A Functional Yeast-Based Screen Identifies the Host Microtubule Cytoskeleton as a Target of Numerous Chlamydia pneumoniae Proteins
Abstract
1. Introduction
2. Results
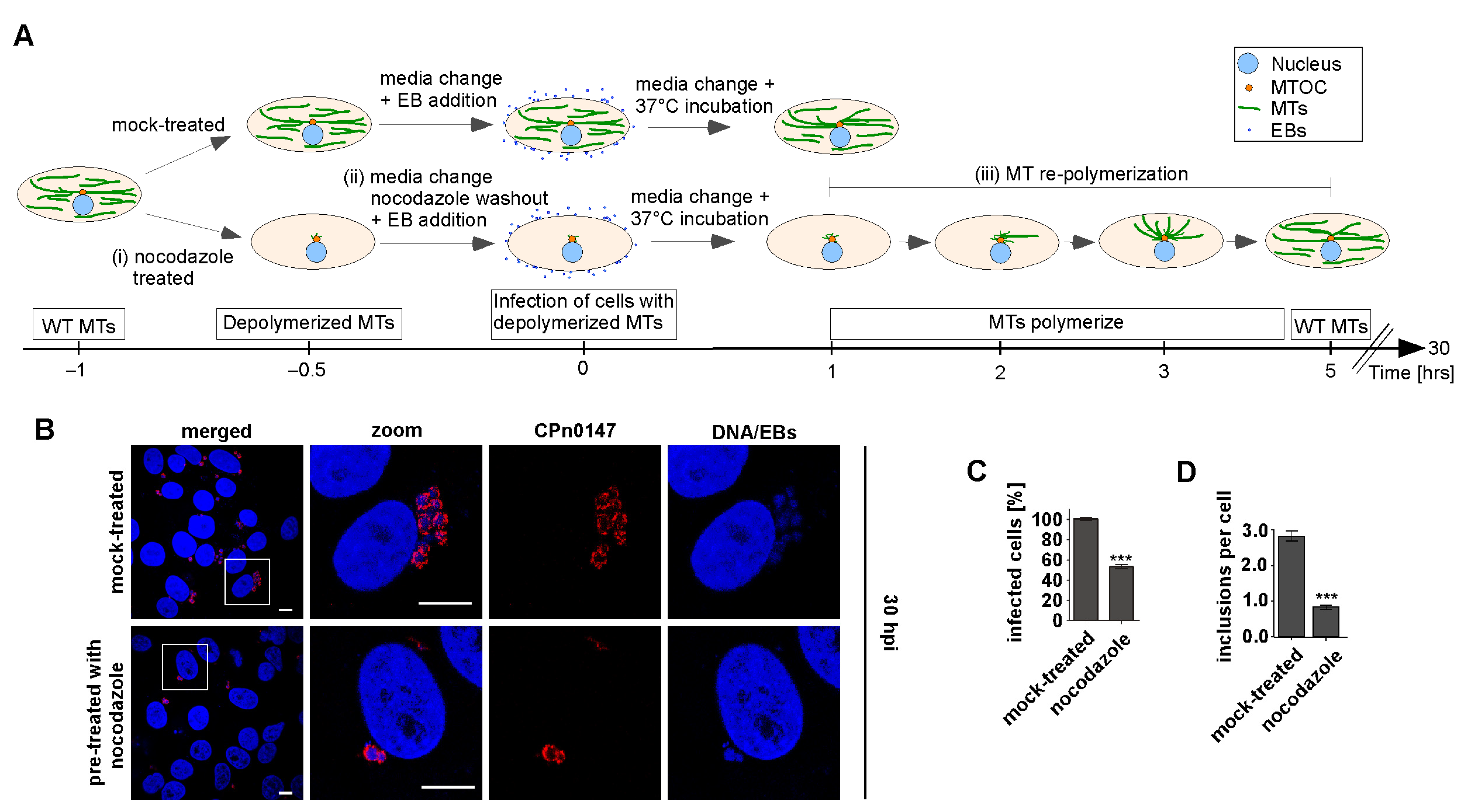
2.1. Early Steps in Cpn Infection Required an Intact MT Cytoskeleton
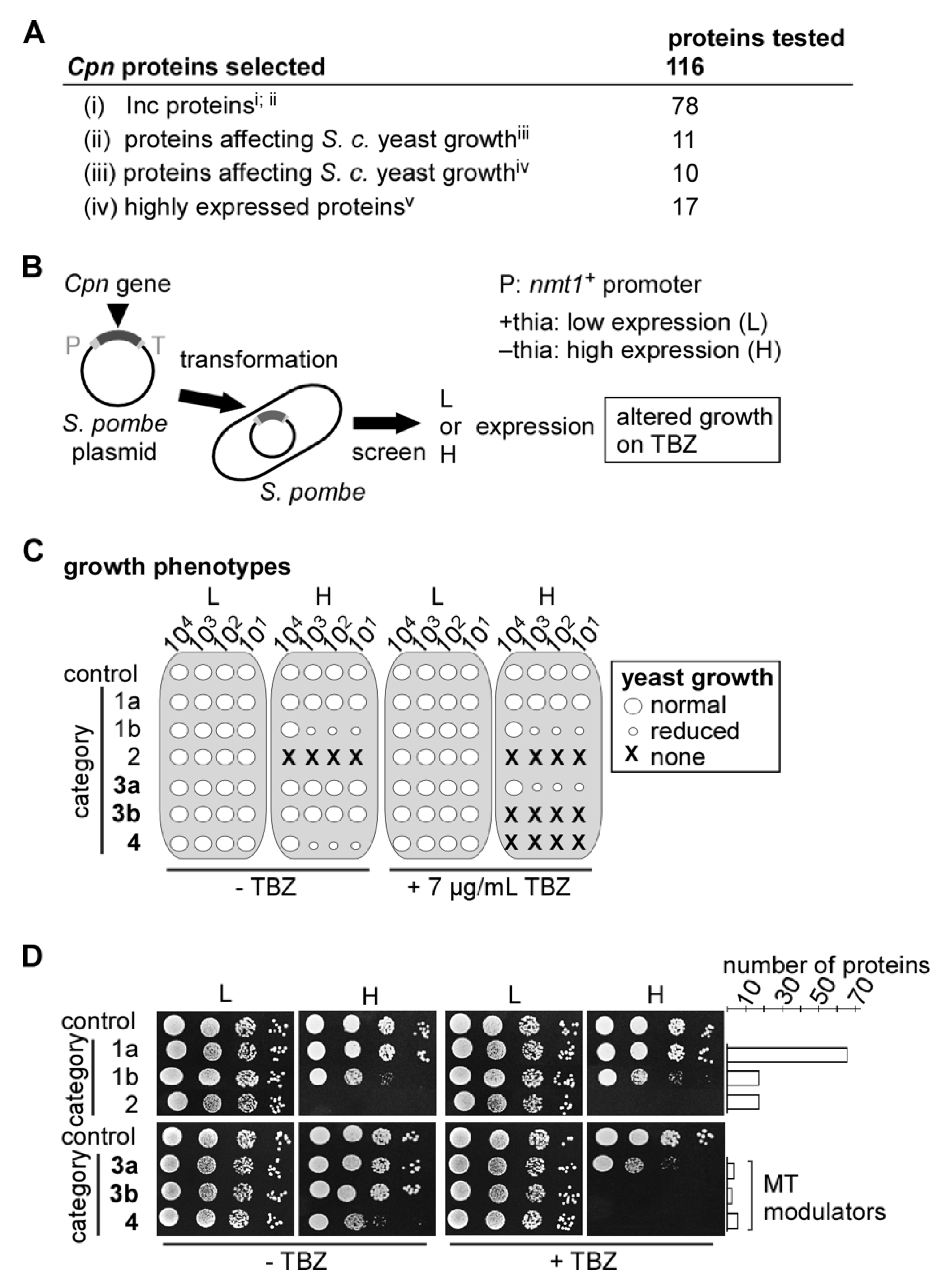
2.2. Identification of Cpn MT-Modulating Proteins via the S. pombe Test System
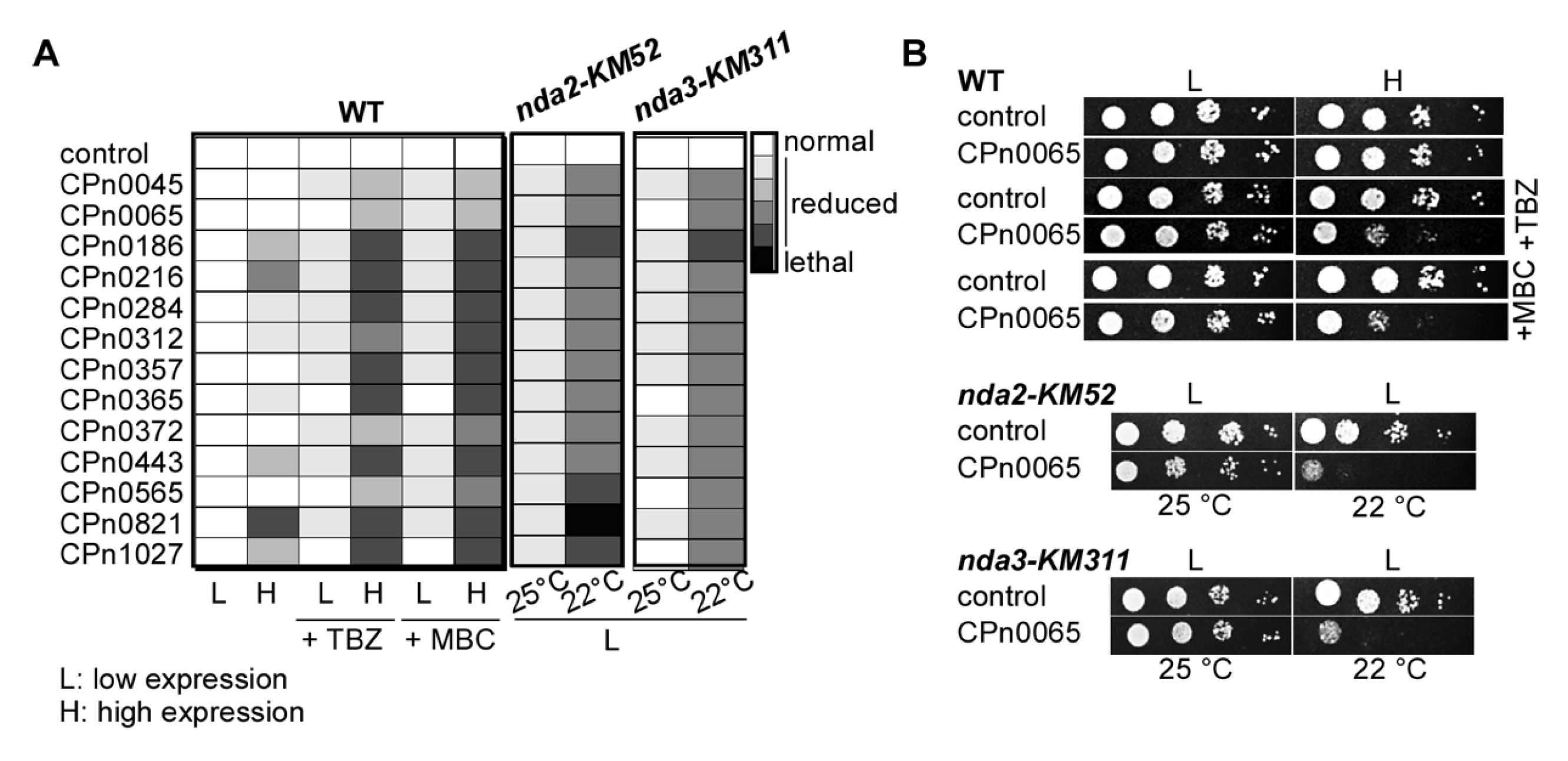
2.3. S. pombe Tubulin Mutants Became Synthetically Lethal upon the Expression of Chlamydial Genes
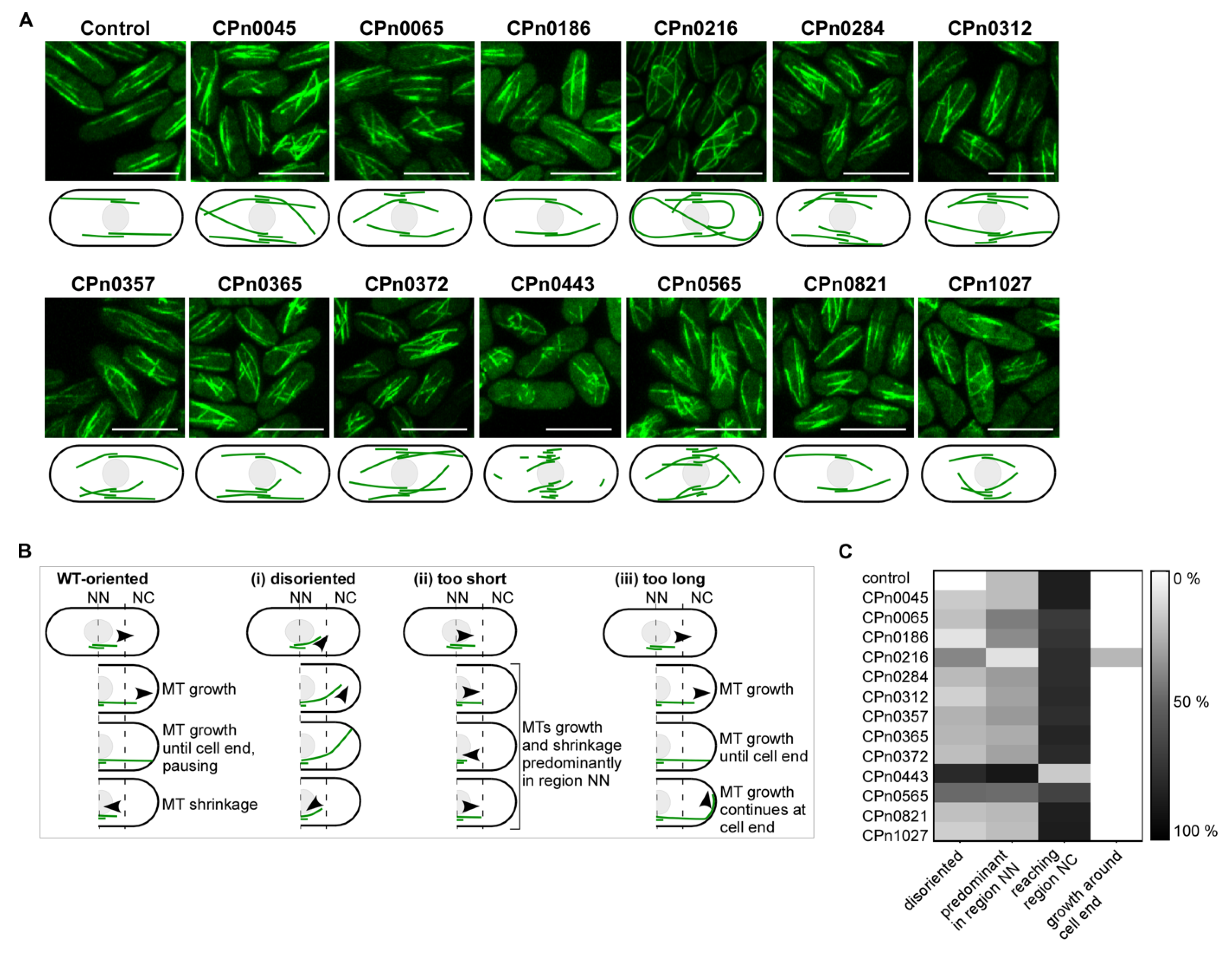
2.4. The S. pombe Interphase MT Cytoskeleton Became Highly Aberrant upon the Expression of Chlamydial Genes
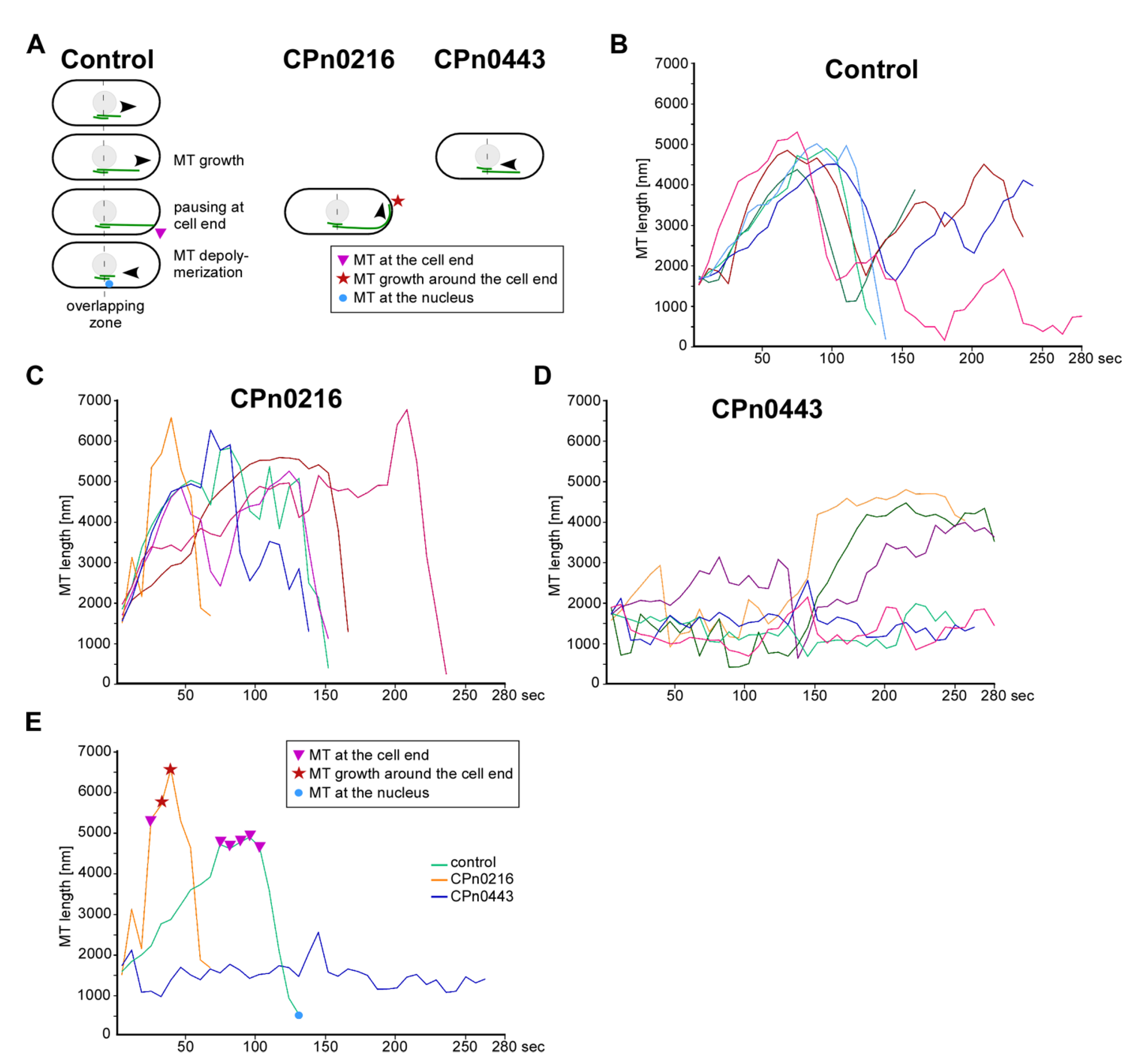
2.5. CPn0216 and CPn0443 Altered the MT Dynamics
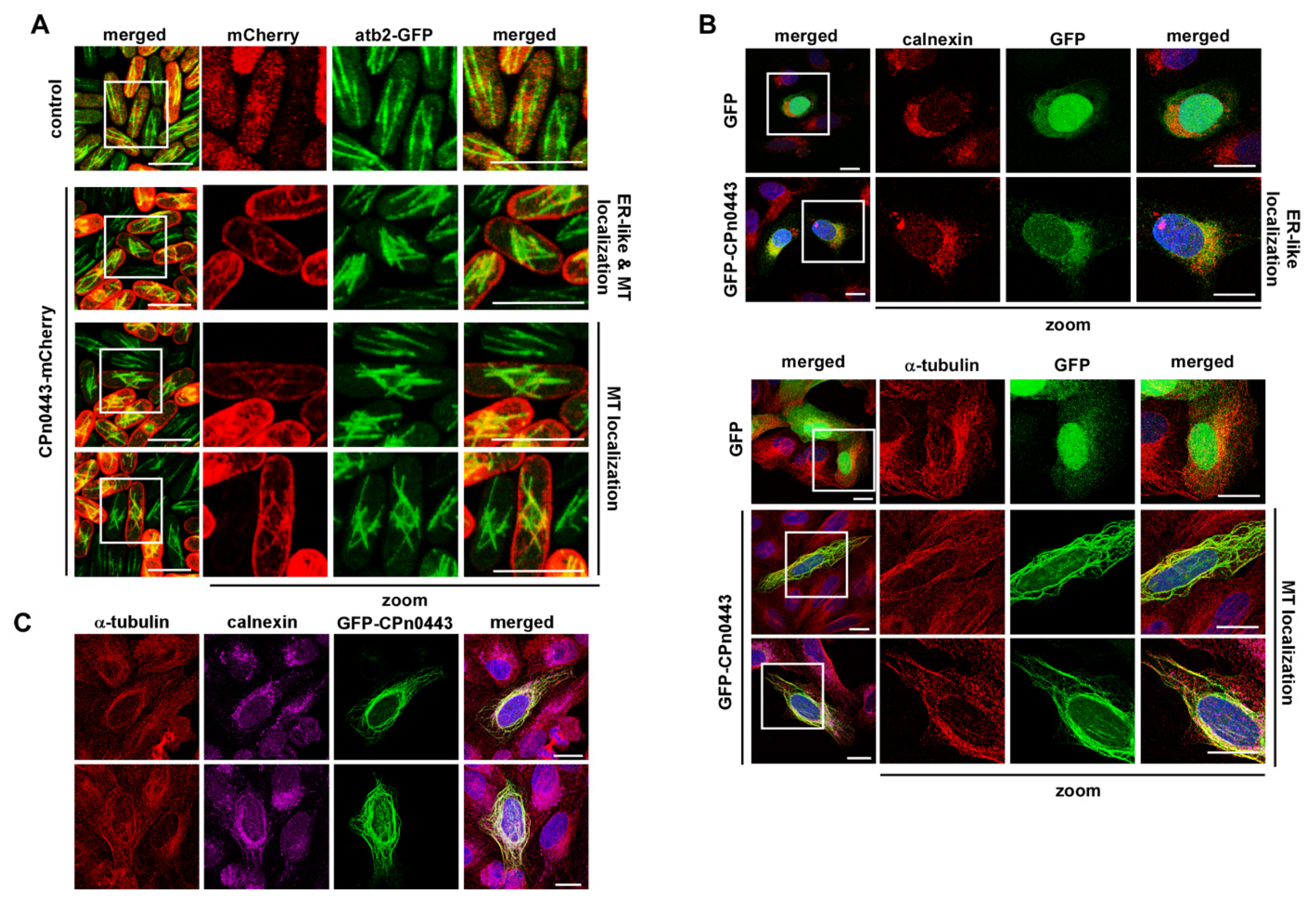
2.6. CPn0443 Associated with MTs in Different Cell Systems
2.7. CPn0443 Had Microtubule-Binding Activity
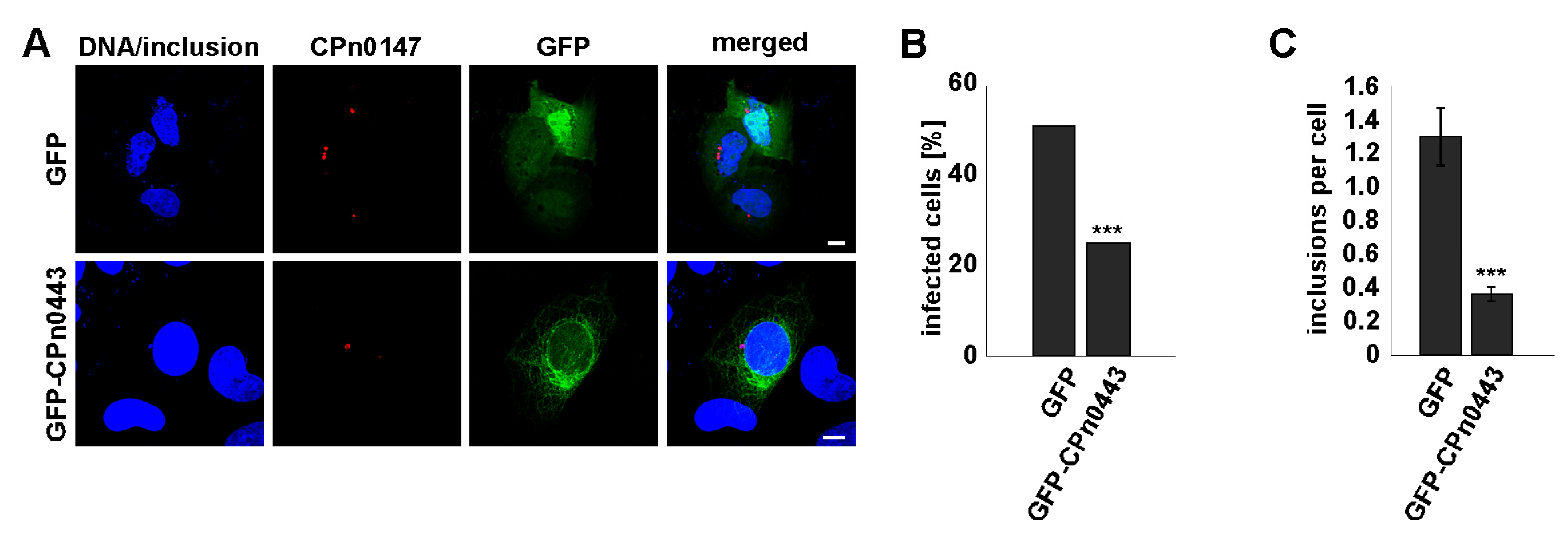
2.8. Ectopically Expressed CPn0443 Attenuated Cpn Infection
3. Discussion
3.1. The MT Cytoskeleton Played a Vital Role in C. pneumoniae Infection
3.2. The Identified MT-Modulating Proteins Were Expressed at Different Stages of the Chlamydial Developmental Cycle
3.3. Eleven of the Identified Proteins Were Inc Proteins
3.4. Inc Proteins and Modulation of the MT Cytoskeleton
4. Material and Methods
4.1. Yeast Strains, Media and Growth Conditions
4.2. Generation of the Cpn Expression Library for Screening in S. pombe
4.3. Yeast Protein Extracts and Western Blot Analysis
4.4. Mammalian Cell Lines and Production of Infectious Cpn EBs
4.5. Drug-Induced MT-Depolymerization and Chlamydia Infection
4.6. Transfection and Staining of Mammalian Cells
4.7. Microscopy and Image Processing
4.8. MT Binding and Bundling Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hohmann, T.; Dehghani, F. The Cytoskeleton-A Complex Interacting Meshwork. Cells 2019, 8, 362. [Google Scholar] [CrossRef] [PubMed]
- Goodson, H.V.; Jonasson, E.M. Microtubules and Microtubule-Associated Proteins. Cold Spring Harb. Perspect. Biol. 2018, 10, a022608. [Google Scholar] [CrossRef] [PubMed]
- Svitkina, T. The Actin Cytoskeleton and Actin-Based Motility. Cold Spring Harb. Perspect. Biol. 2018, 10, a018267. [Google Scholar] [CrossRef]
- Herrmann, H.; Aebi, U. Intermediate Filaments: Structure and Assembly. Cold Spring Harb. Perspect. Biol. 2016, 8, a018242. [Google Scholar] [CrossRef] [PubMed]
- Woods, B.L.; Gladfelter, A.S. The state of the septin cytoskeleton from assembly to function. Curr. Opin. Cell Biol. 2021, 68, 105–112. [Google Scholar] [CrossRef]
