β-Conglutins’ Unique Mobile Arm Is a Key Structural Domain Involved in Molecular Nutraceutical Properties of Narrow-Leafed Lupin (Lupinus angustifolius L.)
Abstract
1. Introduction
2. Results and Discussion
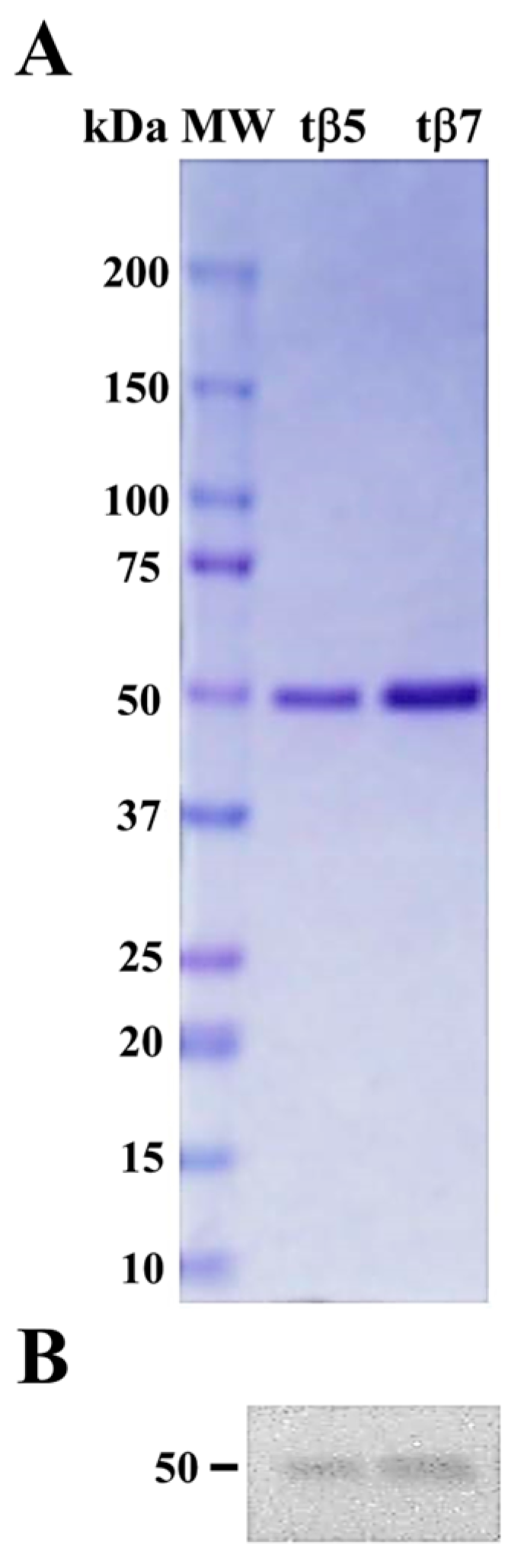
2.1. Purification and Identification of tβ5 and tβ7
2.2. β-Conglutin Proteins Structural Modeling
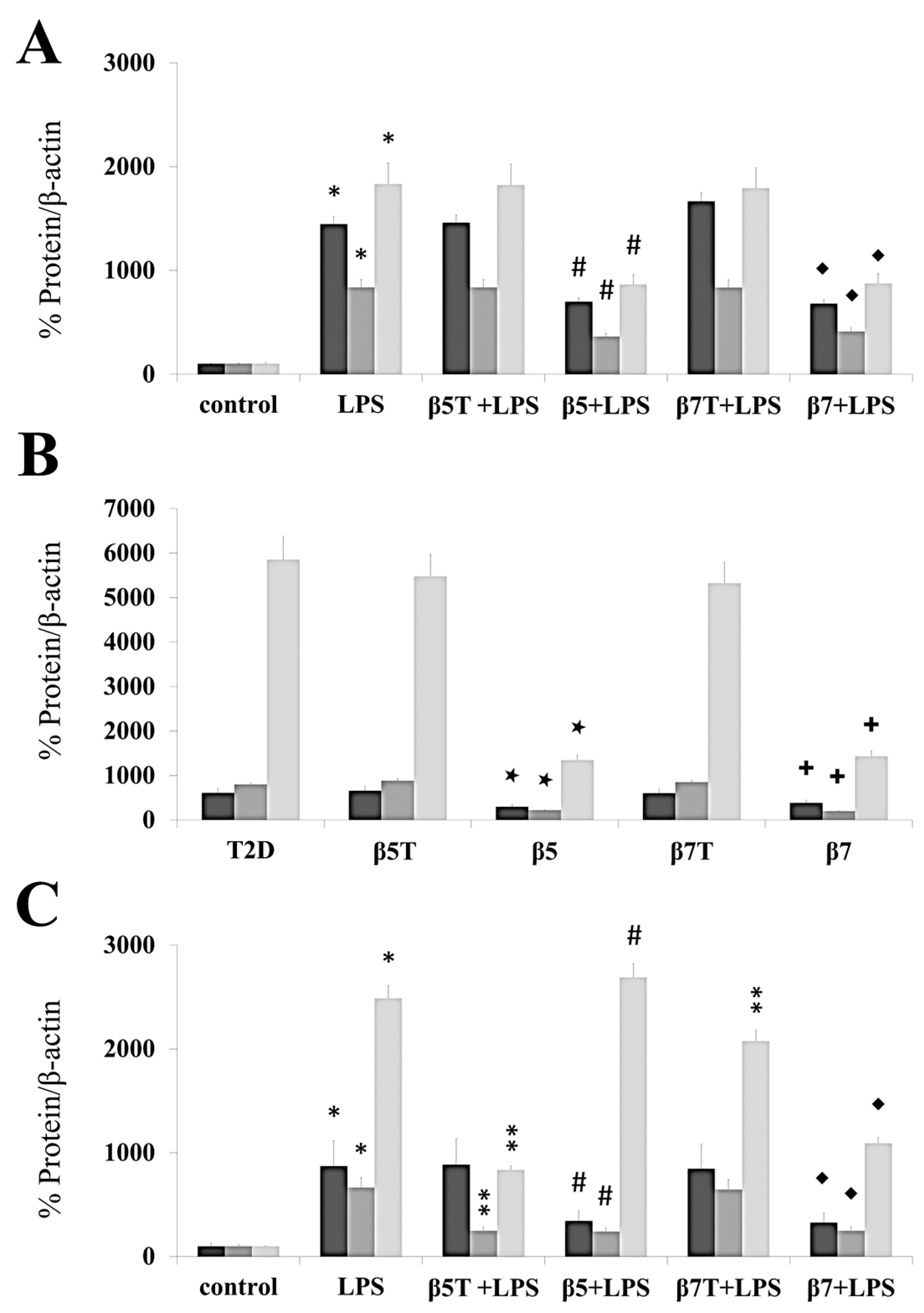
2.3. β-Conglutin Protein Inhibits the Gene Expression of Different Cytokines and Pro-Inflammatory Mediators
2.4. β5 and β7 Conglutins Can Decrease the Production of Pro-Inflammatory Mediators
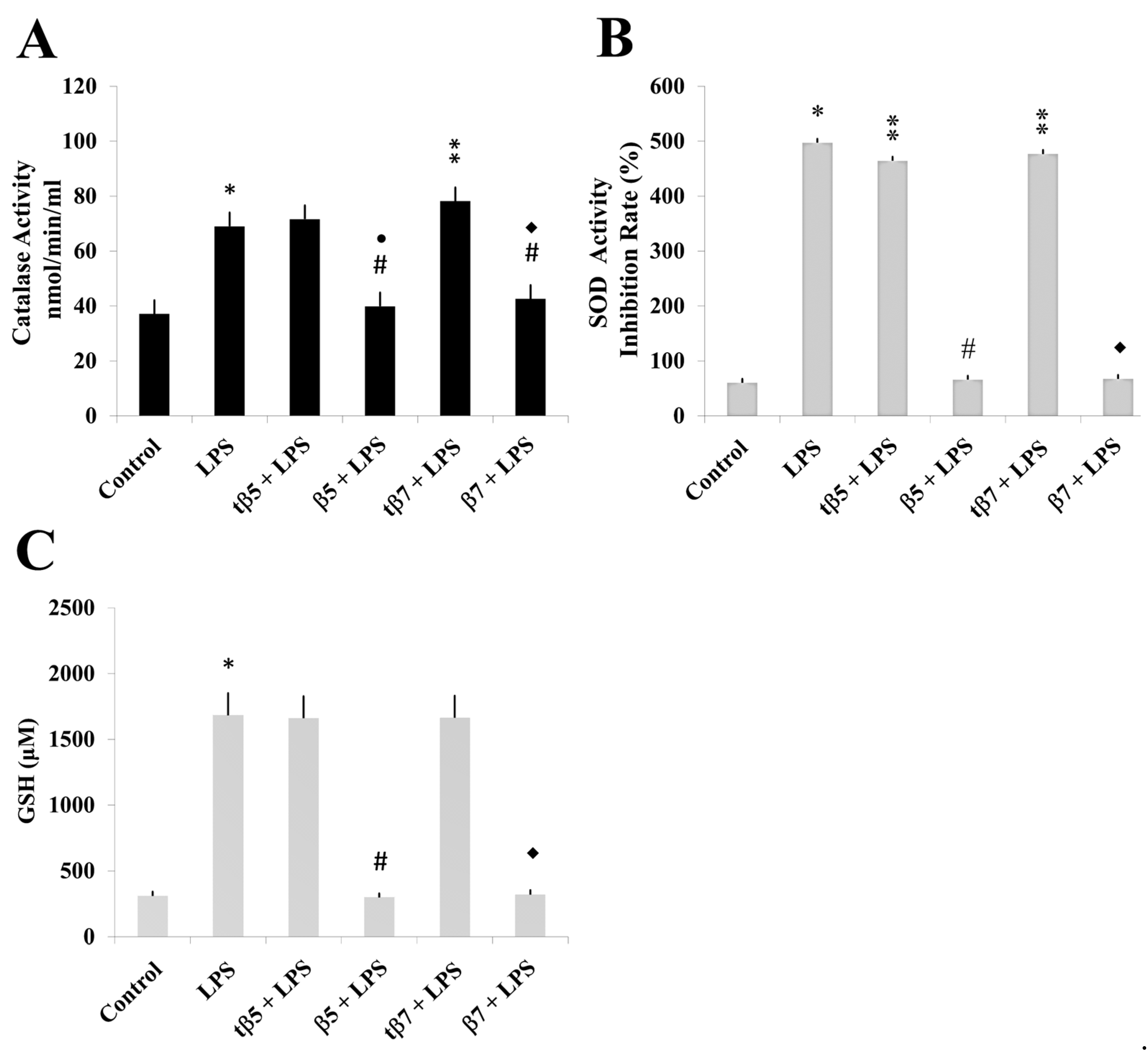
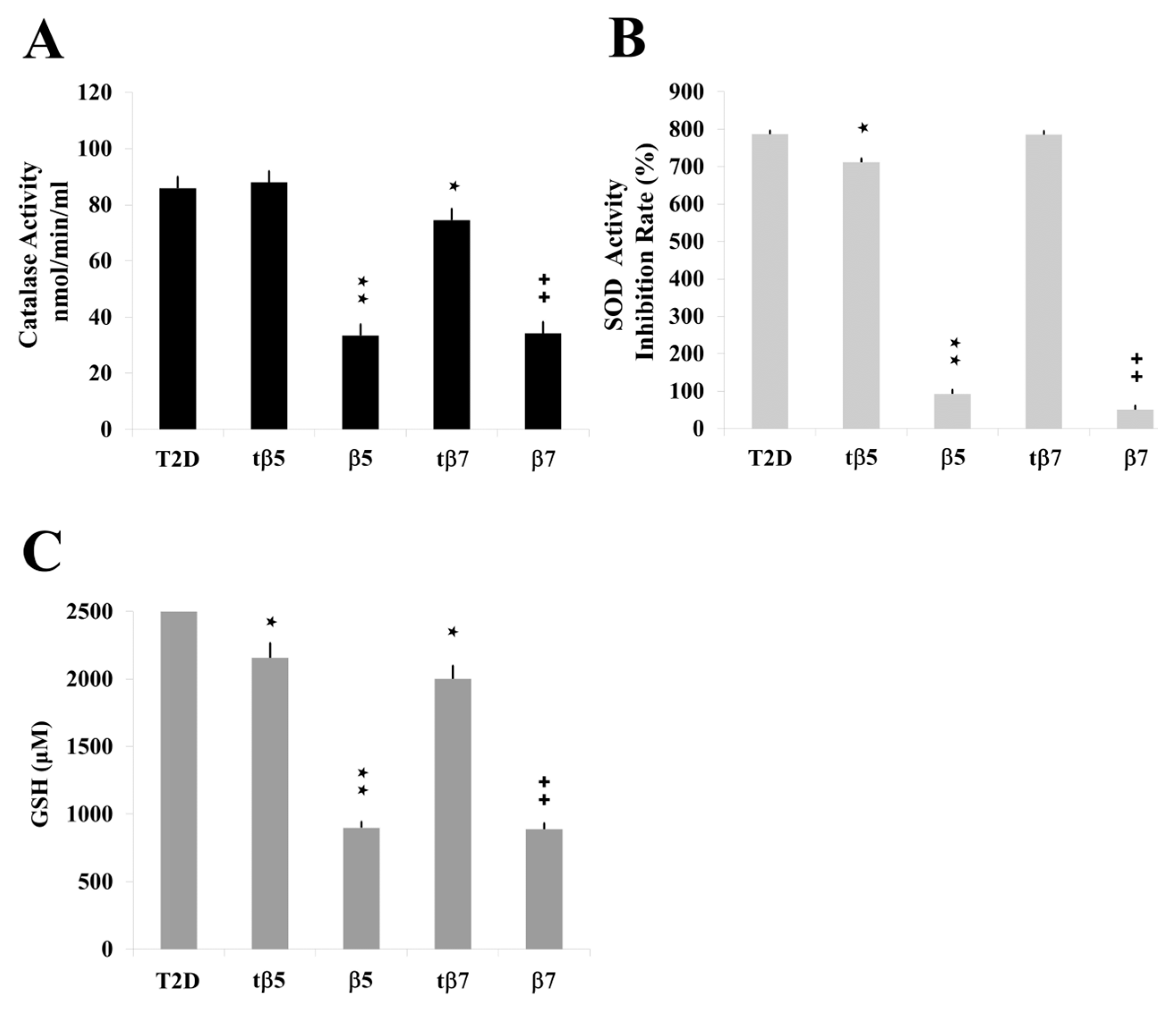
2.5. Assessing the Antioxidant Regulatory Capacity of β5 and β7 Conglutins
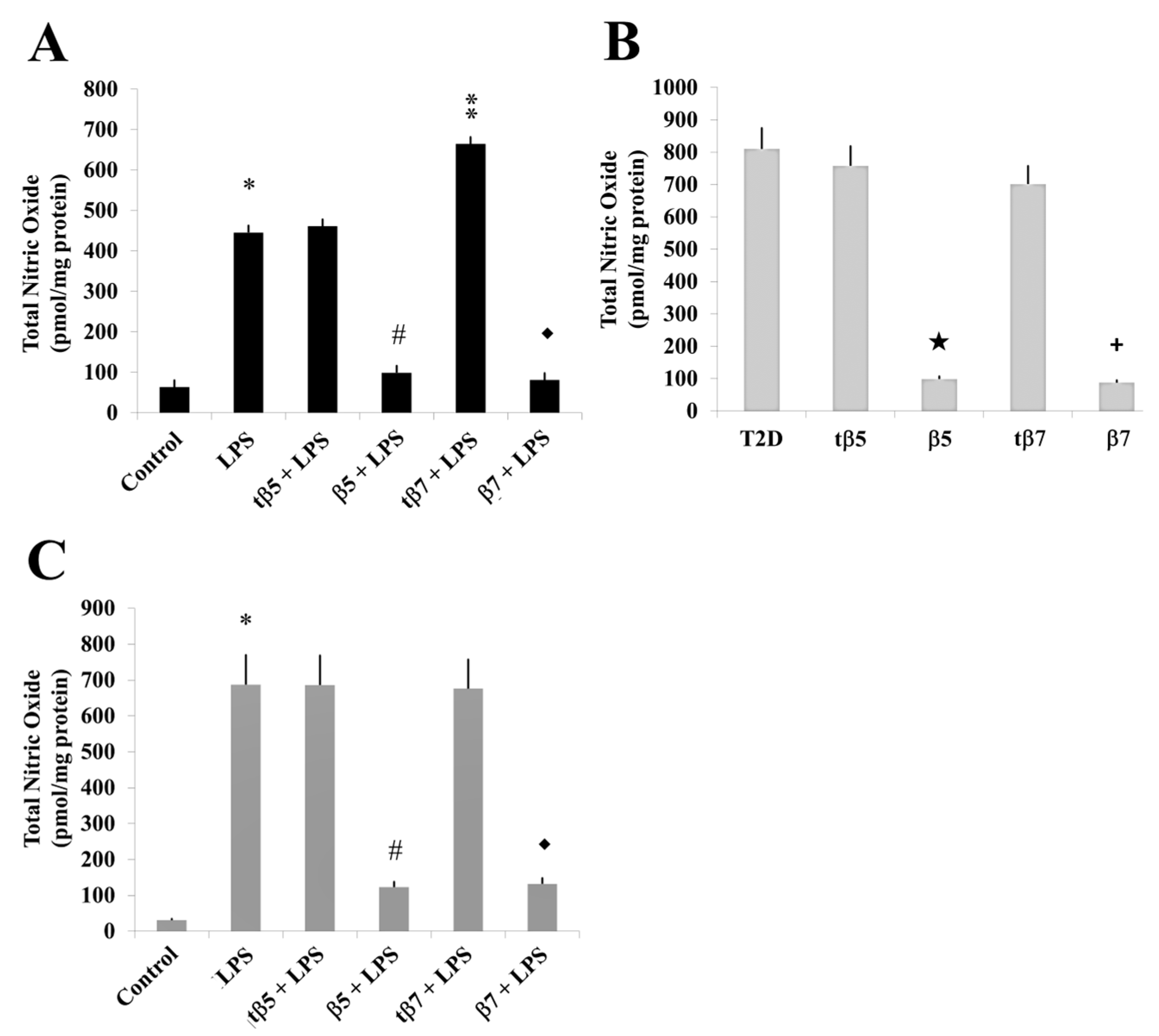
2.6. Effect of β5 and β7 Conglutins on NO Production
3. Materials and Methods
3.1. Construction of Expression Plasmids
3.2. β5 and β7 Conglutin Protein Expression and Purification
3.3. Conglutin Isoforms Antibody Production
3.4. HepG2 Cell Culture and Treatment
3.5. PBMC Isolation and Culture
3.6. Participant Information
3.7. Cell Viability Assay
3.8. SDS–PAGE Protein Separation and Immunoblotting
3.9. Polymerase Chain Reaction (PCR) Array for Inflammatory Response
3.10. ELISA Assays for Cytokines, TNF-α, and Inflammatory Pathway Quantification
3.11. Nitric Oxide Detection
3.12. Antioxidant Enzymatic Activity Assays
3.13. Statistical Analysis
3.14. β-Conglutin Proteins Structural Modeling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ha Pham Thi, H.; Luan Nguyen, T. Legumes research. In Nutraceutical Properties of Legume Seeds: Phytochemical Compounds; Jimenez-Lopez, J.C., Clemente, A., Eds.; InTechOpen: London, UK, 2022; Volume 2. [Google Scholar] [CrossRef]
- Elamine, Y.; Alaiz, M.; Girón-Calle, J.; Guiné, R.P.F.; Vioque, J. Nutritional characteristics of the seed protein in 23 Mediterranean legumes. Agronomy 2022, 12, 400. [Google Scholar] [CrossRef]
- Kiersnowska, K.; Jakubczyk, A. Bioactive peptides obtained from legume seeds as new compounds in metabolic syndrome prevention and diet therapy. Foods 2022, 11, 3300. [Google Scholar] [CrossRef]
- Jimenez-Lopez, J.C.; Clemente, A.; Ochatt, S.J.; Vaz Patto, M.C.; Von Wettberg, E.; Smýkal, P. Biological and genetic basis of agronomical and seed quality traits in legumes. Front. Plant Sci. 2022, 13, 1009980. [Google Scholar] [CrossRef] [PubMed]
