Systemic Analyses of Cuproptosis-Related lncRNAs in Pancreatic Adenocarcinoma, with a Focus on the Molecular Mechanism of LINC00853
Abstract
1. Introduction
2. Results
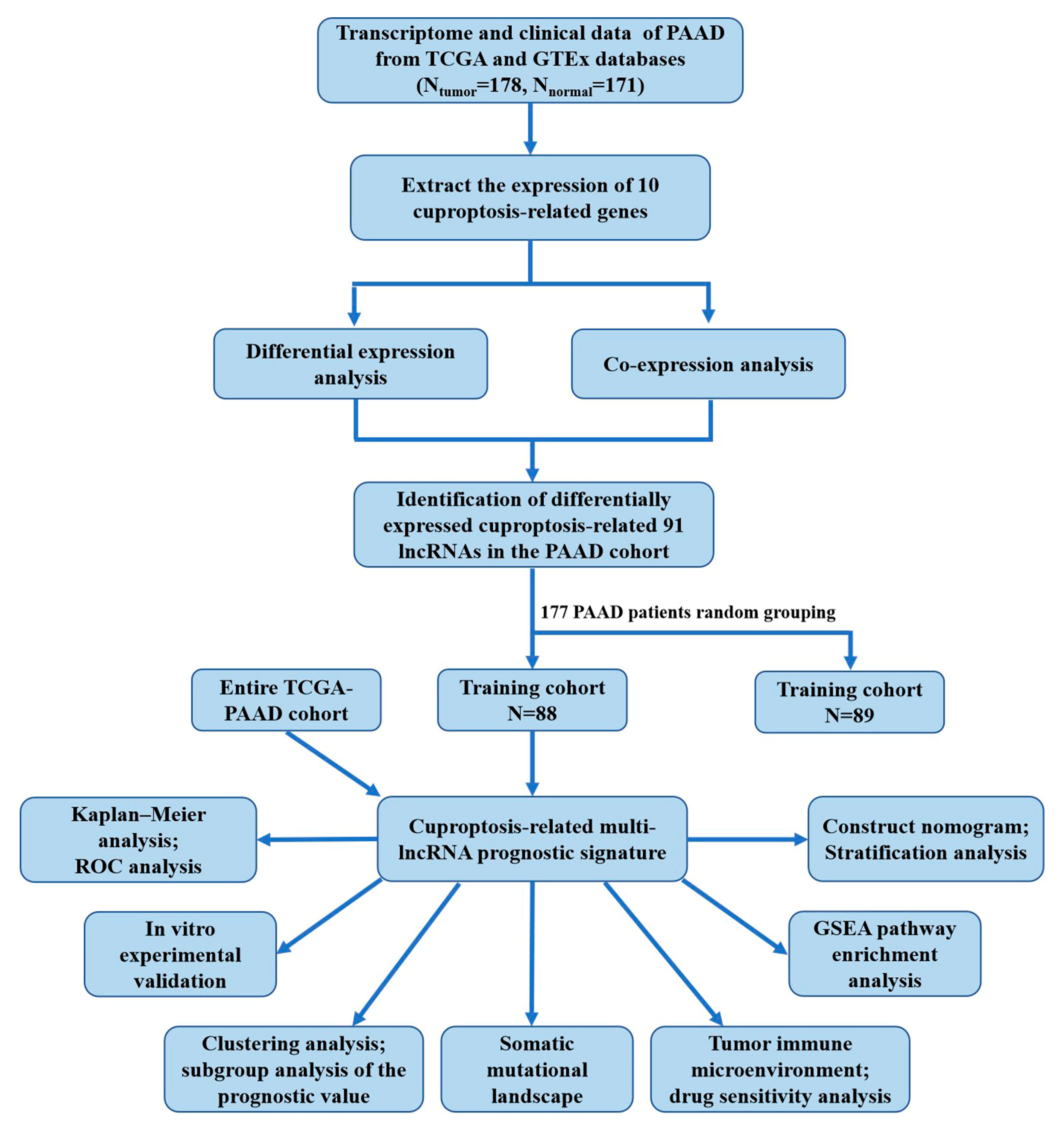
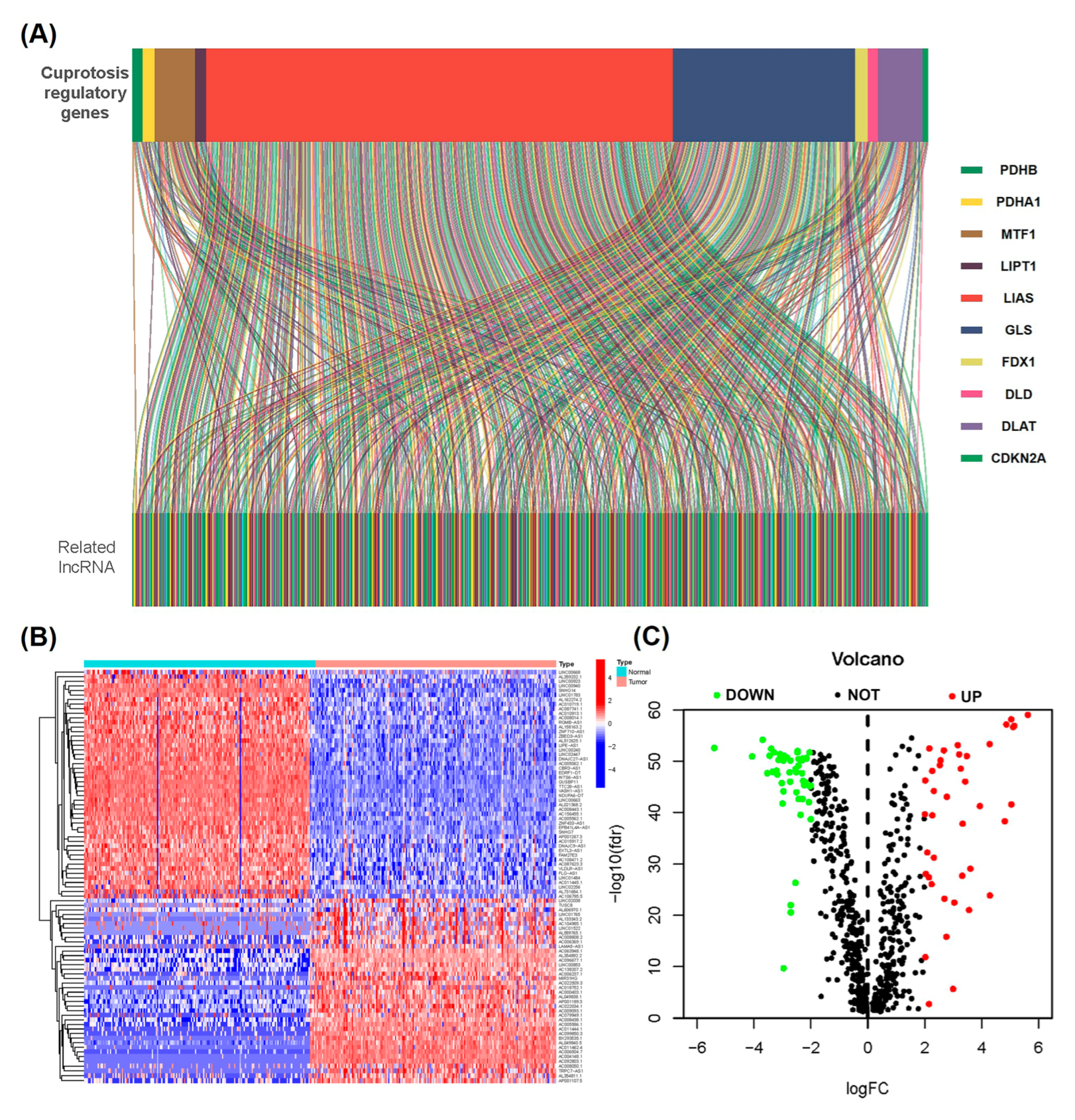
2.1. Determination of Differentially Expressed Cuproptosis-Related lncRNAs in the PAAD Cohort
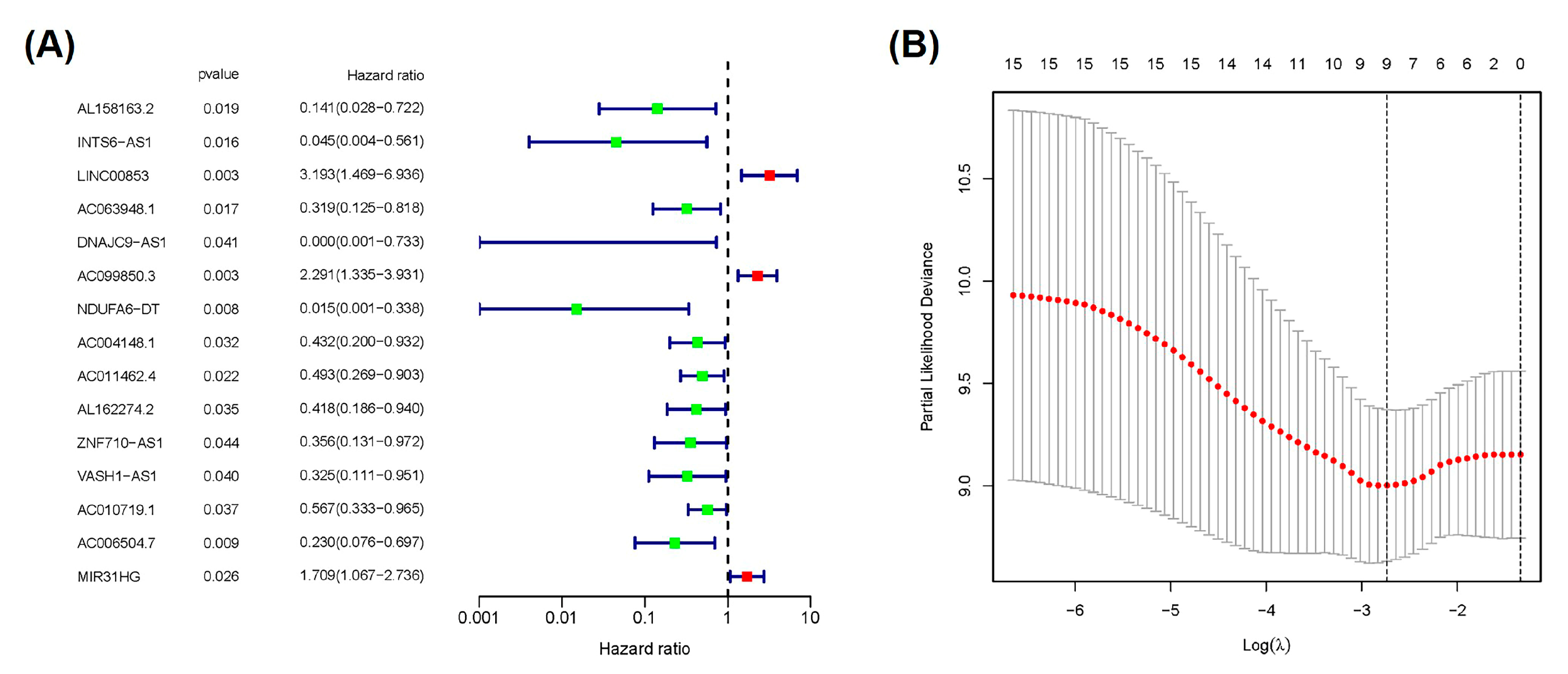
2.2. Construction and Verification of Prognostic Signature According to Cuproptosis-Related lncRNAs
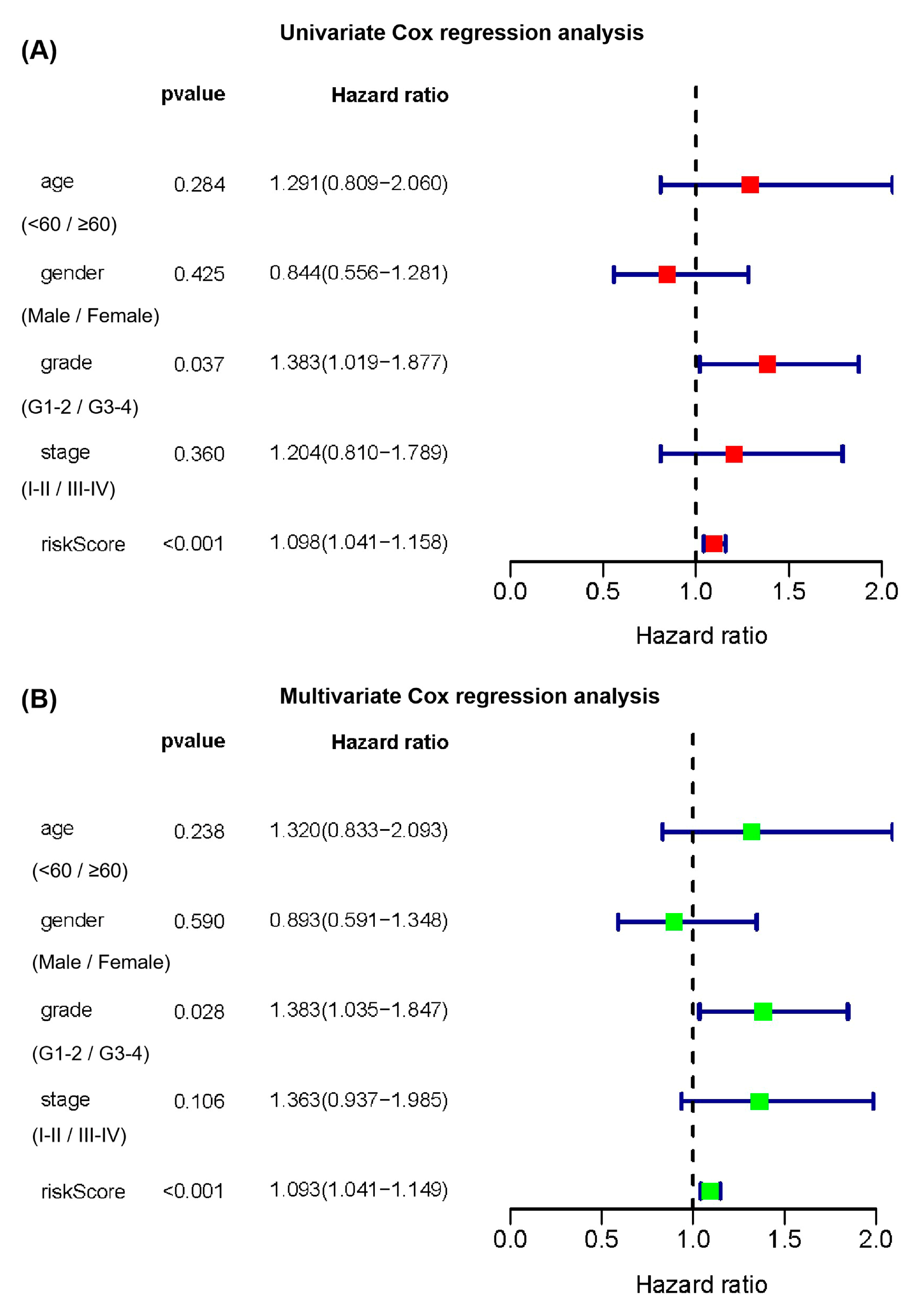
2.3. Subgroup Analysis of the Prognostic Value of the Cuproptosis-Related Multi-lncRNA Signature
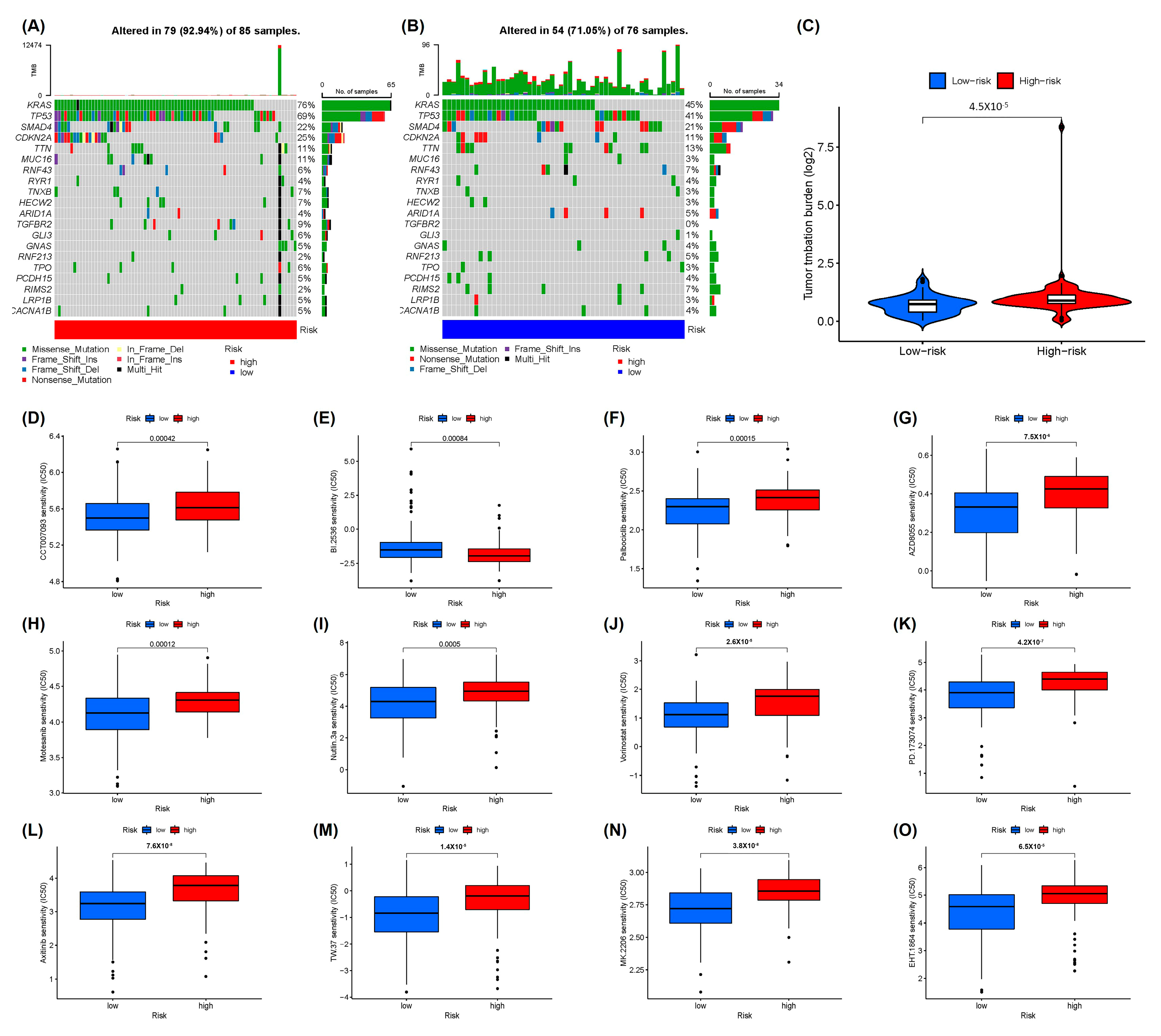
2.4. Somatic Mutational Landscape and Drug Sensitivity Analysis
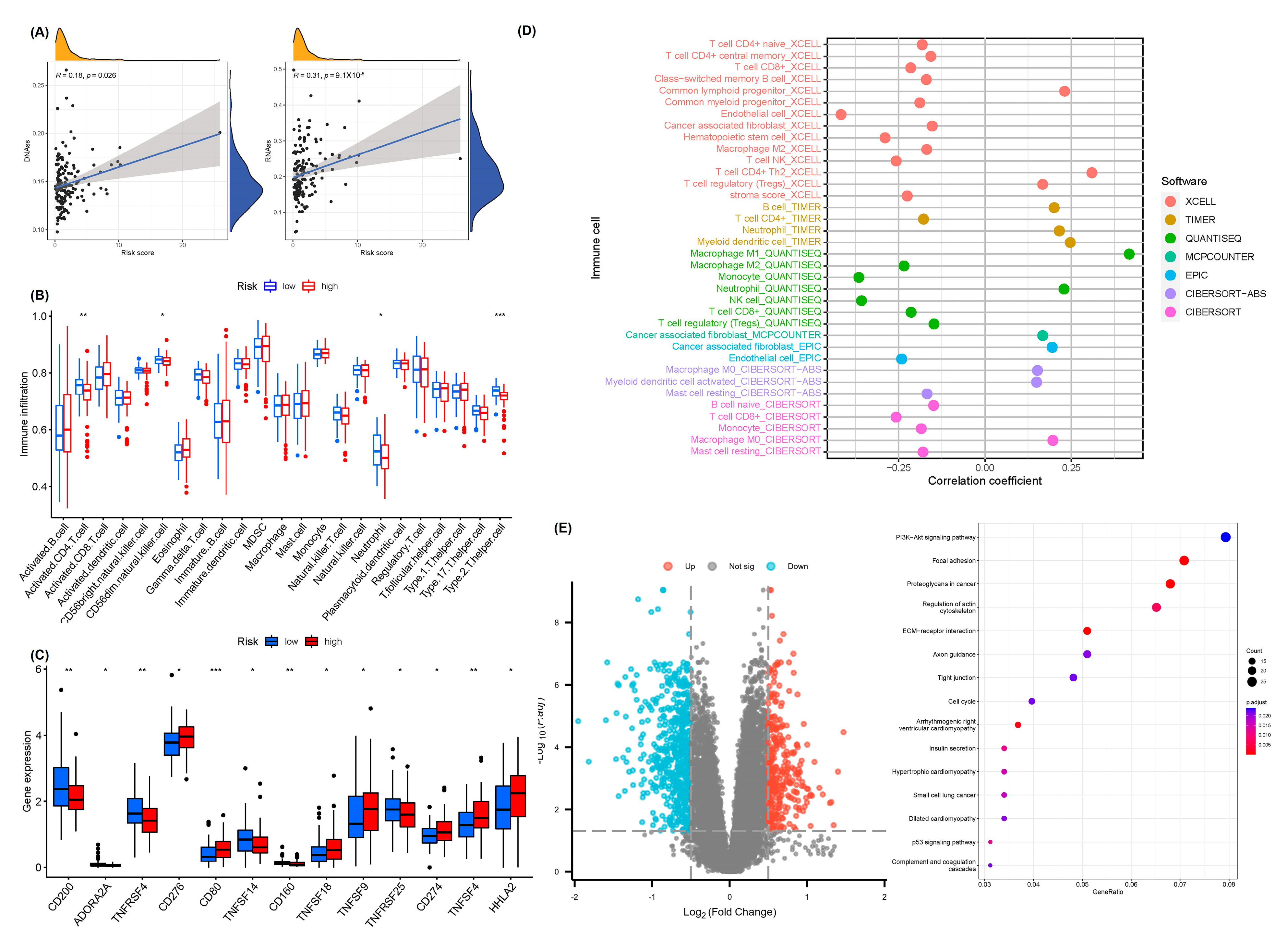
2.5. Analysis of Tumor Immune Microenvironment and Pathways Related to Prognostic Features
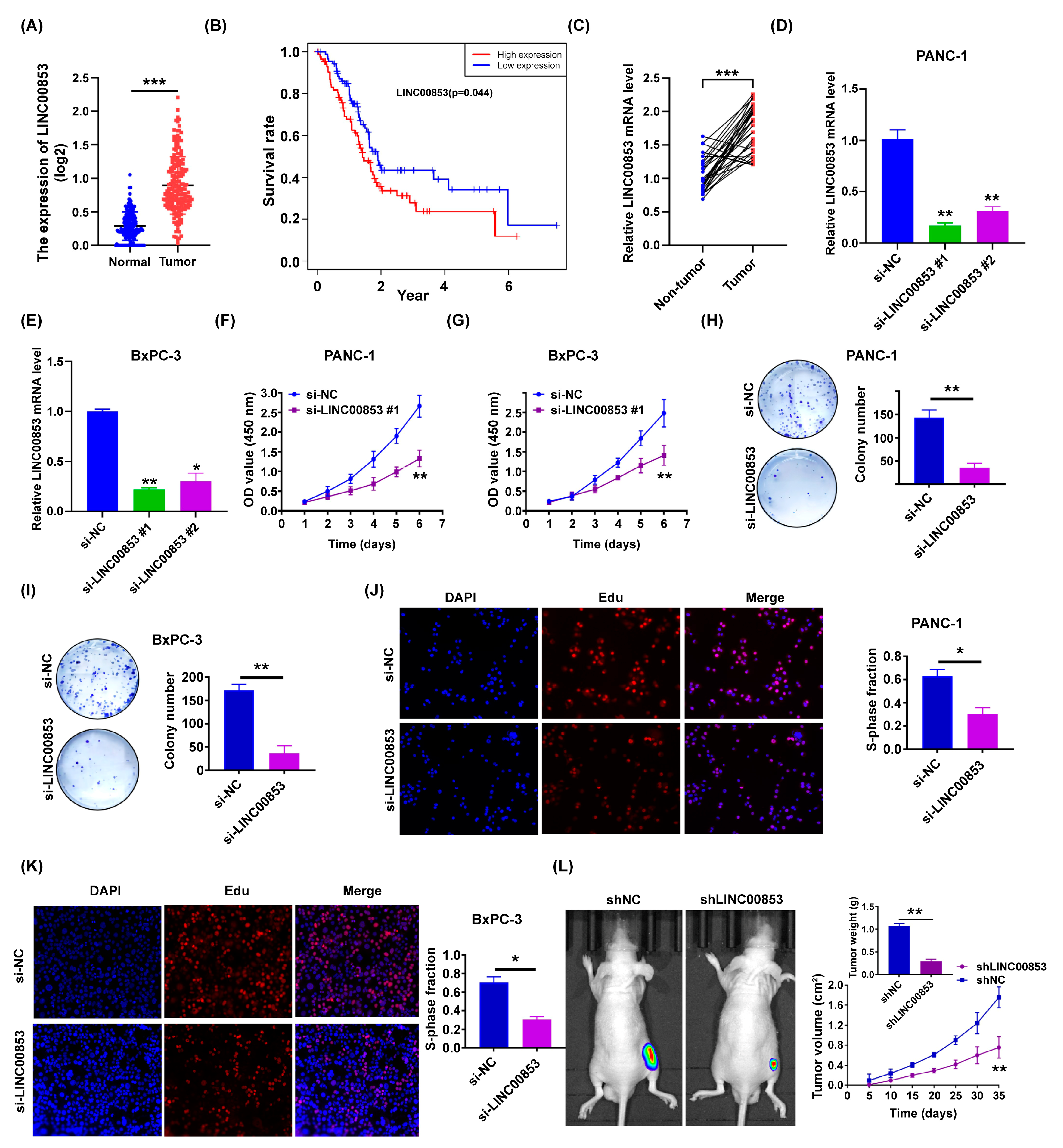
2.6. LINC00853 Is an Oncogene Candidate Gene for PC Progression
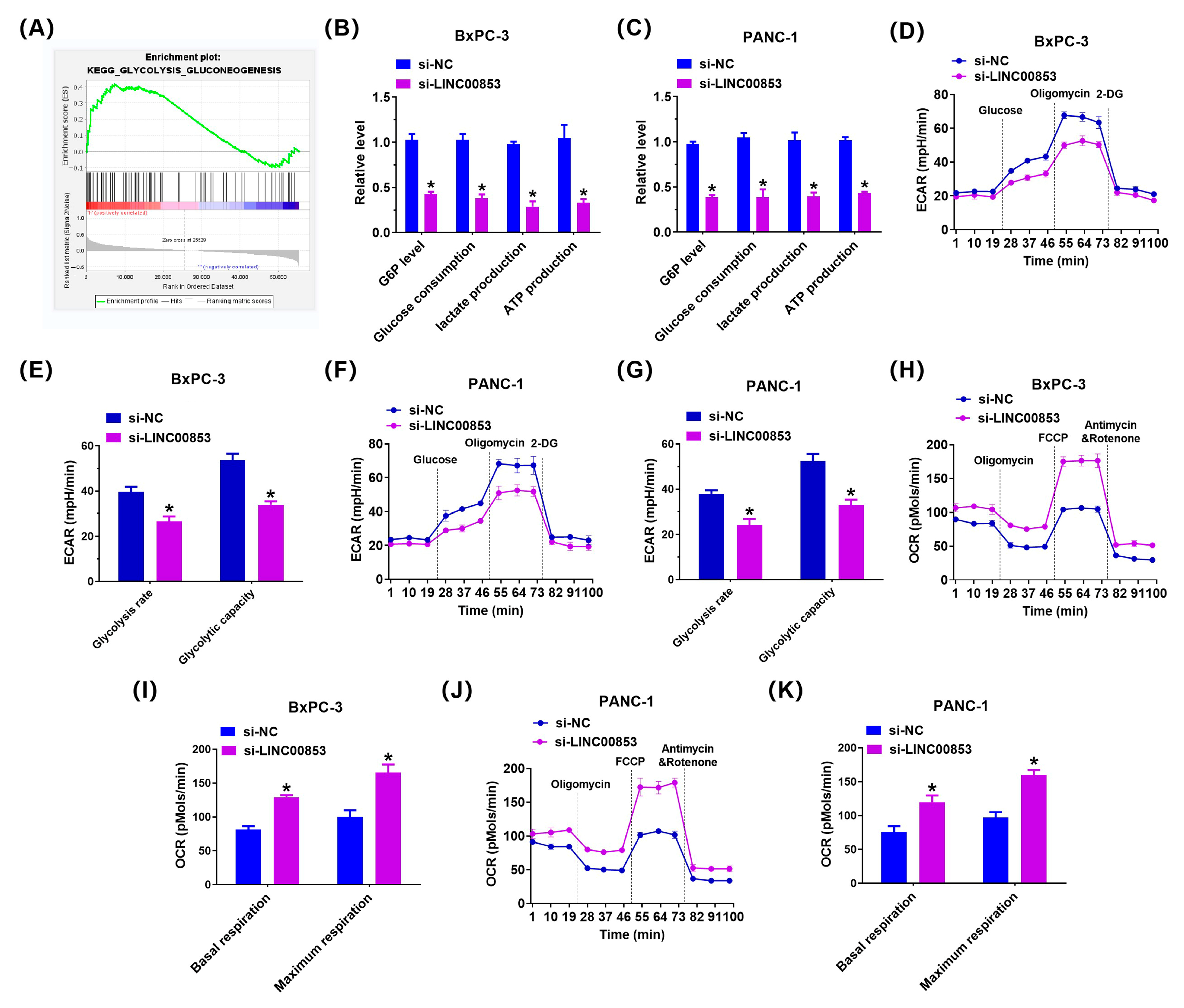
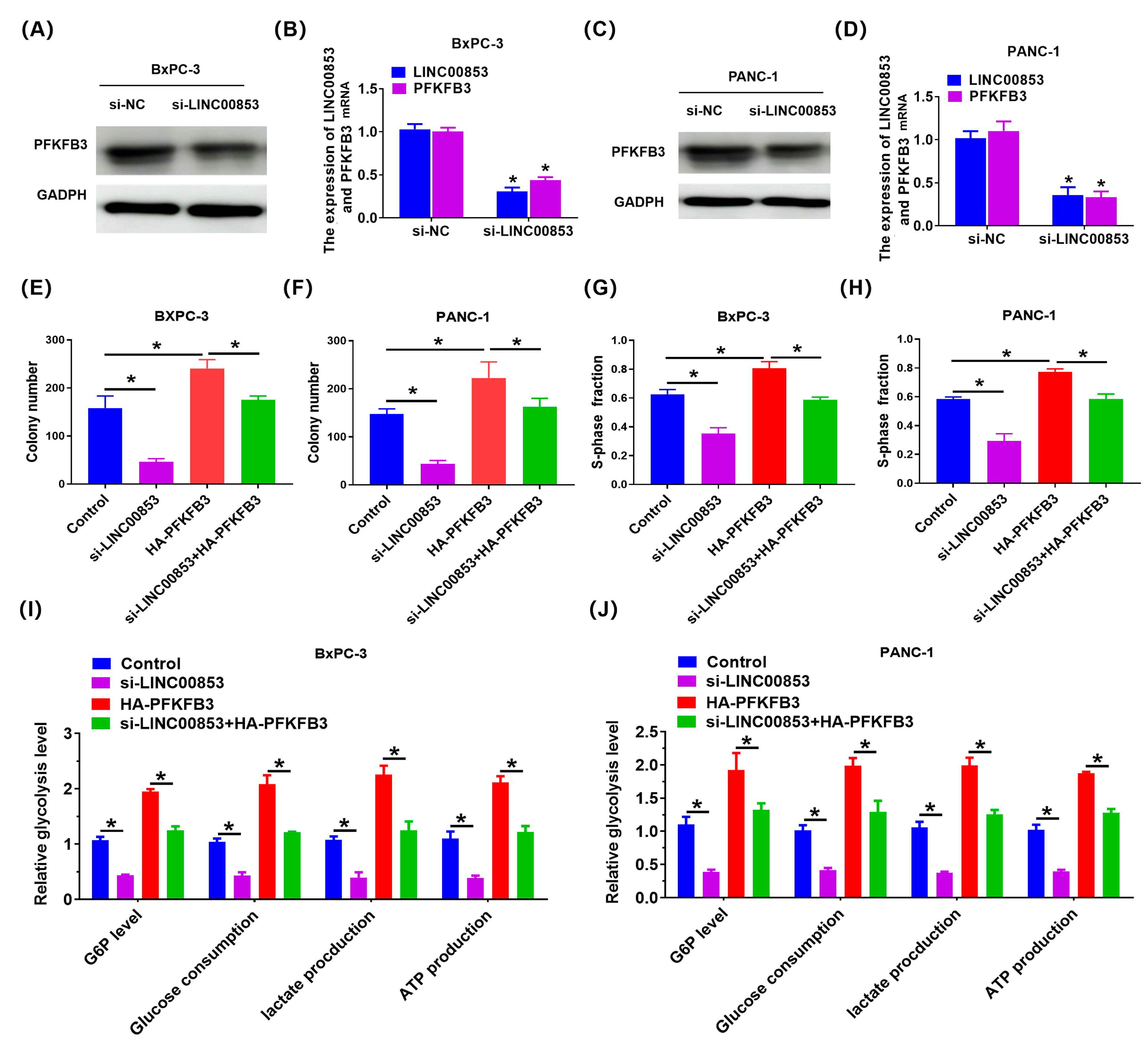
2.7. LINC00853 Enhances Aerobic Glycolysis and Proliferation through PFKFB3 in PC Cells
3. Discussion
4. Materials and Methods
4.1. Acquisition and Processing of Data from PAAD Patients
4.2. Determination of Differentially Expressed Cuproptosis-Associated lncRNAs with Prognostic Value
4.3. Construction and Verification of a Prognostic Gene Signature
4.4. Stemness Index Data Analysis
4.5. Tumor Microenvironment and Clinical Treatment Response Analysis Using the Prognostic Risk Signature
4.6. Gene Set Enrichment Analysis (GSEA) and Functional Enrichment