- Aktories, K. Bacterial protein toxins that modify host regulatory GTPases. Nat. Rev. Microbiol. 2011, 9, 487–498. [Google Scholar] [CrossRef]
- Stradal, T.E.B.; Schelhaas, M. Actin dynamics in host-pathogen interaction. FEBS Lett. 2018, 592, 3658–3669. [Google Scholar] [CrossRef]
- Alqassim, S.S. Functional Mimicry of Eukaryotic Actin Assembly by Pathogen Effector Proteins. Int. J. Mol. Sci. 2022, 23, 11606. [Google Scholar] [CrossRef]
- Radhakrishnan, G.K.; Splitter, G.A. Modulation of host microtubule dynamics by pathogenic bacteria. Biomol. Concepts 2012, 3, 571–580. [Google Scholar] [CrossRef]
- Blasi, F.; Tarsia, P.; Arosio, C.; Fagetti, L.; Allegra, L. Epidemiology of Chlamydia pneumoniae. Clin. Microbiol. Infect. 1998, 4 (Suppl. S4), S1–S6. [Google Scholar] [CrossRef]
- Porritt, R.A.; Crother, T.R. Chlamydia pneumoniae Infection and Inflammatory Diseases. Forum Immunopathol. Dis. Ther. 2016, 7, 237–254. [Google Scholar] [CrossRef]
- Campbell, L.A.; Hahn, D.L. Chlamydia pneumoniae Infections. In Chlamydia Biology: From Genome to Disease; Tran, M., Hegemann, J.H., Sütterlin, C., Eds.; Caister Academic Press: Wymondham, UK, 2020; pp. 31–58. [Google Scholar]
- Schachter, J. Infection and Disease Epidemiology. In Chlamydia: Intracellular Biology, Pathogenesis, and Immunity; Stephens, R.S., Ed.; ASM Press: Washington, DC, USA, 1999; pp. 139–169. [Google Scholar]
- Jordan, S.; Nelson, D.; Geisler, W.M. Chlamydia trachomatis Infections. In Chlamydia Biology: From Genome to Disease; Tran, M., Hegemann, J.H., Sütterlin, C., Eds.; Caister Academic Press: Wymondham, UK, 2020; pp. 1–30. [Google Scholar]
- Elwell, C.; Mirrashidi, K.; Engel, J. Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 2016, 14, 385–400. [Google Scholar] [CrossRef]
- Hybiske, K.; Stephens, R.S. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. USA 2007, 104, 11430–11435. [Google Scholar] [CrossRef]
- Murray, A.; Ward, M.E. Control mechanisms governing the infectivity of Chlamydia trachomatis for HeLa cells: The role of calmodulin. J. Gen. Microbiol. 1984, 130, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Schramm, N.; Wyrick, P.B. Cytoskeletal requirements in Chlamydia trachomatis infection of host cells. Infect. Immun. 1995, 63, 324–332. [Google Scholar] [CrossRef]
- Majeed, M.; Kihlstrom, E. Mobilization of F-actin and clathrin during redistribution of Chlamydia trachomatis to an intracellular site in eucaryotic cells. Infect. Immun. 1991, 59, 4465–4472. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.E.; Murray, A. Control mechanisms governing the infectivity of Chlamydia trachomatis for HeLa cells: Mechanisms of endocytosis. J. Gen. Microbiol. 1984, 130, 1765–1780. [Google Scholar] [CrossRef]