- Juárez-Chairez, M.F.; Meza-Márquez, O.G.; Márquez-Flores, Y.K.; Jiménez-Martínez, C. Potential anti-inflammatory effects of legumes: A review. Br. J. Nutr. 2022, 128, 2158–2169. [Google Scholar] [CrossRef] [PubMed]
- Basson, A.R.; Ahmed, S.; Almutairi, R.; Seo, B.; Cominelli, F. Regulation of intestinal inflammation by soybean and soy-derived compounds. Foods 2021, 10, 774. [Google Scholar] [CrossRef]
- Paterson, S.; Fernández-Tomé, S.; Hernández-Ledesma, B. Modulatory Effects of a lunasin-enriched soybean extract on immune response and oxidative stress-associated biomarkers. Biol. Life Sci. Forum 2022, 12, 10. [Google Scholar] [CrossRef]
- Alaswad, A.A.; Krishnan, H.B. Immunological investigation for the presence of lunasin, a chemopreventive soybean peptide, in the seeds of diverse plants. J. Agric. Food Chem. 2016, 64, 2901. [Google Scholar] [CrossRef] [PubMed]
- González-Montoya, M.; Hernández-Ledesma, B.; Mora-Escobedo, R.; Martínez-Villaluenga, C. Bioactive peptides from germinated soybean with anti-diabetic potential by inhibition of dipeptidyl peptidase-IV, α-amylase, and α-gucosidase enzymes. Int. J. Mol. Sci. 2018, 19, 2883. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, T.; Jiang, B.; Miao, M.; Mu, W. The effects of an antioxidative pentapeptide derived from chickpea protein hydrolysates on oxidative stress in Caco-2 and HT-29 cell lines. J. Funct. Foods 2014, 7, 719. [Google Scholar] [CrossRef]
- Utrilla, M.P.; Peinado, M.J.; Ruiz, R.; Rodriguez-Nogales, A.; Algieri, F.; Rodriguez-Cabezas, M.E.; Clemente, A.; Galvez, J.; Rubio, L.A. Pea (Pisum sativum L.) seed albumin extracts show anti-inflammatory effect in the DSS model of mouse colitis. Mol. Nutr. Food Res. 2015, 59, 807. [Google Scholar] [CrossRef]
- Lima-Cabello, E.; Morales-Santana, S.; Foley, R.C.; Melser, S.; Alché, V.; Siddique, K.H.M.; Singh, K.B.; Alché, J.D.; Jimenez-Lopez, J.C. Ex vivo and in vitro assessment of anti-inflammatory activity of seed β-conglutin proteins from Lupinus angustifolius. J. Funct. Foods 2018, 40, 510. [Google Scholar] [CrossRef]