4.7. Validation of Bioinformatics Results by qRT-PCR
4.8. Subcutaneous Xenograft Model
4.9. Identification of Extracellular Acidification Rate (ECAR) and Oxygen Consumption Rate (OCR)
4.10. Western Blotting
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Chawla, A.; O’Reilly, E.M. Pancreatic Cancer: A Review. JAMA 2021, 326, 851–862. [Google Scholar] [CrossRef]
- Nagtegaal, I.D.; Odze, R.D.; Klimstra, D.; Paradis, V.; Rugge, M.; Schirmacher, P.; Washington, K.M.; Carneiro, F.; Cree, I.A.; WHO Classification of Tumours Editorial Board. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020, 76, 182–188. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Sabater, L.; Muñoz, E.; Roselló, S.; Dorcaratto, D.; Garcés-Albir, M.; Huerta, M.; Roda, D.; Gómez-Mateo, M.C.; Ferrández-Izquierdo, A.; Darder, A.; et al. Borderline resectable pancreatic cancer. Challenges and controversies. Cancer Treat. Rev. 2018, 68, 124–135. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Kleeff, J.; Michl, P.; Costello, E.; Greenhalf, W.; Palmer, D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 333–348. [Google Scholar] [CrossRef] [PubMed]
- Bock, F.J.; Tait, S.W.G. Mitochondria as multifaceted regulators of cell death. Nat. Rev. Mol. Cell Biol. 2020, 21, 85–100. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Berghe, T.V.; Vandenabeele, P.; Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef]
- Tsang, T.; Davis, C.I.; Brady, D.C. Copper biology. Curr. Biol. 2021, 31, R421–R427. [Google Scholar] [CrossRef] [PubMed]