- Grieshaber, S.S.; Grieshaber, N.A.; Hackstadt, T. Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process. J. Cell Sci. 2003, 116, 3793–3802. [Google Scholar] [CrossRef] [PubMed]
- Clausen, J.D.; Christiansen, G.; Holst, H.U.; Birkelund, S. Chlamydia trachomatis utilizes the host cell microtubule network during early events of infection. Mol. Microbiol. 1997, 25, 441–449. [Google Scholar] [CrossRef]
- Kumar, Y.; Valdivia, R.H. Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds. Cell Host Microbe 2008, 4, 159–169. [Google Scholar] [CrossRef]
- Volceanov, L.; Herbst, K.; Biniossek, M.; Schilling, O.; Haller, D.; Nolke, T.; Subbarayal, P.; Rudel, T.; Zieger, B.; Hacker, G. Septins arrange F-actin-containing fibers on the Chlamydia trachomatis inclusion and are required for normal release of the inclusion by extrusion. mBio 2014, 5, e01802–e01814. [Google Scholar] [CrossRef] [PubMed]
- Al-Zeer, M.A.; Al-Younes, H.M.; Kerr, M.; Abu-Lubad, M.; Gonzalez, E.; Brinkmann, V.; Meyer, T.F. Chlamydia trachomatis remodels stable microtubules to coordinate Golgi stack recruitment to the chlamydial inclusion surface. Mol. Microbiol. 2014, 94, 1285–1297. [Google Scholar] [CrossRef]
- Clifton, D.R.; Fields, K.A.; Grieshaber, S.S.; Dooley, C.A.; Fischer, E.R.; Mead, D.J.; Carabeo, R.A.; Hackstadt, T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. USA 2004, 101, 10166–10171. [Google Scholar] [CrossRef] [PubMed]
- Hower, S.; Wolf, K.; Fields, K.A. Evidence that CT694 is a novel Chlamydia trachomatis T3S substrate capable of functioning during invasion or early cycle development. Mol. Microbiol. 2009, 72, 1423–1437. [Google Scholar] [CrossRef]
- Zrieq, R.; Braun, C.; Hegemann, J.H. The Chlamydia pneumoniae Tarp Ortholog CPn0572 Stabilizes Host F-Actin by Displacement of Cofilin. Front. Cell. Infect. Microbiol. 2017, 7, 511. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.D.; Carabeo, R.A. Distinct roles of the Chlamydia trachomatis effectors TarP and TmeA in the regulation of formin and Arp2/3 during entry. J. Cell Sci. 2022, 135, jcs260185. [Google Scholar] [CrossRef]
- Mital, J.; Lutter, E.I.; Barger, A.C.; Dooley, C.A.; Hackstadt, T. Chlamydia trachomatis inclusion membrane protein CT850 interacts with the dynein light chain DYNLT1 (Tctex1). Biochem. Biophys. Res. Commun. 2015, 462, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Mital, J.; Miller, N.J.; Fischer, E.R.; Hackstadt, T. Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network. Cell. Microbiol. 2010, 12, 1235–1249. [Google Scholar] [CrossRef]
