Once upon a Testis: The Tale of Cyclic Nucleotide Phosphodiesterase in Testicular Cancers
Abstract
1. Introduction
2. An Overview of Phosphodiesterase Families in Testis
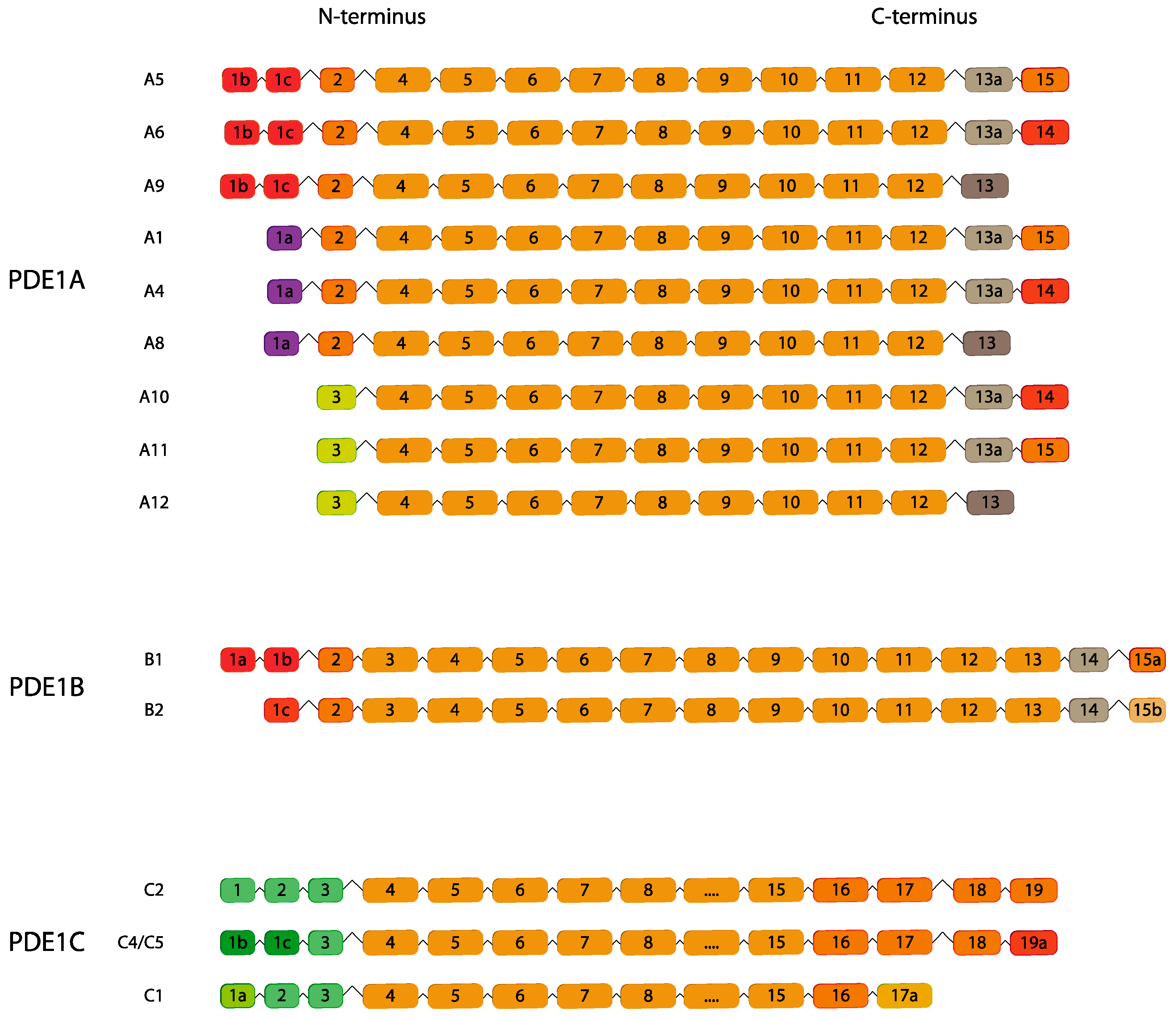
2.1. PDE1
| Gene Family | Gene | Cell Type | References |
|---|---|---|---|
| PDE1 | Un | GCs | [28] |
| PDE1A | rSPT, eSPT, mSPT, SPZ | [22,31,32] | |
| PDE1C | SPC, rSPT, eSPT, mSPT | [32] | |
| PDE2 | PDE2A | SPZ, Un | [31,33] |
| PDE3 | Un | SPZ | [31] |
| PDE3A | SPC, Un | [34] | |
| PDE3B | SPC | [34] | |
| PDE4 | Un | Un, SPZ | [35,36] |
| PDE4A | rSPT, pSPC | [37,38,39,40] | |
| PDE4B | SCs, LCs, SPZ | [31,35,41] | |
| PDE4C | pSPC, SPT, LCs, SPZ | [31,38,39,41] | |
| PDE4D | pSPCs, rSPT, eSPT, mSPT, SPZ, SCs | [31,40,42,43,44,45] | |
| PDE5 | PDE5A | LCs, peritubular cells, Un | [46,47,48,49] |
| PDE6 | PDE6A | SPZ, Un | [31] |
| PDE6C | LCs, SPZ | [31,49] | |
| PDE6D | LCs, SPZ, Un | [31,49] | |
| PDE6G | Un | [31] | |
| PDE6H | Un | [31] | |
| PDE7 | Un | Un | [50] |
| PDE7B | pSPC | [51] | |
| PDE8 | PDE8A | LCs, pSPC, SPZ | [31,41,49,52,53] |
| PDE8B | Un, LCs SPZ | [31,41,49] | |
| PDE9 | PDE9A | Un, LCs | [31,49] |
| PDE10 | PDE10A | SPZ, Un, LCs | [31,49,54,55,56] |
| PDE11 | PDE11A | SPZ | [31,57] |
| Gene Family | Gene | Cell Type | References |
|---|---|---|---|
| PDE1 | PDE1A | SPZ | [36] |
| PDE2 | PDE2A | Un | [64,65] |
| PDE3 | PDE3A | SPZ | [66,67] |
| PDE4 | PDE4A | Un, SPZ | [64,65,66] |
| PDE4B | Un, SPZ | [65,66] | |
| PDE4C | Un, SPZ | [64,66] | |
| PDE4D | Un | [65] | |
| PDE5 | PDE5A | SPZ | [66] |
| PDE6 | PDE6B | Un | [64] |
| PDE7 | PDE7B | Un | [64] |
| PDE8 | PDE8A | Un, LCs | [64,65,68] |
| PDE8B | Un, LCs | [65] | |
| PDE9 | PDE9A | Un | [20,64] |
| PDE10 | PDE10A | Un | [64] |
| PDE11 | PDE11A | SPC, SPT, LCs, Un | [64,65,69] |
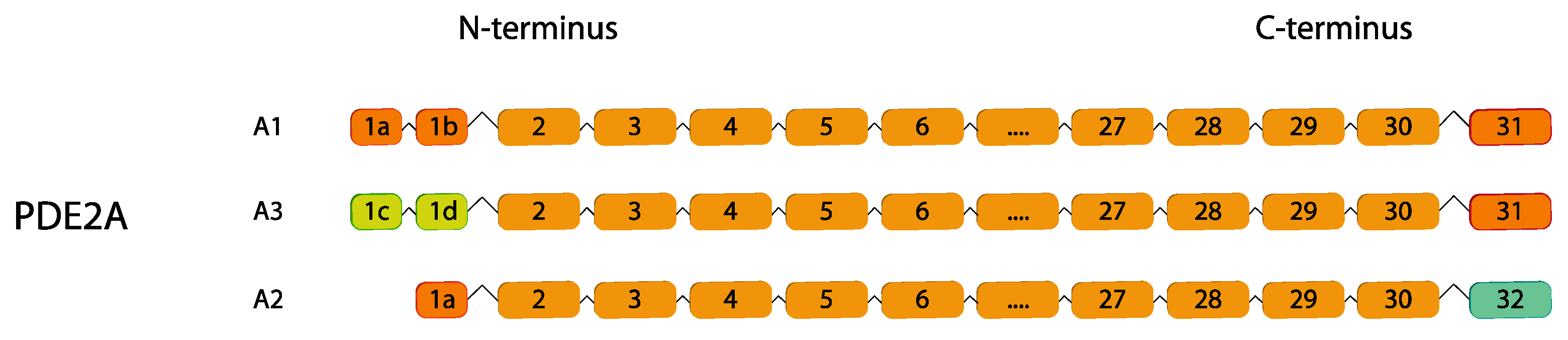
2.2. PDE2
2.3. PDE3
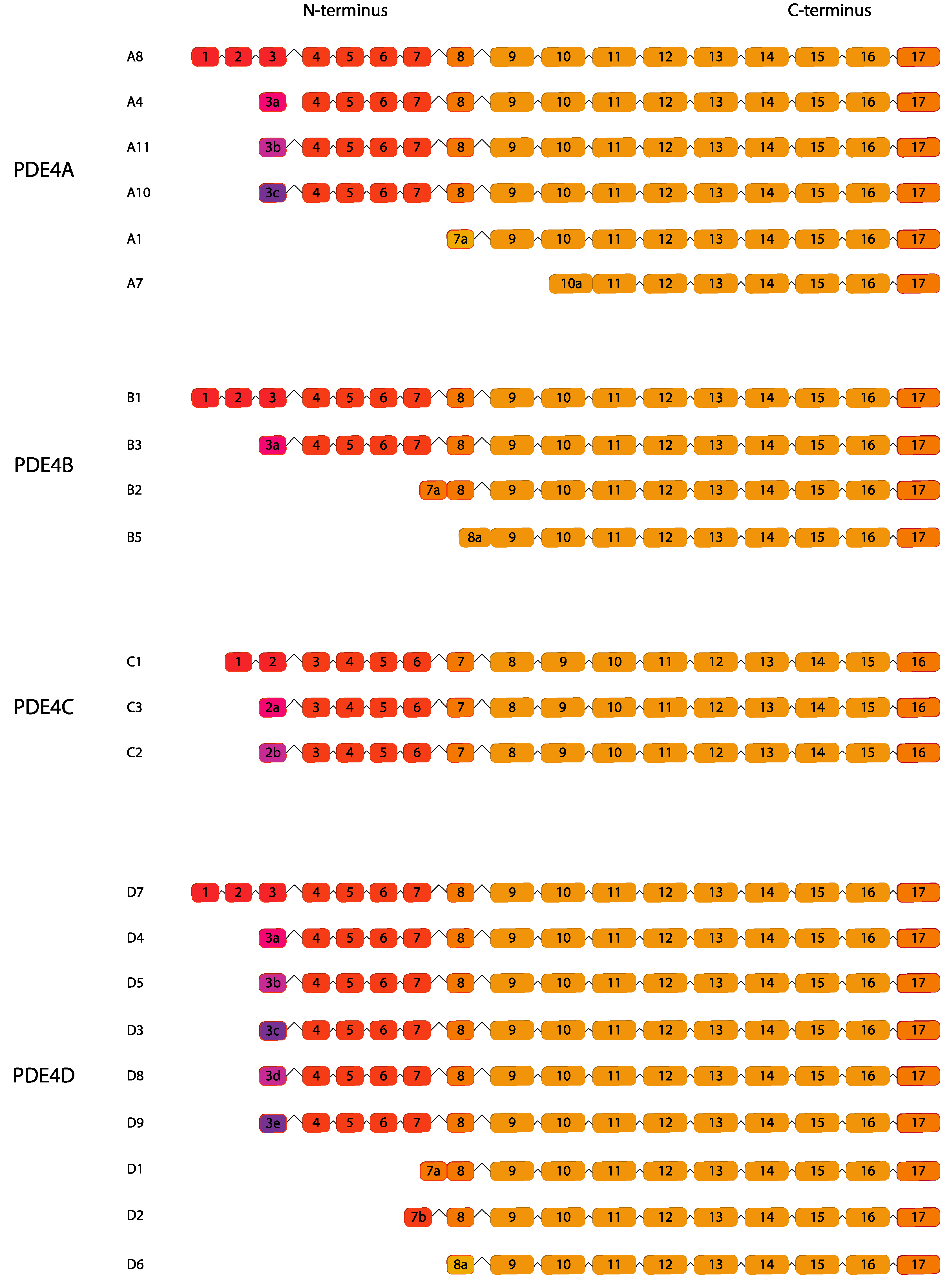
2.4. PDE4
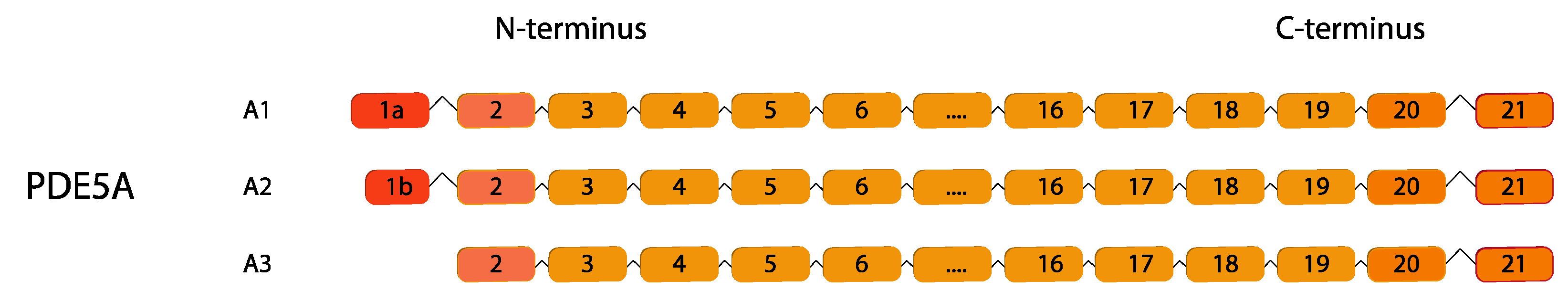
2.5. PDE5
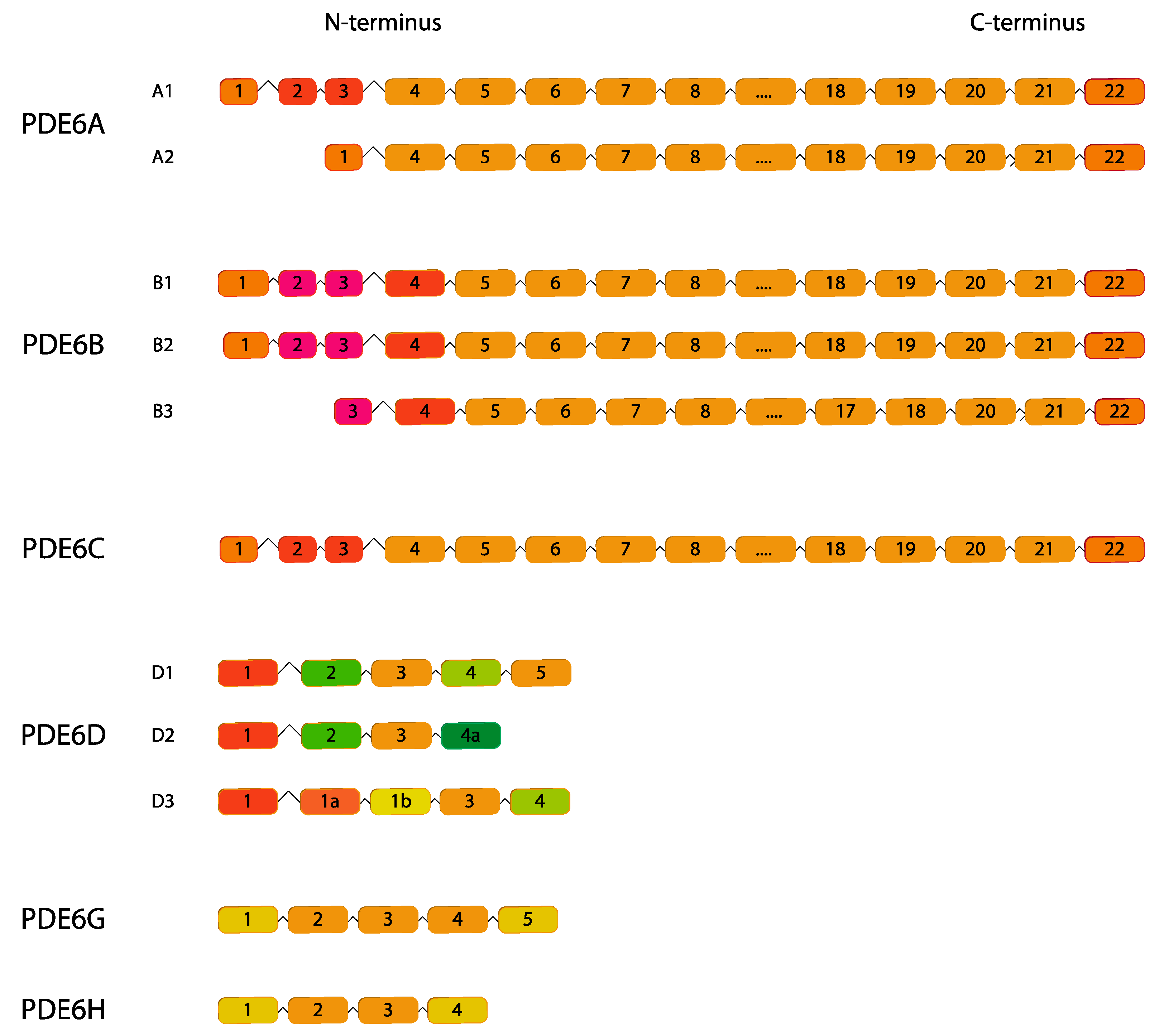
2.6. PDE6
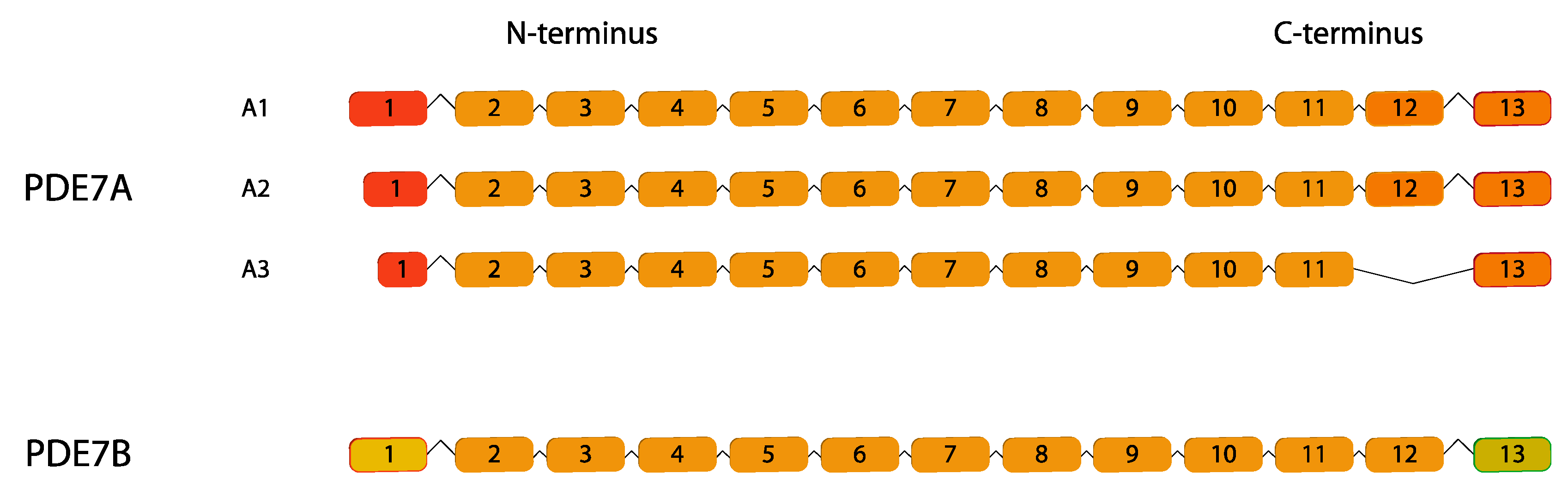
2.7. PDE7
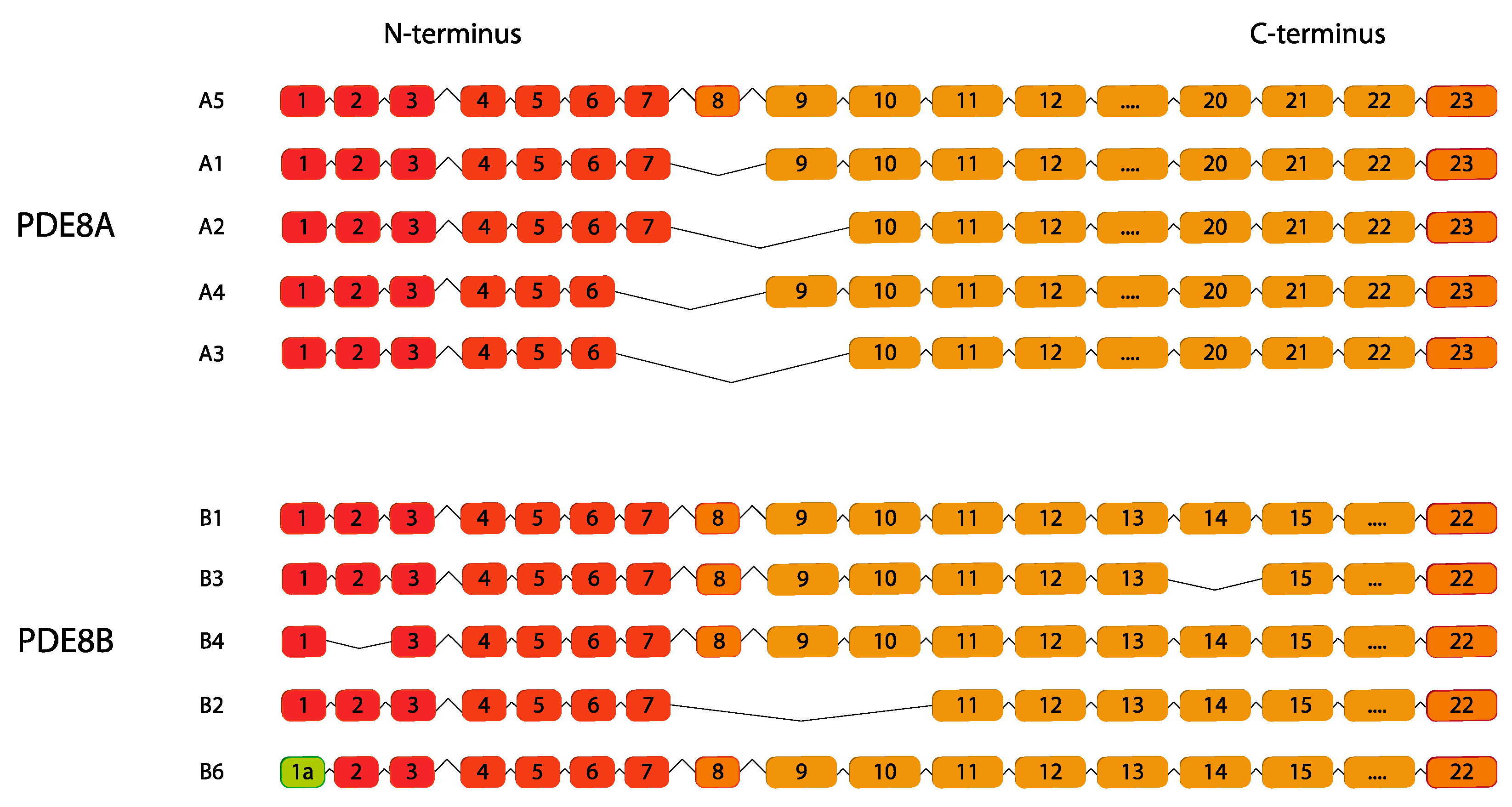
2.8. PDE8
2.9. PDE9
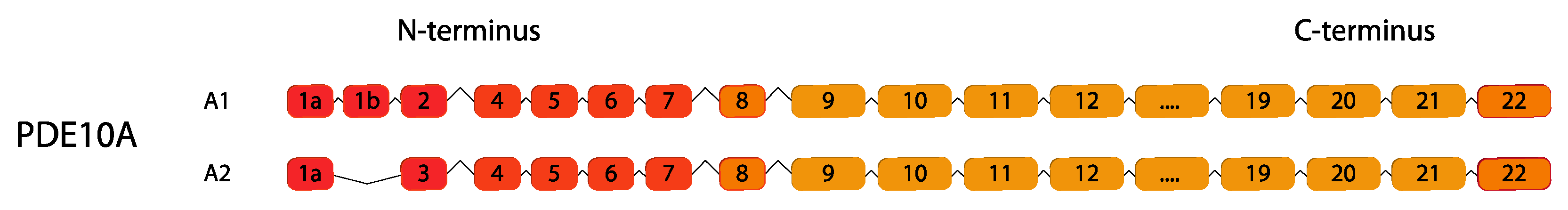
2.10. PDE10
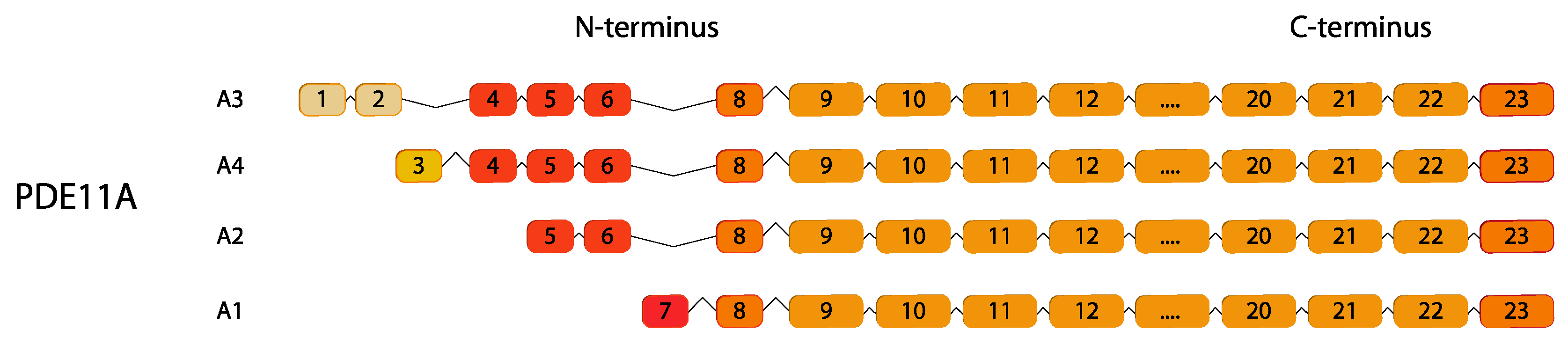
2.11. PDE11
3. An Overview of Phosphodiesterase Families in Testicular Tumors
3.1. Testicular Tumors
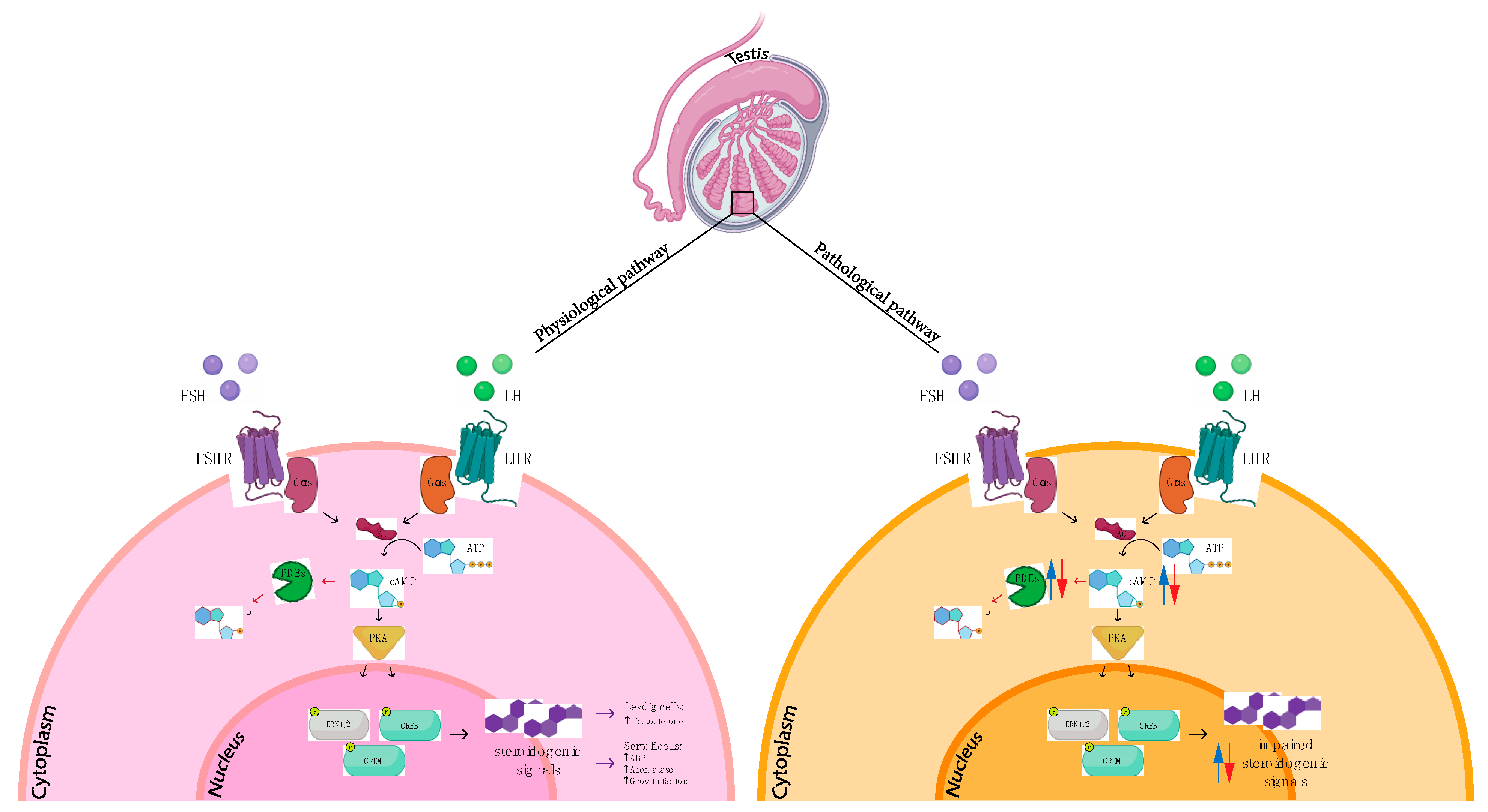
3.2. Phosphodiesterases in Testicular Cancer
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Smolenski, A. Novel roles of cAMP/cGMP-dependent signaling in platelets. J. Thromb. Haemost. 2012, 10, 167–176. [Google Scholar] [CrossRef]
- Gomes, B.; Savignac, M.; Cabral, M.D.; Paulet, P.; Moreau, M.; Leclerc, C.; Feil, R.; Hofmann, F.; Guéry, J.C.; Dietrich, G.; et al. The cGMP/protein kinase G pathway contributes to dihydropyridine-sensitive calcium response and cytokine production in TH2 lymphocytes. J. Biol. Chem. 2006, 281, 12421–12427. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, J.; Ma, P.; Myers, D.E.; Goldberg, I.G.; Sittler, K.J.; Barb, J.J.; Munson, P.J.; del Pilar Cintron, A.; McCoy, J.P.; et al. cGMP-independent nitric oxide signaling and regulation of the cell cycle. BMC Genom. 2005, 6, 151. [Google Scholar] [CrossRef]
- Sassone-Corsi, P. The Cyclic AMP pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011148. [Google Scholar] [CrossRef] [PubMed]
- Francis, S.H.; Busch, J.L.; Corbin, J.D. cGMP-Dependent Protein Kinases and cGMP Phosphodiesterases in Nitric Oxide and cGMP Action. Pharmacol. Rev. 2010, 62, 525–563. [Google Scholar] [CrossRef]
- Garmaroudi, F.S.; Handy, D.E.; Liu, Y.-Y.; Loscalzo, J. Systems Pharmacology and Rational Polypharmacy: Nitric Oxide−Cyclic GMP Signaling Pathway as an Illustrative Example and Derivation of the General Case. PLoS Comput. Biol. 2016, 12, e1004822. [Google Scholar] [CrossRef]
- Conti, M.; Beavo, J. Biochemistry and Physiology of Cyclic Nucleotide Phosphodiesterases: Essential Components in Cyclic Nucleotide Signaling. Annu. Rev. Biochem. 2007, 76, 481–511. [Google Scholar] [CrossRef]
- Torphy, T.J. Phosphodiesterase isozymes molecular targets for novel antiasthma agents. Am. J. Respir. Crit. Care Med. 1998, 157, 351–370. [Google Scholar] [CrossRef]
- Bolger, G.B.; McCahill, A.; Huston, E.; Cheung, Y.F.; McSorley, T.; Baillie, G.S.; Houslay, M.D. The unique amino-terminal region of the PDE4D5 cAMP phosphodiesterase isoform confers preferential interaction with beta-arrestins. J. Biol. Chem. 2003, 278, 49230–49238. [Google Scholar] [CrossRef]
- Kenan, Y.; Murata, T.; Shakur, Y.; Degerman, E.; Manganiello, V.C. Functions of the N-terminal region of cyclic nucleotide phosphodiesterase 3 (PDE 3) isoforms. J. Biol. Chem. 2000, 275, 12331–12338. [Google Scholar] [CrossRef]