- Lima-Cabello, E.; Alche, V.; Foley, R.C.; Andrikopoulos, S.; Morahan, G.; Singh, K.B.; Alche, J.D.; Jimenez-Lopez, J.C. Narrow-leafed lupin (Lupinus angustifolius L.) β-conglutin proteins modulate the insulin signaling pathway as potential type 2 diabetes treatment and inflammatory-related disease amelioration. Mol. Nutr. Food Res. 2017, 61, 1600819. [Google Scholar] [CrossRef] [PubMed]
- Lima-Cabello, E.; Alché, J.D.; Morales-Santana, S.; Clemente, A.; Jimenez-Lopez, J.C. Narrow-leafed lupin (Lupinus angustifolius L.) seeds gamma-conglutin is an anti-inflammatory protein promoting insulin resistance improvement and oxidative stress amelioration in PANC-1 pancreatic cell-line. Antioxidants 2019, 9, 12. [Google Scholar] [CrossRef]
- Montserrat-de la Paz, S.; Villanueva, A.; Pedroche, J.; Millan, F.; Martin, M.E.; Millan-Linares, M.C. Antioxidant and anti-inflammatory properties of bioavailable protein hydrolysates from lupin-derived agri-waste. Biomolecules 2021, 11, 1458. [Google Scholar] [CrossRef] [PubMed]
- Santos-Sánchez, G.; Cruz-Chamorro, I.; Álvarez-Ríos, A.I.; Álvarez-Sánchez, N.; Rodríguez-Ortiz, B.; Álvarez-López, A.I.; Fernández-Pachón, M.S.; Pedroche, J.; Millán, F.; Millán-Linares, M.D.C.; et al. Bioactive peptides from lupin (Lupinus angustifolius) prevent the early stages of atherosclerosis in western diet-fed ApoE-/- mice. J. Agric. Food Chem. 2022, 70, 8243. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Lopez, J.C. Narrow-leafed lupin (Lupinus angustifolius L.) β-conglutin: A multifunctional family of proteins with roles in plant defence, human health benefits, and potential uses as functional food. Legume Sci. 2020, 2, e33. [Google Scholar] [CrossRef]
- Heinzl, G.C.; Tretola, M.; De Benedetti, S.; Silacci, P.; Scarafoni, A. Lupinus albus γ-Conglutin: New findings about its action at the intestinal barrier and a critical analysis of the state of the art on its postprandial glycaemic regulating activity. Nutrients 2022, 14, 3666. [Google Scholar] [CrossRef] [PubMed]
- Tapadia, M.; Johnson, S.; Utikar, R.; Newsholme, P.; Carlessi, R. Antidiabetic effects and mechanisms of action of -conglutin from lupin seeds. J. Funct. Foods 2021, 87, 104786. [Google Scholar] [CrossRef]
- Guerra-Ávila, P.L.; Guzmán, T.J.; Domínguez-Rosales, J.A.; García-López, P.M.; Cervantes-Garduño, A.B.; Wink, M.; Gurrola-Díaz, C.M. Combined gamma conglutin and lupanine treatment exhibits in vivo an enhanced antidiabetic effect by modulating the liver gene expression profile. Pharmaceuticals 2023, 16, 117. [Google Scholar] [CrossRef]