- Böcker, S.; Heurich, A.; Franke, C.; Monajembashi, S.; Sachse, K.; Saluz, H.P.; Hanel, F. Chlamydia psittaci inclusion membrane protein IncB associates with host protein Snapin. Int. J. Med. Microbiol. 2014, 304, 542–553. [Google Scholar] [CrossRef]
- Dumoux, M.; Menny, A.; Delacour, D.; Hayward, R.D. A Chlamydia effector recruits CEP170 to reprogram host microtubule organization. J. Cell Sci. 2015, 128, 3420–3434. [Google Scholar] [CrossRef]
- Wesolowski, J.; Paumet, F. Taking control: Reorganization of the host cytoskeleton by Chlamydia. F1000Res 2017, 6, 2058. [Google Scholar] [CrossRef]
- Haines, A.; Wesolowski, J.; Ryan, N.M.; Monteiro-Bras, T.; Paumet, F. Cross Talk between ARF1 and RhoA Coordinates the Formation of Cytoskeletal Scaffolds during Chlamydia Infection. mBio 2021, 12, e0239721. [Google Scholar] [CrossRef]
- Sherry, J.; Dolat, L.; McMahon, E.; Swaney, D.L.; Bastidas, R.J.; Johnson, J.R.; Valdivia, R.H.; Krogan, N.J.; Elwell, C.A.; Engel, J.N. Chlamydia trachomatis effector Dre1 interacts with dynactin to reposition host organelles during infection. bioRxiv 2022, 2022.2004.2015.488217. [Google Scholar] [CrossRef]
- Sutterlin, C.; Derre, I. Interactions of the Chlamydial Inclusion with the Host Cell. In Chlamydia Biology: From Genome to Disease; Tran, M., Hegemann, J.H., Sütterlin, C., Eds.; Caister Academic Press: Wymondham, UK, 2020; pp. 111–134. [Google Scholar]
- Delevoye, C.; Nilges, M.; Dautry-Varsat, A.; Subtil, A. Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: Oligomerization of IncA mediates interaction between facing membranes. J. Biol. Chem. 2004, 279, 46896–46906. [Google Scholar] [CrossRef]
- Dehoux, P.; Flores, R.; Dauga, C.; Zhong, G.; Subtil, A. Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: The Inc proteins. BMC Genom. 2011, 12, 109. [Google Scholar] [CrossRef] [PubMed]
- Archuleta, T.L.; Du, Y.; English, C.A.; Lory, S.; Lesser, C.; Ohi, M.D.; Ohi, R.; Spiller, B.W. The Chlamydia effector chlamydial outer protein N (CopN) sequesters tubulin and prevents microtubule assembly. J. Biol. Chem. 2011, 286, 33992–33998. [Google Scholar] [CrossRef] [PubMed]
- Nawrotek, A.; Guimaraes, B.G.; Velours, C.; Subtil, A.; Knossow, M.; Gigant, B. Biochemical and Structural Insights into Microtubule Perturbation by CopN from Chlamydia pneumoniae. J. Biol. Chem. 2014, 289, 25199–25210. [Google Scholar] [CrossRef]
- Hoog, J.L.; Schwartz, C.; Noon, A.T.; O’Toole, E.T.; Mastronarde, D.N.; McIntosh, J.R.; Antony, C. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev. Cell 2007, 12, 349–361. [Google Scholar] [CrossRef]
- Loiodice, I.; Janson, M.E.; Tavormina, P.; Schaub, S.; Bhatt, D.; Cochran, R.; Czupryna, J.; Fu, C.; Tran, P.T. Quantifying Tubulin Concentration and Microtubule Number Throughout the Fission Yeast Cell Cycle. Biomolecules 2019, 9, 86. [Google Scholar] [CrossRef]