- Priemel, T.; Palia, G.; Forste, F.; Jehle, F.; Sviben, S.; Mantouvalou, I.; Zaslansky, P.; Bertinetti, L.; Harrington, M.J. Microfluidic-like fabrication of metal ion-cured bioadhesives by mussels. Science 2021, 374, 206–211. [Google Scholar] [CrossRef]
- Kim, B.E.; Nevitt, T.; Thiele, D.J. Mechanisms for copper acquisition, distribution and regulation. Nat. Chem. Biol. 2008, 4, 176–185. [Google Scholar] [CrossRef]
- Ge, E.J.; Bush, A.I.; Casini, A.; Cobine, P.A.; Cross, J.R.; DeNicola, G.M.; Dou, Q.P.; Franz, K.J.; Gohil, V.M.; Gupta, S.; et al. Connecting copper and cancer: From transition metal signalling to metalloplasia. Nat. Rev. Cancer 2022, 22, 102–113. [Google Scholar] [CrossRef]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021, 18, 280–296. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Detappe, A.; Cai, K.; Keys, H.R.; Brune, Z.; Ying, W.; Thiru, P.; Reidy, M.; Kugener, G.; Rossen, J.; et al. Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat. Chem. Biol. 2019, 15, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Hasinoff, B.B.; Yadav, A.A.; Patel, D.; Wu, X. The cytotoxicity of the anticancer drug elesclomol is due to oxidative stress indirectly mediated through its complex with Cu(II). J. Inorg. Biochem. 2014, 137, 22–30. [Google Scholar] [CrossRef]
- Cobine, P.A.; Brady, D.C. Cuproptosis: Cellular and molecular mechanisms underlying copper-induced cell death. Mol. Cell 2022, 82, 1786–1787. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Zhou, F. Cuproptosis: A new form of programmed cell death. Cell Mol. Immunol. 2022, 19, 867–868. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Copper homeostasis: Emerging target for cancer treatment. IUBMB Life 2020, 72, 1900–1908. [Google Scholar] [CrossRef]