- Jimenez-Lopez, J.C.; Melser, S.; DeBoer, K.; Thatcher, L.F.; Kamphuis, L.G.; Foley, R.C.; Singh, K.B. Narrow-Leafed Lupin (Lupinus angustifolius) β1- and β6-conglutin proteins exhibit antifungal activity, protecting plants against necrotrophic pathogen induced damage from Sclerotinia sclerotiorum and Phytophthora nicotianae. Front. Plant Sci. 2016, 7, 1856. [Google Scholar] [CrossRef]
- Mazumder, K.; Biswas, B.; Kerr, P.G.; Blanchard, C.; Nabila, A.; Golder, M.; Aziz, M.G.; Farahnaky, A. Comparative assessment of nutritional, thermal, rheological and functional properties of nine australian lupin cultivars. Sci. Rep. 2021, 11, 21515. [Google Scholar] [CrossRef]
- Escudero-Feliu, J.; García-Costela, M.; Moreno-SanJuan, S.; Puentes-Pardo, J.D.; Arrabal, S.R.; González-Novoa, P.; Núñez, M.I.; Carazo, A.; Jimenez-Lopez, J.C.; León, J. Narrow leafed lupin (Lupinus angustifolius L.) β-conglutin seed proteins as a new natural cytotoxic agents against breast cancer cells. Nutrients 2023, 15, 523. [Google Scholar] [CrossRef]
- Lima-Cabello, E.; Robles-Bolivar, P.; Alché, J.D.; Jimenez-Lopez, J.C. Narrow-leafed lupin β-conglutin proteins epitopes identification and molecular features analysis involved in cross-allergenicity to peanut and other legumes. Genom. Comput. Biol. 2016, 2, e29. [Google Scholar] [CrossRef]
- Jimenez-Lopez, J.C.; Lima-Cabello, E.; Melser, S.; Foley, R.C.; Singh, K.B.; Alché, J.D. Lupin allergy: Uncovering structural features and epitopes of β-conglutin proteins in Lupinus angustifolius L. with a focus on cross-allergenic reactivity to peanut and other legumes. Bioinform. Biomed. Eng. 2015, 9043, 96. [Google Scholar] [CrossRef]
- Jimenez-Lopez, J.C. Lupin (Lupinus angustifolius L.) β-conglutin proteins: Structure functional features, catalytic mechanism modelling and cross-allergenicity identification using protein threading and molecular docking methods. Protein Sci. 2015, 24, 1. [Google Scholar] [CrossRef]
- Lawrence, M.C.; Izard, T.; Beuchat, M.; Blagrove, R.J.; Colman, P.M. Structure of phaseolin at 2.2 A resolution. Implications for a common vicilin/legumin structure and the genetic engineering of seed storage proteins. J. Mol. Biol. 1994, 238, 748. [Google Scholar] [CrossRef]