- Sawin, K.E.; Tran, P.T. Cytoplasmic microtubule organization in fission yeast. Yeast 2006, 23, 1001–1014. [Google Scholar] [CrossRef]
- Lutter, E.I.; Martens, C.; Hackstadt, T. Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comp. Funct. Genom. 2012, 2012, 362104. [Google Scholar] [CrossRef] [PubMed]
- Sisko, J.L.; Spaeth, K.; Kumar, Y.; Valdivia, R.H. Multifunctional analysis of Chlamydia-specific genes in a yeast expression system. Mol. Microbiol. 2006, 60, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Herbst, F. Identifizierung und Charakterisierung Potentieller Neuer Effektorproteine aus Chlamydia pneumoniae; Heinreich-Heine-Universität: Düsseldorf, Germany, 2011. [Google Scholar]
- Maurer, A.P.; Mehlitz, A.; Mollenkopf, H.J.; Meyer, T.F. Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog. 2007, 3, e83. [Google Scholar] [CrossRef]
- Moreno, M.B.; Duran, A.; Ribas, J.C. A family of multifunctional thiamine-repressible expression vectors for fission yeast. Yeast 2000, 16, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Toda, T.; Umesono, K.; Hirata, A.; Yanagida, M. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J. Mol. Biol. 1983, 168, 251–270. [Google Scholar] [CrossRef]
- Garcia, M.A.; Vardy, L.; Koonrugsa, N.; Toda, T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001, 20, 3389–3401. [Google Scholar] [CrossRef]
- Kerres, A.; Jakopec, V.; Fleig, U. The conserved Spc7 protein is required for spindle integrity and links kinetochore complexes in fission yeast. Mol. Biol. Cell 2007, 18, 2441–2454. [Google Scholar] [CrossRef]
- Hagan, I.M.; Hyams, J.S. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 1988, 89 Pt 3, 343–357. [Google Scholar] [CrossRef]
- Hayles, J.; Nurse, P. A journey into space. Nat. Rev. Mol. Cell Biol. 2001, 2, 647–656. [Google Scholar] [CrossRef]
- Beinhauer, J.D.; Hagan, I.M.; Hegemann, J.H.; Fleig, U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J. Cell Biol. 1997, 139, 717–728. [Google Scholar] [CrossRef]
- Nehlig, A.; Molina, A.; Rodrigues-Ferreira, S.; Honore, S.; Nahmias, C. Regulation of end-binding protein EB1 in the control of microtubule dynamics. Cell. Mol. Life Sci. 2017, 74, 2381–2393. [Google Scholar] [CrossRef]
- Erent, M.; Drummond, D.R.; Cross, R.A.S. pombe kinesins-8 promote both nucleation and catastrophe of microtubules. PLoS ONE 2012, 7, e30738. [Google Scholar] [CrossRef] [PubMed]
- Fujita, A.; Vardy, L.; Garcia, M.A.; Toda, T. A fourth component of the fission yeast gamma-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when gamma-tubulin function is compromised. Mol. Biol. Cell 2002, 13, 2360–2373. [Google Scholar] [CrossRef] [PubMed]