- Lelièvre, P.; Sancey, L.; Coll, J.L.; Deniaud, A.; Busser, B. The Multifaceted Roles of Copper in Cancer: A Trace Metal Element with Dysregulated Metabolism, but Also a Target or a Bullet for Therapy. Cancers 2020, 12, 3594. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, L.M.; Libedinsky, A.; Elorza, A.A. Role of Copper on Mitochondrial Function and Metabolism. Front. Mol. Biosci. 2021, 8, 711227. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Kim, H.W.; Nam, J.W. The small peptide world in long noncoding RNAs. Brief. Bioinform. 2019, 20, 1853–1864. [Google Scholar] [CrossRef]
- Nandwani, A.; Rathore, S.; Datta, M. LncRNAs in cancer: Regulatory and therapeutic implications. Cancer Lett. 2021, 501, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.T.; Lin, J.F.; Li, T.; Li, J.J.; Xu, R.H.; Ju, H.Q. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 2021, 41, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, C.; He, Z.; Chen, S.; Li, L.; Sun, C. LncRNA PSMB8-AS1 contributes to pancreatic cancer progression via modulating miR-382-3p/STAT1/PD-L1 axis. J. Exp. Clin. Cancer Res. 2020, 39, 179. [Google Scholar] [CrossRef]
- Huang, X.; Pan, L.; Zuo, Z.; Li, M.; Zeng, L.; Li, R.; Ye, Y.; Zhang, J.; Wu, G.; Bai, R.; et al. LINC00842 inactivates transcription co-regulator PGC-1alpha to promote pancreatic cancer malignancy through metabolic remodelling. Nat. Commun. 2021, 12, 3830. [Google Scholar] [CrossRef]