- Pandit, J.; Forman, M.D.; Fennell, K.F.; Dillman, K.S.; Menniti, F.S. Mechanism for the allosteric regulation of phosphodiesterase 2A deduced from the X-ray structure of a near full-length construct. Proc. Natl. Acad. Sci. USA 2009, 106, 18225–18230. [Google Scholar] [CrossRef]
- Heikaus, C.C.; Pandit, J.; Klevit, R.E. Cyclic Nucleotide Binding GAF Domains from Phosphodiesterases: Structural and Mechanistic Insights. Structure 2009, 17, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Blackman, B.; Scheitrum, C.; Mika, D.; Blanchard, E.; Lei, T.; Conti, M.; Richter, W. The upstream conserved regions (UCRs) mediate homo- and hetero-oligomerization of type 4 cyclic nucleotide phosphodiesterases (PDE4s). Biochem. J. 2014, 459, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wang, P. Per-Arnt-Sim domain-dependent association of cAMP-phosphodiesterase 8A1 with IκB proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 17634–17639. [Google Scholar] [CrossRef] [PubMed]
- Corbin, J.D.; Turko, I.V.; Beasley, A.; Francis, S.H. Phosphorylation of phosphodiesterase-5 by cyclic nucleotide-dependent protein kinase alters its catalytic and allosteric cGMP-binding activities. Eur. J. Biochem. 2000, 267, 2760–2767. [Google Scholar] [CrossRef]
- Goraya, T.A.; Cooper, D.M.F. Ca2+-calmodulin-dependent phosphodiesterase (PDE1): Current perspectives. Cell. Signal. 2005, 17, 789–797. [Google Scholar] [CrossRef]
- Liu, H.; Maurice, D.H. Phosphorylation-mediated Activation and Translocation of the Cyclic AMP-specific Phosphodiesterase PDE4D3 by Cyclic AMP-dependent Protein Kinase and Mitogen-activated Protein Kinases. J. Biol. Chem. 1999, 274, 10557–10565. [Google Scholar] [CrossRef]
- Peng, T.; Gong, J.; Jin, Y.; Zhou, Y.; Tong, R.; Wei, X.; Bai, L.; Shi, J. Inhibitors of phosphodiesterase as cancer therapeutics. Eur. J. Med. Chem. 2018, 150, 742–756. [Google Scholar] [CrossRef]
- Bender, A.T.; Ostenson, C.L.; Wang, E.H.; Beavo, J.A. Selective up-regulation of PDE1B2 upon monocyte-to-macrophage differentiation. Proc. Natl. Acad. Sci. USA 2005, 102, 497–502. [Google Scholar] [CrossRef]
- Bingham, J.; Sudarsanam, S.; Srinivasan, S. Profiling human phosphodiesterase genes and splice isoforms. Biochem. Biophys. Res. Commun. 2006, 350, 25–32. [Google Scholar] [CrossRef]
- Michibata, H.; Yanaka, N.; Kanoh, Y.; Okumura, K.; Omori, K. Human Ca2+/calmodulin-dependent phosphodiesterase PDE1A: Novel splice variants, their specific expression, genomic organization, and chromosomal localization. Biochim. Biophys. Acta Gene Struct. Expr. 2001, 1517, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Vasta, V.; Sonnenburg, W.K.; Yan, C.; Soderling, S.H.; Shimizu-Albergine, M.; Beavo, J.A. Identification of a New Variant of PDE1A Calmodulin-Stimulated Cyclic Nucleotide Phosphodiesterase Expressed in Mouse Sperm1. Biol. Reprod. 2005, 73, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Pekcec, A.; Schülert, N.; Stierstorfer, B.; Deiana, S.; Dorner-Ciossek, C.; Rosenbrock, H. Targeting the dopamine D 1 receptor or its downstream signalling by inhibiting phosphodiesterase-1 improves cognitive performance. Br. J. Pharmacol. 2018, 175, 3021–3033. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Bentley, J.; Sonnenburg, W.; Beavo, J. Differential expression of the 61 kDa and 63 kDa calmodulin-dependent phosphodiesterases in the mouse brain. J. Neurosci. 1994, 14, 973–984. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Zhao, A.Z.; Bentley, J.K.; Beavo, J.A. The Calmodulin-dependent Phosphodiesterase Gene PDE1C Encodes Several Functionally Different Splice Variants in a Tissue-specific Manner. J. Biol. Chem. 1996, 271, 25699–25706. [Google Scholar] [CrossRef] [PubMed]
- Purvis, K.; Hansson, V.; Olsen, A.; Barry, M. Calmodulin Regulation of Testicular Cyclic Nucleotide Phosphodiesterases. Int. J. Androl. 1980, 3, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Purvis, K.; Olsen, A.; Hansson, V. Calmodulin-dependent cyclic nucleotide phosphodiesterases in the immature rat testis. J. Biol. Chem. 1981, 256, 11434–11441. [Google Scholar] [CrossRef]
- Geremia, R.; Rossi, P.; Pezzotti, R.; Conti, M. Cyclic nucleotide phosphodiesterase in developing rat testis identification of somatic and germ-cell forms. Mol. Cell. Endocrinol. 1982, 28, 37–53. [Google Scholar] [CrossRef]
- Geremia, R.; Rossi, P.; Mocini, D.; Pezzotti, R.; Conti, M. Characterization of a calmodulin-dependent high-affinity cyclic AMP and cyclic GMP phosphodiesterase from male mouse germ cells. Biochem. J. 1984, 217, 693–700. [Google Scholar] [CrossRef]