- Ende, R.J.; Murray, R.L.; D’Spain, S.K.; Coppens, I.; Derre, I. Phosphoregulation accommodates Type III secretion and assembly of a tether of ER-Chlamydia inclusion membrane contact sites. eLife 2022, 11, e74535. [Google Scholar] [CrossRef] [PubMed]
- Flor-Parra, I.; Sabido-Bozo, S.; Ikeda, A.; Hanaoka, K.; Aguilera-Romero, A.; Funato, K.; Muniz, M.; Lucena, R. The Ceramide Synthase Subunit Lac1 Regulates Cell Growth and Size in Fission Yeast. Int. J. Mol. Sci. 2021, 23, 303. [Google Scholar] [CrossRef]
- Watanabe, K.; Takao, D.; Ito, K.K.; Takahashi, M.; Kitagawa, D. The Cep57-pericentrin module organizes PCM expansion and centriole engagement. Nat. Commun. 2019, 10, 931. [Google Scholar] [CrossRef]
- Mang, J.; Korzeniewski, N.; Dietrich, D.; Sailer, V.; Tolstov, Y.; Searcy, S.; von Hardenberg, J.; Perner, S.; Kristiansen, G.; Marx, A.; et al. Prognostic Significance and Functional Role of CEP57 in Prostate Cancer. Transl. Oncol. 2015, 8, 487–496. [Google Scholar] [CrossRef]
- Zhou, H.; Zheng, T.; Wang, T.; Li, Q.; Wang, F.; Liang, X.; Chen, J.; Teng, J. CCDC74A/B are K-fiber crosslinkers required for chromosomal alignment. BMC Biol. 2019, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Alberico, E.O.; Duan, A.R.; Goodson, H.V. Measuring Tau-microtubule affinity through cosedimentation assays. Methods Cell Biol. 2017, 141, 115–134. [Google Scholar] [CrossRef] [PubMed]
- Mollinari, C.; Kleman, J.P.; Jiang, W.; Schoehn, G.; Hunter, T.; Margolis, R.L. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 2002, 157, 1175–1186. [Google Scholar] [CrossRef]
- Zhao, Y.; Lieberman, H.B. Schizosaccharomyces pombe: A model for molecular studies of eukaryotic genes. DNA Cell Biol. 1995, 14, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P. A Journey in Science: Cell-Cycle Control. Mol. Med. 2017, 22, 112–119. [Google Scholar] [CrossRef]
- Tuite, M.F. Yeast models of neurodegenerative diseases. Prog. Mol. Biol. Transl. Sci. 2019, 168, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Alfa, C.E.; Hyams, J.S. Microtubules in the fission yeast Schizosaccharomyces pombe contain only the tyrosinated form of alpha-tubulin. Cell Motil. Cytoskelet. 1991, 18, 86–93. [Google Scholar] [CrossRef]
- Wesolowski, J.; Weber, M.M.; Nawrotek, A.; Dooley, C.A.; Calderon, M.; St Croix, C.M.; Hackstadt, T.; Cherfils, J.; Paumet, F. Chlamydia Hijacks ARF GTPases To Coordinate Microtubule Posttranslational Modifications and Golgi Complex Positioning. mBio 2017, 8, e02280-16. [Google Scholar] [CrossRef]