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 2014, 15, 244–253. [Google Scholar] [CrossRef]
- Malta, T.; Sokolov, A.; Gentles, A.; Burzykowski, T.; Poisson, L.; Weinstein, J.; Kaminska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell 2018, 173, 338–354. [Google Scholar] [CrossRef]
- Lee, K.; Hwang, H.; Nam, K.T. Immune response and the tumor microenvironment: How they communicate to regulate gastric cancer. Gut Liver 2014, 8, 131. [Google Scholar] [CrossRef]
- Deng, X.; Yi, X.; Deng, J.; Zou, Y.; Wang, S.; Shan, W.; Liu, P.; Zhang, Z.; Chen, L.; Hao, L. ROCK2 promotes osteosarcoma growth and metastasis by modifying PFKFB3 ubiquitination and degradation. Exp. Cell Res. 2019, 385, 111689. [Google Scholar] [CrossRef]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Gong, Y.; Fan, Z.; Luo, G.; Yang, C.; Huang, Q.; Fan, K.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. The role of necroptosis in cancer biology and therapy. Mol. Cancer 2019, 18, 100. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, Y.; Mao, C.; Liu, S.; Xiao, D.; Huang, J.; Tao, Y. Emerging mechanisms and targeted therapy of ferroptosis in cancer. Mol. Ther. 2021, 29, 2185–2208. [Google Scholar] [CrossRef]
- Nagai, M.; Vo, N.H.; Shin Ogawa, L.; Chimmanamada, D.; Inoue, T.; Chu, J.; Beaudette-Zlatanova, B.C.; Lu, R.; Blackman, R.K.; Barsoum, J.; et al. The oncology drug elesclomol selectively transports copper to the mitochondria to induce oxidative stress in cancer cells. Free Radic. Biol. Med. 2012, 52, 2142–2150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, L.; Lu, J.; Jiang, B.; Liu, C.; Guo, J.; Xiao, G.G. Research progress on long non-coding RNAs and their roles as potential biomarkers for diagnosis and prognosis in pancreatic cancer. Cancer Cell Int. 2020, 20, 457. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Sun, B.F.; Chen, C.Y.; Zhou, J.Y.; Chen, Y.S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G.S.; et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019, 29, 725–738. [Google Scholar] [CrossRef]
- Yap, T.A.; Parkes, E.E.; Peng, W.; Moyers, J.T.; Curran, M.A.; Tawbi, H.A. Development of Immunotherapy Combination Strategies in Cancer. Cancer Discov. 2021, 11, 1368–1397. [Google Scholar] [CrossRef]
- Bear, A.S.; Vonderheide, R.H.; O’Hara, M.H. Challenges and Opportunities for Pancreatic Cancer Immunotherapy. Cancer Cell 2020, 38, 788–802. [Google Scholar] [CrossRef] [PubMed]
- Choueiry, F.; Torok, M.; Shakya, R.; Agrawal, K.; Deems, A.; Benner, B.; Hinton, A.; Shaffer, J.; Blaser, B.W.; Noonan, A.M.; et al. CD200 promotes immunosuppression in the pancreatic tumor microenvironment. J. Immunother. Cancer 2020, 8, e000189. [Google Scholar] [CrossRef]
- Ma, Y.; Li, J.; Wang, H.; Chiu, Y.; Kingsley, C.V.; Fry, D.; Delaney, S.N.; Wei, S.C.; Zhang, J.; Maitra, A.; et al. Combination of PD-1 Inhibitor and OX40 Agonist Induces Tumor Rejection and Immune Memory in Mouse Models of Pancreatic Cancer. Gastroenterology 2020, 159, 306–319.e312. [Google Scholar] [CrossRef]
- Li, G.; Kryczek, I.; Nam, J.; Li, X.; Li, S.; Li, J.; Wei, S.; Grove, S.; Vatan, L.; Zhou, J.; et al. LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy. Nat. Cell Biol. 2021, 23, 526–537. [Google Scholar] [CrossRef]