- Rossi, P.; Pezzotti, R.; Conti, M.; Geremia, R. Cyclic nucleotide phosphodiesterases in somatic and germ cells of mouse seminiferous tubules. Reproduction 1985, 74, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Baxendale, R.W.; Fraser, L.R. Mammalian sperm phosphodiesterases and their involvement in receptor-mediated cell signaling important for capacitation. Mol. Reprod. Dev. 2005, 71, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Zhao, A.Z.; Sonnenburg, W.K.; Beavo, J.A. Stage and Cell-Specific Expression of Calmodulin-Dependent Phosphodiesterases in Mouse Testis1. Biol. Reprod. 2001, 64, 1746–1754. [Google Scholar] [CrossRef]
- Stephenson, D.T.; Coskran, T.M.; Wilhelms, M.B.; Adamowicz, W.O.; O’donnell, M.M.; Muravnick, K.B.; Menniti, F.S.; Kleiman, R.J.; Morton, D. Immunohistochemical Localization of Phosphodiesterase 2A in Multiple Mammalian Species. J. Histochem. Cytochem. 2009, 57, 933–949. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, R.R.; Chin, E.; Zhou, J.; Taira, M.; Murata, T.; Manganiello, V.C.; Bondy, C.A. Distinctive anatomical patterns of gene expression for cGMP-inhibited cyclic nucleotide phosphodiesterases. J. Clin. Investig. 1995, 95, 1528–1538. [Google Scholar] [CrossRef] [PubMed]
- Farooqui, S.M.; Al-Bagdadi, F.; Houslay, M.D.; Bolger, G.B.; Stout, R.; Specian, R.D.; Cherry, J.A.; Conti, M.; O’Donnell, J.M. Surgically Induced Cryptorchidism-Related Degenerative Changes in Spermatogonia Are Associated with Loss of Cyclic Adenosine Monophosphate-Dependent Phosphodiesterases Type 4 in Abdominal Testes of Rats. Biol. Reprod. 2001, 64, 1583–1589. [Google Scholar] [CrossRef]
- Fisch, J.D.; Behr, B.; Conti, M. Enhancement of motility and acrosome reaction in human spermatozoa: Differential activation by type-specific phosphodiesterase inhibitors. Hum. Reprod. 1998, 13, 1248–1254. [Google Scholar] [CrossRef]
- Welch, J.E.; Swinnen, J.V.; O’Brien, D.A.; Eddy, E.M.; Conti, M. Unique Adenosine 3′,5′ Cyclic Monophosphate Phosphodiesterase Messenger Ribonucleic Acids in Rat Spermatogenic Cells: Evidence for Differential Gene Expression during Spermatogenesis1. Biol. Reprod. 1992, 46, 1027–1033. [Google Scholar] [CrossRef] [PubMed]
- Naro, F.; Zhang, R.; Conti, M. Developmental regulation of unique adenosine 3′,5′-monophosphate-specific phosphodiesterase variants during rat spermatogenesis. Endocrinology 1996, 137, 2464–2472. [Google Scholar] [CrossRef]
- Morena, A.R.; Boitani, C.; De Grossi, S.; Stefanini, M.; Conti, M. Erratum: Stage and cell-specific expression of the adenosine 3′,5′-monophosphate-phosphodiesterase genes in the rat seminiferous epithelium (Endocrinology (1995) 136 (687-695)). Endocrinology 1995, 136, 1558. [Google Scholar] [CrossRef]
- Salanova, M.; Chun, S.-Y.; Iona, S.; Puri, C.; Stefanini, M.; Conti, M. Type 4 Cyclic Adenosine Monophosphate-Specific Phosphodiesterases Are Expressed in Discrete Subcellular Compartments during Rat Spermiogenesis*. Endocrinology 1999, 140, 2297–2306. [Google Scholar] [CrossRef]
- Shimizu-Albergine, M.; Tsai, L.-C.L.; Patrucco, E.; Beavo, J.A. cAMP-Specific Phosphodiesterases 8A and 8B, Essential Regulators of Leydig Cell Steroidogenesis. Mol. Pharmacol. 2012, 81, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Conti, M.; Iona, S.; Cuomo, M.; Swinnen, J.V.; Odeh, J.; Svoboda, M.E. Characterization of a hormone-inducible, high affinity adenosine 3′,5′-cyclic monophosphate phosphodiesterase from the rat Sertoli cell. Biochemistry 1995, 34, 7979–7987. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, J.V.; Tsikalas, K.E.; Conti, M. Properties and hormonal regulation of two structurally related cAMP phosphodiesterases from the rat Sertoli cell. J. Biol. Chem. 1991, 266, 18370–18377. [Google Scholar] [CrossRef] [PubMed]
- Vicini, E. Characterization of an Intronic Promoter of a Cyclic Adenosine 3′,5′-Monophosphate (cAMP)-Specific Phosphodiesterase Gene that Confers Hormone and cAMP Inducibility. Mol. Endocrinol. 1997, 11, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Levallet, G.; Levallet, J.; Bouraïma-Lelong, H.; Bonnamy, P.-J. Expression of the cAMP-Phosphodiesterase PDE4D Isoforms and Age-Related Changes in Follicle-Stimulating Hormone-Stimulated PDE4 Activities in Immature Rat Sertoli Cells1. Biol. Reprod. 2007, 76, 794–803. [Google Scholar] [CrossRef]
- Scipioni, A.; Stefanini, S.; Santone, R.; Giorgi, M. Immunohistochemical localisation of PDE5 in Leydig and myoid cells of prepuberal and adult rat testis. Histochem. Cell Biol. 2005, 124, 401–407. [Google Scholar] [CrossRef]
- Campolo, F.; Zevini, A.; Cardarelli, S.; Monaco, L.; Barbagallo, F.; Pellegrini, M.; Cornacchione, M.; Di Grazia, A.; De Arcangelis, V.; Gianfrilli, D.; et al. Identification of murine phosphodiesterase 5A isoforms and their functional characterization in HL-1 cardiac cell line. J. Cell. Physiol. 2018, 233, 325–337. [Google Scholar] [CrossRef]