- Albrecht, M.; Sharma, C.M.; Dittrich, M.T.; Muller, T.; Reinhardt, R.; Vogel, J.; Rudel, T. The transcriptional landscape of Chlamydia pneumoniae. Genome Biol. 2011, 12, R98. [Google Scholar] [CrossRef]
- Murra, G. Identifizierung und Charakterisierung von Adhäsionsproteinen des Humanpathogenen Erregers Chlamydia pneumoniae; Heinrich-Heine-Universität Düsseldorf: Düsseldorf, Germany, 2009. [Google Scholar]
- Luo, J.; Liu, G.; Zhong, Y.; Jia, T.; Liu, K.; Chen, D.; Zhong, G. Characterization of hypothetical proteins Cpn0146, 0147, 0284 & 0285 that are predicted to be in the Chlamydia pneumoniae inclusion membrane. BMC Microbiol. 2007, 7, 38. [Google Scholar] [CrossRef]
- Flores, R.; Luo, J.; Chen, D.; Sturgeon, G.; Shivshankar, P.; Zhong, Y.; Zhong, G. Characterization of the hypothetical protein Cpn1027, a newly identified inclusion membrane protein unique to Chlamydia pneumoniae. Microbiology 2007, 153, 777–786. [Google Scholar] [CrossRef]
- Meadows, J.C.; Messin, L.J.; Kamnev, A.; Lancaster, T.C.; Balasubramanian, M.K.; Cross, R.A.; Millar, J.B. Opposing kinesin complexes queue at plus tips to ensure microtubule catastrophe at cell ends. EMBO Rep. 2018, 19, e46196. [Google Scholar] [CrossRef]
- Stanhope, R.; Flora, E.; Bayne, C.; Derre, I. IncV, a FFAT motif-containing Chlamydia protein, tethers the endoplasmic reticulum to the pathogen-containing vacuole. Proc. Natl. Acad. Sci. USA 2017, 114, 12039–12044. [Google Scholar] [CrossRef]
- Zheng, P.; Obara, C.J.; Szczesna, E.; Nixon-Abell, J.; Mahalingan, K.K.; Roll-Mecak, A.; Lippincott-Schwartz, J.; Blackstone, C. ER proteins decipher the tubulin code to regulate organelle distribution. Nature 2022, 601, 132–138. [Google Scholar] [CrossRef]
- Jakopec, V.; Walla, E.; Fleig, U. Versatile use of Schizosaccharomyces pombe plasmids in Saccharomyces cerevisiae. FEMS Yeast Res. 2011, 11, 653–655. [Google Scholar] [CrossRef]
- Subtil, A.; Blocker, A.; Dautry-Varsat, A. Type III secretion system in Chlamydia species: Identified members and candidates [letter]. Microbes Infect. 2000, 2, 367–369. [Google Scholar] [CrossRef]
- Jantos, C.A.; Heck, S.; Roggendorf, R.; Sen-Gupta, M.; Hegemann, J.H. Antigenic and molecular analyses of different Chlamydia pneumoniae strains. J. Clin. Microbiol. 1997, 35, 620–623. [Google Scholar] [CrossRef]
- Weinmaier, T.; Hoser, J.; Eck, S.; Kaufhold, I.; Shima, K.; Strom, T.M.; Rattei, T.; Rupp, J. Genomic factors related to tissue tropism in Chlamydia pneumoniae infection. BMC Genom. 2015, 16, 268. [Google Scholar] [CrossRef]
