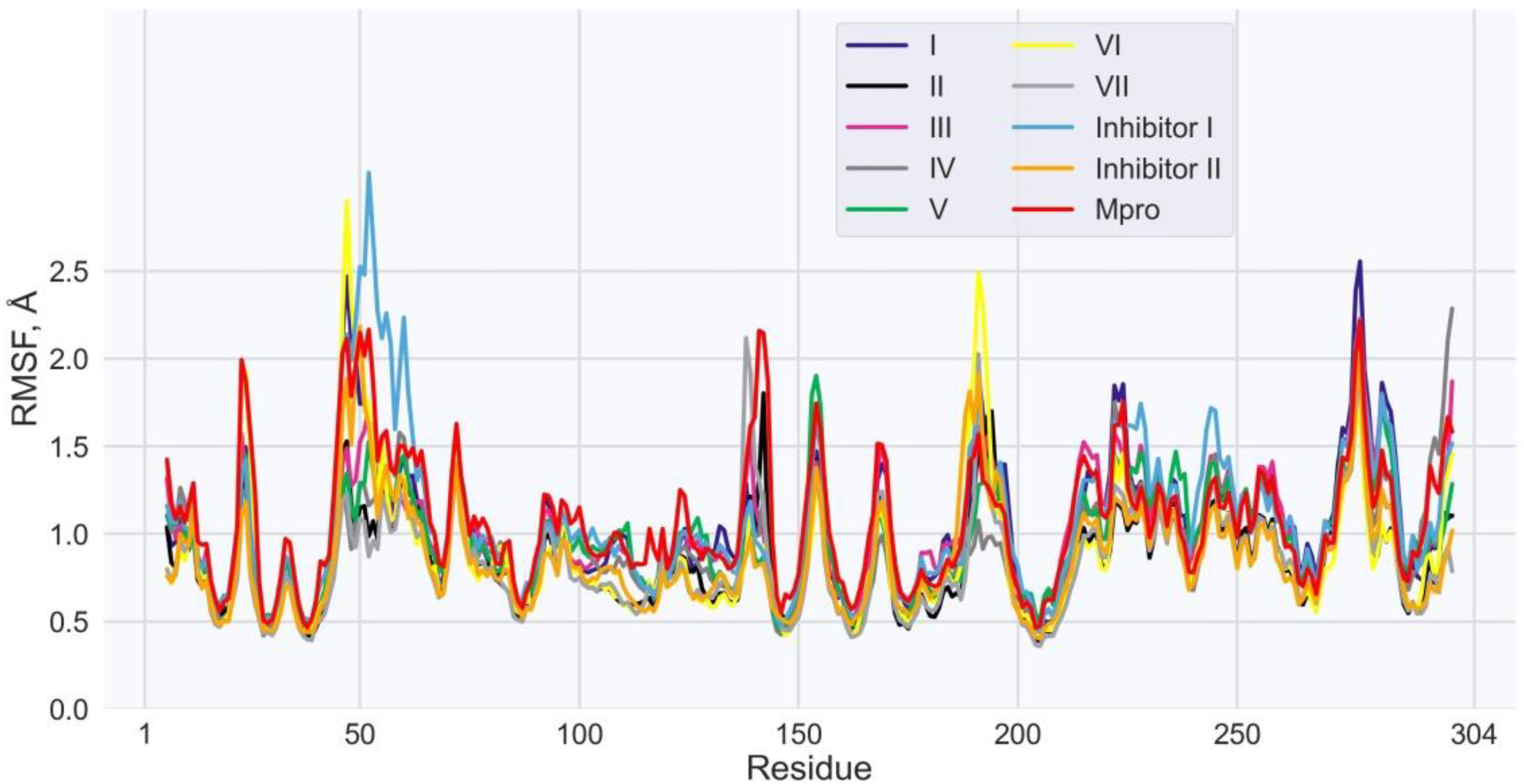
| Values of RMSF (Å) for the Individual Residues of Mpro | |||||||||
| His41 | 0.5 | 0.5 | 0.6 | 0.6 | 0.6 | 0.6 | 0.5 | 0.7 | 0.6 |
| Met49 | 2.0 | 1.0 | 1.3 | 1.1 | 1.1 | 2.0 | 0.9 | 2.2 | 2.0 |
| Asn142 | 1.2 | 1.8 | 1.0 | 1.2 | 0.9 | 0.9 | 1.1 | 0.9 | 0.8 |
| Gly143 | 1.0 | 0.9 | 0.8 | 0.7 | 0.8 | 0.9 | 0.9 | 0.7 | 0.7 |
| Cys145 | 0.6 | 0.6 | 0.5 | 0.4 | 0.5 | 0.6 | 0.5 | 0.5 | 0.5 |
| Met165 | 0.7 | 0.6 | 0.7 | 0.6 | 0.5 | 0.6 | 0.5 | 0.6 | 0.6 |
| Glu166 | 0.8 | 0.7 | 0.8 | 0.6 | 0.6 | 0.8 | 0.6 | 0.7 | 0.7 |
| Gln189 | 1.3 | 1.1 | 1.4 | 0.9 | 1.0 | 1.7 | 1.0 | 1.5 | 1.8 |
| Model | Generation Starting Point Description | Generation Process Description |
|---|---|---|
| Unsupervised (embeddings model) | Random number vectors drawn from fitted normal distributions | Random numbers are used as embeddings and fed to the decoder |
| Unsupervised (embeddings model) | Compounds with binding free energy less than −9 kcal/mol, sampled from the test set | Embeddings for these compounds are calculated, distortion is then added, and updated embeddings are fed to the decoder |
| Semi-supervised (energy model) | Random number vectors drawn from fitted normal distributions and a preset binding free energy value | Random vectors are used as embeddings and are passed as latent layer inputs along with a preset binding free energy value |