- Mahmoud, S.A.; El-Saeid, M.E.; Mohy Eldin, A.; Abdel Atty, A.M. Conglutin gamma, a lupin seed protein reduces oxidative stress in diabetics. J. Glob. Sci. Res. 2023, 8, 2866. [Google Scholar] [CrossRef]
- Galicia-Garcia, U.; Benito-Vicente, A.; Jebari, S.; Larrea-Sebal, A.; Siddiqi, H.; Uribe, K.B.; Ostolaza, H.; Martín, C. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 2020, 21, 6275. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, Q.; Yuan, H.; Zhou, L.; Li, F.F.; Wang, S.M.; Shi, G.; Wang, M. Nitric oxide mediates inflammation in type ii diabetes mellitus through the PPARγ/eNOS signaling pathway. PPAR Res. 2020, 2020, 8889612. [Google Scholar] [CrossRef]
- Nakamura, A.; Terauchi, Y. Impaired insulin secretion and related factors in East Asian individuals. J. Diabetes Investig. 2022, 13, 233. [Google Scholar] [CrossRef]
- Kany, S.; Vollrath, J.T.; Relja, B. Cytokines in inflammatory disease. Int. J. Mol. Sci. 2019, 20, 6008. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Torres, M.; Carazo-Gallego, A.; Jimenez-Lopez, J.C.; Avilés-Pérez, M.D.; Díaz-Arco, S.; Lozano-Alonso, S.; Lima-Cabello, E.; Alché, J.D.; Reyes-García, R.; Morales-Santana, S. Differential inflammatory environment in patients with osteoporosis and type 2 diabetes mellitus. Rev. Osteoporos Metab. Min. 2022, 14, 34. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2017, 9, 7204. [Google Scholar] [CrossRef] [PubMed]
- La Manna, S.; Di Natale, C.; Florio, D.; Marasco, D. Peptides as therapeutic agents for inflammatory-related diseases. Int. J. Mol. Sci. 2018, 19, 2714. [Google Scholar] [CrossRef] [PubMed]
- Forcina, L.; Miano, C.; Scicchitano, B.M.; Rizzuto, E.; Berardinelli, M.G.; De Benedetti, F.; Pelosi, L.; Musarò, A. Increased circulating levels of interleukin-6 affect the redox balance in skeletal muscle. Oxidative Med. Cell. Longev. 2019, 2019, 3018584. [Google Scholar] [CrossRef]
- Cantaert, T.; Baeten, D.; Tak, P.P.; van Baarsen, L.G. Type I IFN and TNFα cross-regulation in immune-mediated inflammatory disease: Basic concepts and clinical relevance. Arthritis Res. Ther. 2010, 12, 219. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Chung, D.H. TLR4-mediated IL-12 production enhances IFN-γ and IL-1β production, which inhibits TGF-β production and promotes antibody-induced joint inflammation. Arthritis Res. Ther. 2012, 14, R210. [Google Scholar] [CrossRef]