- Saraiva, K.L.A.; Silva, A.K.S.E.; Wanderley, M.I.; De Araújo, A.A.; De Souza, J.R.B.; Peixoto, C.A. Chronic treatment with sildenafil stimulates Leydig cell and testosterone secretion. Int. J. Exp. Pathol. 2009, 90, 454–462. [Google Scholar] [CrossRef]
- Andric, S.A.; Janjic, M.M.; Stojkov, N.J.; Kostic, T.S. Sildenafil treatment in vivo stimulates Leydig cell steroidogenesis via the cAMP/cGMP signaling pathway. Am. J. Physiol. Metab. 2010, 299, E544–E550. [Google Scholar] [CrossRef]
- Bloom, T.J.; Beavo, J.A. Identification and tissue-specific expression of PDE7 phosphodiesterase splice variants. Proc. Natl. Acad. Sci. USA 1996, 93, 14188–14192. [Google Scholar] [CrossRef]
- Sasaki, T.; Kotera, J.; Omori, K. Novel alternative splice variants of rat phosphodiesterase 7B showing unique tissue-specific expression and phosphorylation. Biochem. J. 2002, 361, 211. [Google Scholar] [CrossRef] [PubMed]
- Vasta, V.; Shimizu-Albergine, M.; Beavo, J.A. Modulation of Leydig cell function by cyclic nucleotide phosphodiesterase 8A. Proc. Natl. Acad. Sci. USA 2006, 103, 19925–19930. [Google Scholar] [CrossRef]
- Soderling, S.H.; Bayuga, S.J.; Beavo, J.A. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc. Natl. Acad. Sci. USA 1998, 95, 8991–8996. [Google Scholar] [CrossRef]
- Soderling, S.H.; Bayuga, S.J.; Beavo, J.A. Isolation and characterization of a dual-substrate phosphodiesterase gene family: PDE10A. Proc. Natl. Acad. Sci. USA 1999, 96, 7071–7076. [Google Scholar] [CrossRef] [PubMed]
- Fujishige, K.; Kotera, J.; Michibata, H.; Yuasa, K.; Takebayashi, S.; Okumura, K.; Omori, K. Cloning and Characterization of a Novel Human Phosphodiesterase That Hydrolyzes Both cAMP and cGMP (PDE10A). J. Biol. Chem. 1999, 274, 18438–18445. [Google Scholar] [CrossRef]
- Maréchal, L.; Guillemette, C.; Goupil, S.; Blondin, P.; Leclerc, P.; Richard, F.J. Cyclic nucleotide phosphodiesterases in human spermatozoa and seminal fluid: Presence of an active PDE10A in human spermatozoa. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 147–156. [Google Scholar] [CrossRef]
- Wayman, C.; Phillips, S.; Lunny, C.; Webb, T.; Fawcett, L.; Baxendale, R.; Burgess, G. Phosphodiesterase 11 (PDE11) regulation of spermatozoa physiology. Int. J. Impot. Res. 2005, 17, 216–223. [Google Scholar] [CrossRef]
- Bentley, J.K.; Kadlecek, A.; Sherbert, C.H.; Seger, D.; Sonnenburg, W.K.; Charbonneau, H.; Novack, J.P.; Beavo, J.A. Molecular cloning of cDNA encoding a “63”-kDa calmodulin-stimulated phosphodiesterase from bovine brain. J. Biol. Chem. 1992, 267, 18676–18682. [Google Scholar] [CrossRef] [PubMed]
- Kincaid, R.L.; Balaban, C.D.; Billingsley, M.L. Regional and developmental expression of calmodulin-dependent cyclic nucleotide phosphodiesterase in rat brain. Adv. Second. Messenger Phosphoprot. Res. 1992, 25, 111–122. [Google Scholar]
- Repaske, D.R.; Swinnen, J.V.; Jin, S.L.; Van Wyk, J.J.; Conti, M. A polymerase chain reaction strategy to identify and clone cyclic nucleotide phosphodiesterase cDNAs. Molecular cloning of the cDNA encoding the 63-kDa calmodulin-dependent phosphodiesterase. J. Biol. Chem. 1992, 267, 18683–18688. [Google Scholar] [CrossRef]
- Sonnenburg, W.K.; Seger, D.; Beavo, J.A. Molecular cloning of a cDNA encoding the “61-kDa” calmodulin-stimulated cyclic nucleotide phosphodiesterase. Tissue-specific expression of structurally related isoforms. J. Biol. Chem. 1993, 268, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Loughney, K.; Martins, T.J.; Harris, E.A.S.; Sadhu, K.; Hicks, J.B.; Sonnenburg, W.K.; Beavo, J.A.; Ferguson, K. Isolation and Characterization of cDNAs Corresponding to Two Human Calcium, Calmodulin-regulated, 3′,5′-Cyclic Nucleotide Phosphodiesterases. J. Biol. Chem. 1996, 271, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Zhao, A.Z.; Bentley, J.K.; Loughney, K.; Ferguson, K.; Beavo, J.A. Molecular cloning and characterization of a calmodulin-dependent phosphodiesterase enriched in olfactory sensory neurons. Proc. Natl. Acad. Sci. USA 1995, 92, 9677–9681. [Google Scholar] [CrossRef]
- Azevedo, M.F.; Faucz, F.R.; Bimpaki, E.; Horvath, A.; Levy, I.; de Alexandre, R.B.; Ahmad, F.; Manganiello, V.; Stratakis, C.A. Clinical and Molecular Genetics of the Phosphodiesterases (PDEs). Endocr. Rev. 2014, 35, 195–233. [Google Scholar] [CrossRef]
- Campolo, F.; Capponi, C.; Tarsitano, M.G.; Tenuta, M.; Pozza, C.; Gianfrilli, D.; Magliocca, F.; Venneri, M.A.; Vicini, E.; Lenzi, A.; et al. cAMP-specific phosphodiesterase 8A and 8B isoforms are differentially expressed in human testis and Leydig cell tumor. Front. Endocrinol. 2022, 13, 1010924. [Google Scholar] [CrossRef]