| Semi-supervised (energy model) | Compounds with binding free energy less than −8 kcal/mol were sampled from the test set and improved binding free energy values | Embeddings for these compounds are calculated, then distortion is added, and updated embeddings along with improved binding free energy values are passed to the decoder |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrianov, A.M.; Shuldau, M.A.; Furs, K.V.; Yushkevich, A.M.; Tuzikov, A.V. AI-Driven De Novo Design and Molecular Modeling for Discovery of Small-Molecule Compounds as Potential Drug Candidates Targeting SARS-CoV-2 Main Protease. Int. J. Mol. Sci. 2023, 24, 8083. https://doi.org/10.3390/ijms24098083
Andrianov AM, Shuldau MA, Furs KV, Yushkevich AM, Tuzikov AV. AI-Driven De Novo Design and Molecular Modeling for Discovery of Small-Molecule Compounds as Potential Drug Candidates Targeting SARS-CoV-2 Main Protease. International Journal of Molecular Sciences. 2023; 24(9):8083. https://doi.org/10.3390/ijms24098083
Chicago/Turabian StyleAndrianov, Alexander M., Mikita A. Shuldau, Konstantin V. Furs, Artsemi M. Yushkevich, and Alexander V. Tuzikov. 2023. "AI-Driven De Novo Design and Molecular Modeling for Discovery of Small-Molecule Compounds as Potential Drug Candidates Targeting SARS-CoV-2 Main Protease" International Journal of Molecular Sciences 24, no. 9: 8083. https://doi.org/10.3390/ijms24098083
APA StyleAndrianov, A. M., Shuldau, M. A., Furs, K. V., Yushkevich, A. M., & Tuzikov, A. V. (2023). AI-Driven De Novo Design and Molecular Modeling for Discovery of Small-Molecule Compounds as Potential Drug Candidates Targeting SARS-CoV-2 Main Protease. International Journal of Molecular Sciences, 24(9), 8083. https://doi.org/10.3390/ijms24098083