- Wiley, S.R.; Winkles, J.A. TWEAK, a member of the TNF superfamily, is a multifunctional cytokine that binds the TweakR/Fn14 receptor. Cytokine Growth Factor Rev. 2003, 14, 241. [Google Scholar] [CrossRef]
- Frangie, C.; Daher, J. Role of myeloperoxidase in inflammation and atherosclerosis. Biomed. Rep. 2022, 16, 53. [Google Scholar] [CrossRef]
- Carmo de Carvalho e Martins, M.D.; da Silva Santos Oliveira, A.S.; da Silva, L.A.A.; Primo, M.G.S.; de Carvalho Lira, V.B. Biological indicators of oxidative stress [Malondialdehyde, Catalase, Glutathione Peroxidase, and Superoxide Dismutase] and their application in nutrition. In Biomarkers in Nutrition. Biomarkers in Disease: Methods, Discoveries and Applications; Patel, V.B., Preedy, V.R., Eds.; Springer: Cham, Singapore, 2022. [Google Scholar] [CrossRef]
- Ighodaro, M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287. [Google Scholar] [CrossRef]
- Sharma, J.N.; Al-Omran, A.; Parvathy, S.S. Role of nitric oxide in inflammatory diseases. Inflammopharmacology 2007, 15, 252. [Google Scholar] [CrossRef] [PubMed]
- Stancill, J.S.; Kasmani, M.Y.; Khatun, A.; Cui, W.; Corbett, J.A. Cytokine and nitric oxide-dependent gene regulation in islet endocrine and nonendocrine cells. Function 2022, 3, zqab063. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.C.; Chou, M.J.; Wang, C.H. Lunasin attenuates obesity-related inflammation in RAW264.7 cells and 3T3-L1 adipocytes by inhibiting inflammatory cytokine production. PLoS ONE 2017, 12, e0171969. [Google Scholar] [CrossRef] [PubMed]
- Garmidolova, A.; Desseva, I.; Mihaylova, D.; Lante, A. Bioactive peptides from lupinus spp. seed proteins-state-of-the-art and perspectives. Appl. Sci. 2022, 12, 3766. [Google Scholar] [CrossRef]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell viability assays. In Assay Guidance Manual; Markossian, S., Grossman, A., Brimacombe, K., Arkin, M., Auld, D., Austin, C., Baell, J., Chung, T.D.Y., Coussens, N.P., Dahlin, J.L., et al., Eds.; Bethesda, Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2013. Available online: https://pubmed.ncbi.nlm.nih.gov/23805433/ (accessed on 1 July 2016).