- Richter, W. Detection of mRNA transcripts of cyclic nucleotide phosphodiesterase subtypes in ejaculated human spermatozoa. Mol. Hum. Reprod. 1999, 5, 732–736. [Google Scholar] [CrossRef]
- Lefiévre, L.; de Lamirande, E.; Gagnon, C. Presence of Cyclic Nucleotide Phosphodiesterases PDE1A, Existing as a Stable Complex with Calmodulin, and PDE3A in Human Spermatozoa1. Biol. Reprod. 2002, 67, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wu, P.; Egan, R.W.; Billah, M.M. Human phosphodiesterase 8A splice variants: Cloning, gene organization, and tissue distribution. Gene 2001, 280, 183–194. [Google Scholar] [CrossRef]
- D’Andrea, M.R.; Qiu, Y.; Haynes-Johnson, D.; Bhattacharjee, S.; Kraft, P.; Lundeen, S. Expression of PDE11A in Normal and Malignant Human Tissues. J. Histochem. Cytochem. 2005, 53, 895–903. [Google Scholar] [CrossRef]
- Isidori, A.M.; Cornacchione, M.; Barbagallo, F.; Di Grazia, A.; Barrios, F.; Fassina, L.; Monaco, L.; Giannetta, E.; Gianfrilli, D.; Garofalo, S.; et al. Inhibition of type 5 phosphodiesterase counteracts β2-adrenergic signalling in beating cardiomyocytes. Cardiovasc. Res. 2015, 106, 408–420. [Google Scholar] [CrossRef]
- Acin-Perez, R.; Russwurm, M.; Günnewig, K.; Gertz, M.; Zoidl, G.; Ramos, L.; Buck, J.; Levin, L.R.; Rassow, J.; Manfredi, G.; et al. A Phosphodiesterase 2A Isoform Localized to Mitochondria Regulates Respiration. J. Biol. Chem. 2011, 286, 30423–30432. [Google Scholar] [CrossRef]
- Ding, B.; Abe, J.; Wei, H.; Huang, Q.; Walsh, R.A.; Molina, C.A.; Zhao, A.; Sadoshima, J.; Blaxall, B.C.; Berk, B.C.; et al. Functional Role of Phosphodiesterase 3 in Cardiomyocyte Apoptosis. Circulation 2005, 111, 2469–2476. [Google Scholar] [CrossRef] [PubMed]
- Bender, A. Differentiation of human monocytes in vitro with granulocyte–macrophage colony-stimulating factor and macrophage colony-stimulating factor produces distinct changes in cGMP phosphodiesterase expression. Cell. Signal. 2004, 16, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, F.; Rotilio, V.; Assenza, M.R.; Aguanno, S.; Orsini, T.; Putti, S.; Isidori, A.M.; Lenzi, A.; Naro, F.; De Angelis, L.; et al. PDE2A Is Indispensable for Mouse Liver Development and Hematopoiesis. Int. J. Mol. Sci. 2020, 21, 2902. [Google Scholar] [CrossRef]
- Farmer, R.; Burbano, S.D.; Patel, N.S.; Sarmiento, A.; Smith, A.J.; Kelly, M.P. Phosphodiesterases PDE2A and PDE10A both change mRNA expression in the human brain with age, but only PDE2A changes in a region-specific manner with psychiatric disease. Cell. Signal. 2020, 70, 109592. [Google Scholar] [CrossRef]
- Assenza, M.R.; Barbagallo, F.; Barrios, F.; Cornacchione, M.; Campolo, F.; Vivarelli, E.; Gianfrilli, D.; Auletta, L.; Soricelli, A.; Isidori, A.M.; et al. Critical role of phosphodiesterase 2A in mouse congenital heart defects. Cardiovasc. Res. 2018, 114, 830–845. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Taira, M.; Hockman, S.; Shimada, F.; Lieman, J.; Napolitano, M.; Ward, D.; Taira, M.; Makino, H.; Manganiello, V.C. Characterization of the cDNA and Gene Encoding Human PDE3B, the cGIP1 Isoform of the Human Cyclic GMP-Inhibited Cyclic Nucleotide Phosphodiesterase Family. Genomics 1996, 36, 476–485. [Google Scholar] [CrossRef]
- Meacci, E.; Taira, M.; Moos, M.; Smith, C.J.; Movsesian, M.A.; Degerman, E.; Belfrage, P.; Manganiello, V. Molecular cloning and expression of human myocardial cGMP-inhibited cAMP phosphodiesterase. Proc. Natl. Acad. Sci. USA 1992, 89, 3721–3725. [Google Scholar] [CrossRef]
- Shakur, Y.; Takeda, K.; Kenan, Y.; Yu, Z.-X.; Rena, G.; Brandt, D.; Houslay, M.D.; Degerman, E.; Ferrans, V.J.; Manganiello, V.C. Membrane Localization of Cyclic Nucleotide Phosphodiesterase 3 (PDE3). J. Biol. Chem. 2000, 275, 38749–38761. [Google Scholar] [CrossRef]
- Hambleton, R.; Krall, J.; Tikishvili, E.; Honeggar, M.; Ahmad, F.; Manganiello, V.C.; Movsesian, M.A. Isoforms of Cyclic Nucleotide Phosphodiesterase PDE3 and Their Contribution to cAMP Hydrolytic Activity in Subcellular Fractions of Human Myocardium. J. Biol. Chem. 2005, 280, 39168–39174. [Google Scholar] [CrossRef]
- Movsesian, M.; Ahmad, F.; Hirsch, E. Functions of PDE3 Isoforms in Cardiac Muscle. J. Cardiovasc. Dev. Dis. 2018, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Omori, K.; Kotera, J. Overview of PDEs and Their Regulation. Circ. Res. 2007, 100, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.W.; Ahmad, F.; Tang, Y.; Hockman, S.C.; Kee, H.J.; Berger, K.; Guirguis, E.; Choi, Y.H.; Schimel, D.M.; Aponte, A.M.; et al. White to beige conversion in PDE3B KO adipose tissue through activation of AMPK signaling and mitochondrial function. Sci. Rep. 2017, 7, 40445. [Google Scholar] [CrossRef] [PubMed]