- Xu, J.; Shao, T.; Song, M.; Xie, Y.; Zhou, J.; Yin, J.; Ding, N.; Zou, H.; Li, Y.; Zhang, J. MIR22HG acts as a tumor suppressor via TGFbeta/SMAD signaling and facilitates immunotherapy in colorectal cancer. Mol. Cancer 2020, 19, 51. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef]
- Liu, L.; Bai, X.; Wang, J.; Tang, X.R.; Wu, D.H.; Du, S.S.; Du, X.J.; Zhang, Y.W.; Zhu, H.B.; Fang, Y.; et al. Combination of TMB and CNA Stratifies Prognostic and Predictive Responses to Immunotherapy across Metastatic Cancer. Clin. Cancer Res. 2019, 25, 7413–7423. [Google Scholar] [CrossRef]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef]
- Chen, L.; Yuan, R.; Wen, C.; Liu, T.; Feng, Q.; Deng, X.; Du, Y.; Peng, X. E3 ubiquitin ligase UBR5 promotes pancreatic cancer growth and aerobic glycolysis by downregulating FBP1 via destabilization of C/EBPalpha. Oncogene 2021, 40, 262–276. [Google Scholar] [CrossRef]
- Shin, E.; Koo, J.S. Glucose Metabolism and Glucose Transporters in Breast Cancer. Front. Cell Dev. Biol. 2021, 9, 728759. [Google Scholar] [CrossRef]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Bradburn, M.J.; Clark, T.G.; Love, S.B.; Altman, D.G. Survival analysis part II: Multivariate data analysis—An introduction to concepts and methods. Br. J. Cancer 2003, 89, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Therneau, T.; Lumley, T. Survival: Survival Analysis, Including Penalised Likelihood. R package Version 2.35-7. R Found. Stat. Comput. 2009. Available online: http://cran.r-project.org/web/packages/survival/index.html (accessed on 31 March 2022).