| pg/mL | HepG2 Control | LPS | LPS+tβ5 1 | LPS+β5 | LPS+tβ71 | LPS+β7 |
| Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | |
| IL-17 | 14.30 ± 0.06 | 140.00 ± 0.02 * | 153.00 ± 0.05 | 11.20 ± 0.05 # | 156.00 ± 0.04 | 18.30 ± 0.01 ♦ |
| IL-12 | 9.27 ± 0.02 | 241.00 ± 0.05 * | 243.00 ± 0.02 | 76.00 ± 0.04 # | 271.00 ± 0.09 | 82.00 ± 0.02 ♦ |
| IL-27 | 11.20 ± 0.03 | 198.00 ± 0.05 * | 192.00 ± 0.01 | 87.00 ± 0.02 # | 176.00 ± 0.05 | 100.00 ± 0.04 ♦ |
| IL-6 | 23.50 ± 0.05 | 287.00 ± 0.04 * | 301.00 ± 0.01 | 178.00 ± 0.05 # | 262.00 ± 0.01 | 177.00 ± 0.06 ♦ |
| IL-2 | 13.20 ± 0.04 | 56.00 ± 0.05 * | 49.00 ± 0.01 | 32.30 ± 0.05 # | 52.00 ± 0.03 | 21.30. ± 0.05 ♦ |
| IL-8 | 8.30 ± 0.02 | 79.00 ± 0.03 * | 82.00 ± 0.01 | 41.00 ± 0.03 # | 80.00 ± 0.05 | 32.00 ± 0.01 ♦ |
| MPO | 9.30 ± 0.01 | 321.00 ± 0.10 * | 439.00 ± 0.02 | 173.00 ± 0.05 # | 501.00 ± 0.03 | 178.00 ± 0.06 ♦ |
| INFγ | 44.90 ± 0.01 | 1907.00 ± 0.10 * | 1947.00 ± 0.01 | 653.00 ± 0.02 # | 1883.00 ± 0.05 | 631.00 ± 0.02 ♦ |
| S-TNF-R1 | 812.00 ± 0.01 | 2091.00 ± 0.06 * | 1982.00 ± 0.04 | 932.00 ± 0.04 # | 1922.00 ± 0.02 | 921.00 ± 0.05 ♦ |
| S-TNF-R2 | 712.00 ± 0.03 | 2931.00 ± 0.07 * | 2786.00 ± 0.05 | 1023.00 ± 0.02 # | 2891.00 ± 0.05 | 1234.00 ± 0.05 ♦ |
| TWEAK | 87.00 ± 0.02 | 638.00 ± 0.06 * | 676.00 ± 0.05 | 213.00 ± 0.01 # | 711.00 ± 0.03 | 345.00 ± 0.01 ♦ |
| pg/mL | T2D | tβ5 1 | β5 | tβ7 1 | β7 | |
| Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | ||
| IL-17 | 150.30 ± 0.03 | 153.00 ± 0.01 | 11.20 ± 0.02 ◊ | 156.00 ± 0.03 | 18.30 ± 0.01 ◊ | |
| IL-12 | 219.27 ± 0.03 | 240.32 ± 0.05 | 76.31 ± 0.02 ◊ | 231.30 ± 0.01 | 81.32 ± 0.04 ◊ | |
| IL-27 | 43.20 ± 0.03 | 47.40 ± 0.03 | 23.50 ± 0.02 ◊ | 46.30 ± 0.05 | 26.70 ± 0.02 ◊ | |
| IL-6 | 65.50 ± 0.01 | 64.30 ± 0.02 | 36.40 ± 0.02 ◊ | 70.10 ± 0.05 | 41.40 ± 0.09 ◊ | |
| IL-2 | 43.00 ± 0.01 | 42.00 ± 0.01 | 22.80 ± 0.02 ◊ | 48.70 ± 0.01 | 29.80 ± 0.05 ◊ | |
| IL-8 | 67.00 ± 0.05 | 67.30 ± 0.03 | 36.80 ± 0.02 ◊ | 63.90 ± 0.03 | 35.80 ± 0.05 ◊ | |
| MPO | 121.00 ± 0.01 | 119.30 ± 0.04 | 54.80 ± 0.02 ◊ | 128.60 ± 0.06 | 56.40 ± 0.06 ◊ | |
| INFγ | 231.90 ± 0.05 | 242.30 ± 0.03 | 108.30 ± 0.02 ◊ | 227.30 ± 0.09 | 96.30 ± 0.02 ◊ | |
| S-TNF-R1 | 1843.00 ± 0.01 | 1797.30 ± 0.1 | 646.30 ± 0.02 ◊ | 1811.10 ± 0.09 | 701.30 ± 0.02 ◊ | |
| S-TNF-R2 | 1615.00 ± 0.05 | 1573.40 ± 0.20 | 936.32 ± 0.02 ◊ | 1678.30 ± 0.01 | 927.34 ± 0.05 ◊ | |
| TWEAK | 198.00 ± 0.01 | 201.30 ± 0.04 | 76.30 ± 0.02 ◊ | 189.30 ± 0.09 | 67.90 ± 0.30 ◊ | |
| pg/mL | Healthy Control | LPS | LPS+tβ5 1 | LPS+β5 | LPS+tβ7 1 | LPS+β7 |
| Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | Mean ± SEM | |
| IL-17 | 1.66 ± 0.02 | 143.00 ± 0.02 * | 153.00 ± 0.04 | 11.20 ± 0.01 # | 156.00 ± 0.03 | 18.30 ± 0.01 ♦ |