- Paes, D.; Schepers, M.; Rombaut, B.; van den Hove, D.; Vanmierlo, T.; Prickaerts, J. The Molecular Biology of Phosphodiesterase 4 Enzymes as Pharmacological Targets: An Interplay of Isoforms, Conformational States, and Inhibitors. Pharmacol. Rev. 2021, 73, 1016–1049. [Google Scholar] [CrossRef] [PubMed]
- Huacuja, L.; Delgado, N.M.; Merchant, H.; Pancardo, R.M.; Rosado, A. Cyclic AMP Induced Incorporation of 33Pi into Human Spermatozoa Membrane Components. Biol. Reprod. 1977, 17, 89–96. [Google Scholar] [CrossRef]
- Wang, G.; Guo, Y.; Zhou, T.; Shi, X.; Yu, J.; Yang, Y.; Wu, Y.; Wang, J.; Liu, M.; Chen, X.; et al. In-depth proteomic analysis of the human sperm reveals complex protein compositions. J. Proteom. 2013, 79, 114–122. [Google Scholar] [CrossRef]
- Golkowski, M.; Shimizu-Albergine, M.; Suh, H.W.; Beavo, J.A.; Ong, S.-E. Studying mechanisms of cAMP and cyclic nucleotide phosphodiesterase signaling in Leydig cell function with phosphoproteomics. Cell. Signal. 2016, 28, 764–778. [Google Scholar] [CrossRef]
- Francis, S.H.; Lincoln, T.M.; Corbin, J.D. Characterization of a novel cGMP binding protein from rat lung. J. Biol. Chem. 1980, 255, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Hamet, P.; Coquil, J.F. Cyclic GMP binding and cyclic GMP phosphodiesterase in rat platelets. J. Cyclic Nucleotide Res. 1978, 4, 281–290. [Google Scholar]
- Lin, C.-S.; Chow, S.; Lau, A.; Tu, R.; Lue, T.F. Human PDE5A gene encodes three PDE5 isoforms from two alternate promoters. Int. J. Impot. Res. 2002, 14, 15–24. [Google Scholar] [CrossRef]
- Lin, C.-S.; Lau, A.; Tu, R.; Lue, T.F. Expression of Three Isoforms of cGMP-Binding cGMP-Specific Phosphodiesterase (PDE5) in Human Penile Cavernosum. Biochem. Biophys. Res. Commun. 2000, 268, 628–635. [Google Scholar] [CrossRef]
- West, T.M.; Wang, Q.; Deng, B.; Zhang, Y.; Barbagallo, F.; Reddy, G.R.; Chen, D.; Phan, K.S.; Xu, B.; Isidori, A.; et al. Phosphodiesterase 5 Associates with β2 Adrenergic Receptor to Modulate Cardiac Function in Type 2 Diabetic Hearts. J. Am. Heart Assoc. 2019, 8, e012273. [Google Scholar] [CrossRef]
- De Arcangelis, V.; De Angelis, L.; Barbagallo, F.; Campolo, F.; de Oliveira do Rego, A.G.; Pellegrini, M.; Naro, F.; Giorgi, M.; Monaco, L. Phosphodiesterase 5a Signalling in Skeletal Muscle Pathophysiology. Int. J. Mol. Sci. 2022, 24, 703. [Google Scholar] [CrossRef]
- Kotera, J.; Yanaka, N.; Fujishige, K.; Imai, Y.; Akatsuka, H.; Ishizuka, T.; Kawashima, K.; Omori, K. Expression of Rat cGMP-Binding cGMP-Specific Phosphodiesterase mRNA in Purkinje Cell Layers during Postnatal Neuronal Development. Eur. J. Biochem. 1997, 249, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Baillie, G.S.; Tejeda, G.S.; Kelly, M.P. Therapeutic targeting of 3′,5′-cyclic nucleotide phosphodiesterases: Inhibition and beyond. Nat. Rev. Drug Discov. 2019, 18, 770–796. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.S.; Geethakumari, A.M.; Biswas, K.H. Phosphodiesterase 5 (PDE5): Structure-function regulation and therapeutic applications of inhibitors. Biomed. Pharmacother. 2021, 134, 111128. [Google Scholar] [CrossRef]
- Pofi, R.; Fiore, D.; De Gaetano, R.; Panio, G.; Gianfrilli, D.; Pozza, C.; Barbagallo, F.; Xiang, Y.K.; Giannakakis, K.; Morano, S.; et al. Phosphodiesterase-5 inhibition preserves renal hemodynamics and function in mice with diabetic kidney disease by modulating miR-22 and BMP7. Sci. Rep. 2017, 7, 44584. [Google Scholar] [CrossRef]
- Cesarini, V.; Guida, E.; Campolo, F.; Crescioli, C.; Di Baldassarre, A.; Pisano, C.; Balistreri, C.R.; Ruvolo, G.; Jannini, E.A.; Dolci, S. Type 5 phosphodiesterase (PDE5) and the vascular tree: From embryogenesis to aging and disease. Mech. Ageing Dev. 2020, 190, 111311. [Google Scholar] [CrossRef]
- Yanaka, N.; Kotera, J.; Ohtsuka, A.; Akatsuka, H.; Imai, Y.; Michibata, H.; Fujishige, K.; Kawai, E.; Takebayashi, S.-I.; Okumura, K.; et al. Expression, structure and chromosomal localization of the human cGMP-binding cGMP-specific phosphodiesterase PDE5A gene. Eur. J. Biochem. 1998, 255, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Andric, S.A.; Janjic, M.M.; Stojkov, N.J.; Kostic, T.S. Protein kinase G-mediated stimulation of basal Leydig cell steroidogenesis. Am. J. Physiol. Metab. 2007, 293, E1399–E1408. [Google Scholar] [CrossRef]