- Masumoto, H.; Matsuyama, S. The combination of NAD+-dependent deacetylase gene deletion and the interruption of gluconeogenesis causes increased glucose metabolism in budding yeast. PLoS ONE 2018, 13, e0194942. [Google Scholar] [CrossRef] [PubMed]
- Molleken, K.; Hegemann, J.H. Acquisition of Rab11 and Rab11-Fip2-A novel strategy for Chlamydia pneumoniae early survival. PLoS Pathog. 2017, 13, e1006556. [Google Scholar] [CrossRef]
- Moreno, S.; Klar, A.; Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991, 194, 795–823. [Google Scholar] [PubMed]
- Tran, P.T.; Paoletti, A.; Chang, F. Imaging green fluorescent protein fusions in living fission yeast cells. Methods 2004, 33, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Subtil, A.; Parsot, C.; Dautry-Varsat, A. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 2001, 39, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Bannantine, J.P.; Griffiths, R.S.; Viratyosin, W.; Brown, W.J.; Rockey, D.D. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell. Microbiol. 2000, 2, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Subtil, A.; Delevoye, C.; Balana, M.E.; Tastevin, L.; Perrinet, S.; Dautry-Varsat, A. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol Microbiol 2005, 56, 1636–1647. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Jia, T.; Flores, R.; Chen, D.; Zhong, G. Hypothetical Protein Cpn0308 Is Localized in the Chlamydia pneumoniae Inclusion Membrane. Infect. Immun. 2007, 75, 497–503. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wevers, C.; Höhler, M.; Alcázar-Román, A.R.; Hegemann, J.H.; Fleig, U. A Functional Yeast-Based Screen Identifies the Host Microtubule Cytoskeleton as a Target of Numerous Chlamydia pneumoniae Proteins. Int. J. Mol. Sci. 2023, 24, 7618. https://doi.org/10.3390/ijms24087618
Wevers C, Höhler M, Alcázar-Román AR, Hegemann JH, Fleig U. A Functional Yeast-Based Screen Identifies the Host Microtubule Cytoskeleton as a Target of Numerous Chlamydia pneumoniae Proteins. International Journal of Molecular Sciences. 2023; 24(8):7618. https://doi.org/10.3390/ijms24087618
Chicago/Turabian StyleWevers, Carolin, Mona Höhler, Abel R. Alcázar-Román, Johannes H. Hegemann, and Ursula Fleig. 2023. "A Functional Yeast-Based Screen Identifies the Host Microtubule Cytoskeleton as a Target of Numerous Chlamydia pneumoniae Proteins" International Journal of Molecular Sciences 24, no. 8: 7618. https://doi.org/10.3390/ijms24087618
APA StyleWevers, C., Höhler, M., Alcázar-Román, A. R., Hegemann, J. H., & Fleig, U. (2023). A Functional Yeast-Based Screen Identifies the Host Microtubule Cytoskeleton as a Target of Numerous Chlamydia pneumoniae Proteins. International Journal of Molecular Sciences, 24(8), 7618. https://doi.org/10.3390/ijms24087618