| IL-12 | 27.76 ± 0.05 | 241.00 ± 0.03 * | 243.00 ± 0.09 | 76.00 ± 0.05 # | 251.00 ± 0.05 | 81.30 ± 0.01 ♦ |
| IL-27 | 256.70 ± 0.08 | 473.30 ± 0.04 * | 421.20 ± 0.06 | 238.40 ± 0.09 # | 491.30 ± 0.05 | 298.40 ± 0.05 ♦ |
| IL-6 | 16.80 ± 0.03 | 198.30 ± 0.05 * | 187.40 ± 0.10 | 57.40 ± 0.04 # | 194.30 ± 0.04 | 67.40 ± 0.06 ♦ |
| IL-2 | 14.50 ± 0.02 | 79.40 ± 0.09 * | 78.40 ± 0.01 | 34.60 ± 0.06 # | 81.30 ± 0.05 | 36.60 ± 0.05 ♦ |
| IL-8 | 9.40 ± 0.06 | 145.60 ± 0.09 * | 152.40 ± 0.02 | 24.50 ± 0.04 # | 148.40 ± 0.10 | 32.40 ± 0.08 ♦ |
| MPO | 5.80 ± 0.02 | 349.40 ± 0.09 * | 374.50 ± 0.04 | 163.40 ± 0.02 # | 356.40 ± 0.02 | 174.40 ± 0.03 ♦ |
| INFγ | 37.50 ± 0.04 | 284.40 ± 0.01 * | 278.40 ± 0.08 | 87.40 ± 0.04 # | 291.40 ± 0.09 | 79.40 ± 0.05 ♦ |
| S-TNF-R1 | 801.30 ± 0.09 | 1489.40 ± 0.10 * | 1456.80 ± 0.01 | 783.40 ± 0.01 # | 1581.10 ± 0.08 | 900.00 ± 0.02 ♦ |
| S-TNF-R2 | 765.30 ± 0.05 | 1932.30 ± 0.05 * | 1911.30 ± 0.05 | 609.90 ± 0.01 # | 1900.30 ± 0.04 | 1239.90 ± 0.06 ♦ |
| TWEAK | 86.30 ± 0.06 | 684.30 ± 0.06 * | 711.20 ± 0.03 | 211.30 ± 0.03 # | 765.40 ± 0.09 | 309.30 ± 0.05 ♦ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lima-Cabello, E.; Escudero-Feliu, J.; Peralta-Leal, A.; Garcia-Fernandez, P.; Siddique, K.H.M.; Singh, K.B.; Núñez, M.I.; León, J.; Jimenez-Lopez, J.C. β-Conglutins’ Unique Mobile Arm Is a Key Structural Domain Involved in Molecular Nutraceutical Properties of Narrow-Leafed Lupin (Lupinus angustifolius L.). Int. J. Mol. Sci. 2023, 24, 7676. https://doi.org/10.3390/ijms24087676
Lima-Cabello E, Escudero-Feliu J, Peralta-Leal A, Garcia-Fernandez P, Siddique KHM, Singh KB, Núñez MI, León J, Jimenez-Lopez JC. β-Conglutins’ Unique Mobile Arm Is a Key Structural Domain Involved in Molecular Nutraceutical Properties of Narrow-Leafed Lupin (Lupinus angustifolius L.). International Journal of Molecular Sciences. 2023; 24(8):7676. https://doi.org/10.3390/ijms24087676
Chicago/Turabian StyleLima-Cabello, Elena, Julia Escudero-Feliu, Andreina Peralta-Leal, Pedro Garcia-Fernandez, Kadambot H. M. Siddique, Karam B. Singh, Maria I. Núñez, Josefa León, and Jose C. Jimenez-Lopez. 2023. "β-Conglutins’ Unique Mobile Arm Is a Key Structural Domain Involved in Molecular Nutraceutical Properties of Narrow-Leafed Lupin (Lupinus angustifolius L.)" International Journal of Molecular Sciences 24, no. 8: 7676. https://doi.org/10.3390/ijms24087676
APA StyleLima-Cabello, E., Escudero-Feliu, J., Peralta-Leal, A., Garcia-Fernandez, P., Siddique, K. H. M., Singh, K. B., Núñez, M. I., León, J., & Jimenez-Lopez, J. C. (2023). β-Conglutins’ Unique Mobile Arm Is a Key Structural Domain Involved in Molecular Nutraceutical Properties of Narrow-Leafed Lupin (Lupinus angustifolius L.). International Journal of Molecular Sciences, 24(8), 7676. https://doi.org/10.3390/ijms24087676