- Casarini, L.; Riccetti, L.; Limoncella, S.; Lazzaretti, C.; Barbagallo, F.; Pacifico, S.; Guerrini, R.; Tagliavini, S.; Trenti, T.; Simoni, M.; et al. Probing the Effect of Sildenafil on Progesterone and Testosterone Production by an Intracellular FRET/BRET Combined Approach. Biochemistry 2019, 58, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Limoncella, S.; Lazzaretti, C.; Paradiso, E.; D’Alessandro, S.; Barbagallo, F.; Pacifico, S.; Guerrini, R.; Tagliavini, S.; Trenti, T.; Santi, D.; et al. Phosphodiesterase (PDE) 5 inhibitors sildenafil, tadalafil and vardenafil impact cAMP-specific PDE8 isoforms-linked second messengers and steroid production in a mouse Leydig tumor cell line. Mol. Cell. Endocrinol. 2022, 542, 111527. [Google Scholar] [CrossRef] [PubMed]
- Sokanovic, S.J.; Capo, I.; Medar, M.M.; Andric, S.A.; Kostic, T.S. Long-term inhibition of PDE5 ameliorates aging-induced changes in rat testis. Exp. Gerontol. 2018, 108, 139–148. [Google Scholar] [CrossRef]
- Dimitriadis, F.; Tsampalas, S.; Tsounapi, P.; Giannakis, D.; Chaliasos, N.; Baltogiannis, D.; Miyagawa, I.; Saito, M.; Takenaka, A.; Sofikitis, N. Effects of phosphodiesterase-5 inhibitor vardenafil on testicular androgen-binding protein secretion, the maintenance of foci of advanced spermatogenesis and the sperm fertilising capacity in azoospermic men. Andrologia 2012, 44, 144–153. [Google Scholar] [CrossRef]
- Aversa, A.; Mazzilli, F.; Rossi, T.; Delfino, M.; Isidori, A.M.; Fabbri, A. Effects of sildenafil (ViagraTM) administration on seminal parameters and post-ejaculatory refractory time in normal males*. Hum. Reprod. 2000, 15, 131–134. [Google Scholar] [CrossRef]
- du Plessis, S.S.; de Jongh, P.S.; Franken, D.R. Effect of acute in vivo sildenafil citrate and in vitro 8-bromo-cGMP treatments on semen parameters and sperm function. Fertil. Steril. 2004, 81, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Jannini, E.A.; Lombardo, F.; Salacone, P.; Gandini, L.; Lenzi, A. Treatment of sexual dysfunctions secondary to male infertility with sildenafil citrate. Fertil. Steril. 2004, 81, 705–707. [Google Scholar] [CrossRef]
- Burger, M.; Sikka, S.; Bivalacqua, T.; Lamb, D.; Hellstrom, W. The effect of sildenafil on human sperm motion and function from normal and infertile men. Int. J. Impot. Res. 2000, 12, 229–234. [Google Scholar] [CrossRef]
- Andrade, J.R.; Traboulsi, A.; Hussain, A.; Dubin, N.H. In vitro effects of sildenafil and phentolamine, drugs used for erectile dysfunction, on human sperm motility. Am. J. Obstet. Gynecol. 2000, 182, 1093–1095. [Google Scholar] [CrossRef]
- Mostafa, T. In vitro sildenafil citrate use as a sperm motility stimulant. Fertil. Steril. 2007, 88, 994–996. [Google Scholar] [CrossRef]
- Lefièvre, L.; De Lamirande, E.; Gagnon, C. The cyclic GMP-specific phosphodiesterase inhibitor, sildenafil, stimulates human sperm motility and capacitation but not acrosome reaction. J. Androl. 2000, 21, 929–937. [Google Scholar]
- Cuadra, D.L.; Chan, P.J.; Patton, W.C.; Stewart, S.C.; King, A. Type 5 phosphodiesterase regulation of human sperm motility. Am. J. Obstet. Gynecol. 2000, 182, 1013–1015. [Google Scholar] [CrossRef]
- Cote, R.H. Characteristics of Photoreceptor PDE (PDE6): Similarities and differences to PDE5. Int. J. Impot. Res. 2004, 16, S28–S33. [Google Scholar] [CrossRef] [PubMed]
- Tcheudji, J.F.K.; Lebeau, L.; Virmaux, N.; Maftei, C.G.; Cote, R.H.; Lugnier, C.; Schultz, P. Molecular organization of bovine rod cGMP-phosphodiesterase 6. J. Mol. Biol. 2001, 310, 781–791. [Google Scholar] [CrossRef]
- Wang, P.; Wu, P.; Egan, R.W.; Billah, M.M. Cloning, Characterization, and Tissue Distribution of Mouse Phosphodiesterase 7A1. Biochem. Biophys. Res. Commun. 2000, 276, 1271–1277. [Google Scholar] [CrossRef]
- Giembycz, M.A.; Corrigan, C.J.; Seybold, J.; Newton, R.; Barnes, P.J. Identification of cyclic AMP phosphodiesterases 3, 4 and 7 in human CD4+ and CD8+ T-lymphocytes: Role in regulating proliferation and the biosynthesis of interleukin-2. Br. J. Pharmacol. 1996, 118, 1945–1958. [Google Scholar] [CrossRef]
- Fisher, D.A.; Smith, J.F.; Pillar, J.S.; St. Denis, S.H.; Cheng, J.B. Isolation and Characterization of PDE8A, a Novel Human cAMP-Specific Phosphodiesterase. Biochem. Biophys. Res. Commun. 1998, 246, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Matsushima, K.; Ohashi, H.; Tsunoda, H.; Murase, S.; Kawarada, Y.; Tanaka, T. Molecular Cloning and Characterization of Human PDE8B, a Novel Thyroid-Specific Isozyme of 3′,5′-Cyclic Nucleotide Phosphodiesterase. Biochem. Biophys. Res. Commun. 1998, 250, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Jorde, R.; Schirmer, H.; Wilsgaard, T.; Joakimsen, R.M.; Mathiesen, E.B.; Njølstad, I.; Løchen, M.-L.; Figenschau, Y.; Svartberg, J.; Hutchinson, M.S.; et al. The Phosphodiesterase 8B Gene rs4704397 is Associated with Thyroid Function, Risk of Myocardial Infarction, and Body Height: The Tromsø Study. Thyroid 2014, 24, 215–222. [Google Scholar] [CrossRef]