- Kassambara, A.; Kosinski, M. Survminer: Drawing Survival Curves Using ‘ggplot2’. 2016, R Package Version 0.3; CRAN: 2016; Volume 1. Available online: https://cran.r-project.org/package=survminer (accessed on 31 March 2022).
- Heagerty, P.; Saha, P. time-dependent ROC curve estimation from censored survival data. Biometrics 2013, 66, 999–1011. [Google Scholar]
- Blanche, P.; Dartigues, J.F.; Jacqmin-Gadda, H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 2013, 32, 5381–5397. [Google Scholar] [CrossRef] [PubMed]
- Krijthe, J.; van der Maaten, L.; Krijthe, M.J. Package ‘Rtsne’, R Package Version 0.13; CRAN: 2017; Available online: https://github.com/jkrijthe/Rtsne (accessed on 31 March 2022).
- Wickham, H. Elegant Graphics for Data Analysis (ggplot2). Appl. Spat. Data Anal. R 2009. Available online: https://cran.r-project.org/web/packages/ggplot2/citation.html (accessed on 31 March 2022).
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar]
- Wei, T.; Simko, V.; Levy, M.; Xie, Y.; Jin, Y.; Zemla, J. Corrplot: Visualization of a Correlation Matrix, R. Package Version 0.73; CRAN: 2013; Volume 230. Available online: http://cran.r-project.org/package=corrplot (accessed on 31 March 2022).
- Kassambara, A. ggpubr: “ggplot2” Based Publication Ready Plots, R Package Version 0.4.0; CRAN: 2020; Available online: https://cran.rproject.org/web/packages/ggpubr/ggpubr.pdf (accessed on 31 March 2022).
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]

| lncRNA | Coefficient |
|---|---|
| LINC00853 | 1.33435227976852 |
| AC099850.3 | 0.794073987427955 |
| AC010719.1 | −0.766590583753927 |
| AC006504.7 | −1.51899571329944 |
| Variable | Group | Entire Cohort (n = 177) | Train Cohort (n = 88) | Test Cohort (n = 89) | p-Value |
|---|---|---|---|---|---|
| Age | ≤65 | 91 (51.41%) | 46 (52.27%) | 45 (50.56%) | 0.9384 |
| >65 | 86 (48.59%) | 42 (47.73%) | 44 (49.44%) | ||
| Gender | Female | 80 (45.2%) | 41 (46.59%) | 39 (43.82%) | 0.8264 |
| Male | 97 (54.8%) | 47 (53.41%) | 50 (56.18%) | ||
| Grade | G1 | 28 (15.82%) | 18 (20.45%) | 10 (11.24%) | 0.3202 |
| G2 | 93 (52.54%) | 42 (47.73%) | 51 (57.3%) | ||
| G3 | 51 (28.81%) | 25 (28.41%) | 26 (29.21%) | ||
| G4 | 3 (1.69%) | 2 (2.27%) | 1 (1.12%) | ||
| unknow | 2 (1.13%) | 1 (1.14%) | 1 (1.12%) | ||
| Stage | Stage I | 20 (11.3%) | 7 (7.95%) | 13 (14.61%) | 0.4895 |
| Stage II | 145 (81.92%) | 73 (82.95%) | 72 (80.9%) | ||
| Stage III | 2 (1.13%) | 1 (1.14%) | 1 (1.12%) | ||
| Stage IV | 6 (3.39%) | 4 (4.55%) | 2 (2.25%) | ||
| unknow | 4 (2.26%) | 3 (3.41%) | 1 (1.12%) | ||
| T stage | T1 | 5 (2.82%) | 2 (2.27%) | 3 (3.37%) | 0.3211 |
| T2 | 27 (15.25%) | 9 (10.23%) | 18 (20.22%) | ||
| T3 | 141 (79.66%) | 74 (84.09%) | 67 (75.28%) | ||
| T4 | 2 (1.13%) | 1 (1.14%) | 1 (1.12%) | ||
| unknow | 2 (1.13%) | 2 (2.27%) | 0 (0%) | ||
| M stage | M0 | 81 (45.76%) | 40 (45.45%) | 41 (46.07%) | 0.6936 |
| M1 | 6 (3.39%) | 4 (4.55%) | 2 (2.25%) | ||
| unknow | 90 (50.85%) | 44 (50%) | 46 (51.69%) | ||
| N stage | N0 | 50 (28.25%) | 20 (22.73%) | 30 (33.71%) | 0.172 |
| N1 | 121 (68.36%) | 64 (72.73%) | 57 (64.04%) | ||
| unknow | 6 (3.39%) | 4 (4.55%) | 2 (2.25%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Zhang, L.; He, H.; Shao, F.; Gao, Y.; He, J. Systemic Analyses of Cuproptosis-Related lncRNAs in Pancreatic Adenocarcinoma, with a Focus on the Molecular Mechanism of LINC00853. Int. J. Mol. Sci. 2023, 24, 7923. https://doi.org/10.3390/ijms24097923
Chen L, Zhang L, He H, Shao F, Gao Y, He J. Systemic Analyses of Cuproptosis-Related lncRNAs in Pancreatic Adenocarcinoma, with a Focus on the Molecular Mechanism of LINC00853. International Journal of Molecular Sciences. 2023; 24(9):7923. https://doi.org/10.3390/ijms24097923
Chicago/Turabian StyleChen, Leifeng, Lin Zhang, Haihua He, Fei Shao, Yibo Gao, and Jie He. 2023. "Systemic Analyses of Cuproptosis-Related lncRNAs in Pancreatic Adenocarcinoma, with a Focus on the Molecular Mechanism of LINC00853" International Journal of Molecular Sciences 24, no. 9: 7923. https://doi.org/10.3390/ijms24097923
APA StyleChen, L., Zhang, L., He, H., Shao, F., Gao, Y., & He, J. (2023). Systemic Analyses of Cuproptosis-Related lncRNAs in Pancreatic Adenocarcinoma, with a Focus on the Molecular Mechanism of LINC00853. International Journal of Molecular Sciences, 24(9), 7923. https://doi.org/10.3390/ijms24097923