- Patrucco, E.; Albergine, M.S.; Santana, L.F.; Beavo, J.A. Phosphodiesterase 8A (PDE8A) regulates excitation–contraction coupling in ventricular myocytes. J. Mol. Cell. Cardiol. 2010, 49, 330–333. [Google Scholar] [CrossRef]
- Tsai, L.-C.; Beavo, J. Regulation of Adrenal Steroidogenesis by the High-affinity Phosphodiesterase 8 Family. Horm. Metab. Res. 2012, 44, 790–794. [Google Scholar] [CrossRef]
- Tsai, L.-C.L.; Shimizu-Albergine, M.; Beavo, J.A. The High-Affinity cAMP-Specific Phosphodiesterase 8B Controls Steroidogenesis in the Mouse Adrenal Gland. Mol. Pharmacol. 2011, 79, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Rentero, C.; Monfort, A.; Puigdomènech, P. Identification and distribution of different mRNA variants produced by differential splicing in the human phosphodiesterase 9A gene. Biochem. Biophys. Res. Commun. 2003, 301, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Shao, Y.; Hou, J.; Cui, W.; Liang, B.; Huang, Y.; Li, Z.; Wu, Y.; Zhu, X.; Liu, P.; et al. Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor. Mol. Pharmacol. 2015, 88, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wu, P.; Egan, R.W.; Billah, M.M. Identification and characterization of a new human type 9 cGMP-specific phosphodiesterase splice variant (PDE9A5). Gene 2003, 314, 15–27. [Google Scholar] [CrossRef]
- Andreeva, S.G.; Dikkes, P.; Epstein, P.M.; Rosenberg, P.A. Expression of cGMP-Specific Phosphodiesterase 9A mRNA in the Rat Brain. J. Neurosci. 2001, 21, 9068–9076. [Google Scholar] [CrossRef]
- Van Staveren, W.C.G.; Glick, J.; Markerink-Van Ittersum, M.; Shimizu, M.; Beavo, J.A.; Steinbusch, H.W.M.; De Vente, J. Cloning and localization of the cGMP-specific phosphodiesterase type 9 in the rat brain. J. Neurocytol. 2002, 31, 729–741. [Google Scholar] [CrossRef]
- Schmidt, C. Phosphodiesterase Inhibitors as Potential Cognition Enhancing Agents. Curr. Top. Med. Chem. 2010, 10, 222–230. [Google Scholar] [CrossRef]
- Bender, A.T.; Beavo, J.A. Cyclic Nucleotide Phosphodiesterases: Molecular Regulation to Clinical Use. Pharmacol. Rev. 2006, 58, 488–520. [Google Scholar] [CrossRef]
- MacMullen, C.M.; Vick, K.; Pacifico, R.; Fallahi-Sichani, M.; Davis, R.L. Novel, primate-specific PDE10A isoform highlights gene expression complexity in human striatum with implications on the molecular pathology of bipolar disorder. Transl. Psychiatry 2016, 6, e742. [Google Scholar] [CrossRef]
- Reneerkens, O.A.H.; Rutten, K.; Bollen, E.; Hage, T.; Blokland, A.; Steinbusch, H.W.M.; Prickaerts, J. Inhibition of phoshodiesterase type 2 or type 10 reverses object memory deficits induced by scopolamine or MK-801. Behav. Brain Res. 2013, 236, 16–22. [Google Scholar] [CrossRef]
- Fujishige, K.; Kotera, J.; Omori, K. Striatum- and testis-specific phosphodiesterase PDE10A. Eur. J. Biochem. 1999, 266, 1118–1127. [Google Scholar] [CrossRef]
- Siuciak, J.A.; McCarthy, S.A.; Chapin, D.S.; Fujiwara, R.A.; James, L.C.; Williams, R.D.; Stock, J.L.; McNeish, J.D.; Strick, C.A.; Menniti, F.S.; et al. Genetic deletion of the striatum-enriched phosphodiesterase PDE10A: Evidence for altered striatal function. Neuropharmacology 2006, 51, 374–385. [Google Scholar] [CrossRef]
- Fawcett, L.; Baxendale, R.; Stacey, P.; McGrouther, C.; Harrow, I.; Soderling, S.; Hetman, J.; Beavo, J.A.; Phillips, S.C. Molecular cloning and characterization of a distinct human phosphodiesterase gene family: PDE11A. Proc. Natl. Acad. Sci. USA 2000, 97, 3702–3707. [Google Scholar] [CrossRef]
- Makhlouf, A.; Kshirsagar, A.; Niederberger, C. Phosphodiesterase 11: A brief review of structure, expression and function. Int. J. Impot. Res. 2006, 18, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Kelly, M. Does Phosphodiesterase 11A (PDE11A) Hold Promise as a Future Therapeutic Target? Curr. Pharm. Des. 2014, 21, 389–416. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, K.; Kotera, J.; Fujishige, K.; Michibata, H.; Sasaki, T.; Omori, K. Isolation and Characterization of Two Novel Phosphodiesterase PDE11A Variants Showing Unique Structure and Tissue-specific Expression. J. Biol. Chem. 2000, 275, 31469–31479. [Google Scholar] [CrossRef]
- De Felici, M.; Klinger, F.; Campolo, F.; Balistreri, C.; Barchi, M.; Dolci, S. To Be or Not to Be a Germ Cell: The Extragonadal Germ Cell Tumor Paradigm. Int. J. Mol. Sci. 2021, 22, 5982. [Google Scholar] [CrossRef]
- Dolci, S.; Campolo, F.; De Felici, M. Gonadal development and germ cell tumors in mouse and humans. Semin. Cell Dev. Biol. 2015, 45, 114–123. [Google Scholar] [CrossRef]
- Litchfield, K.; Levy, M.; Huddart, R.A.; Shipley, J.; Turnbull, C. The genomic landscape of testicular germ cell tumours: From susceptibility to treatment. Nat. Rev. Urol. 2016, 13, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Isidori, A.M.; Pozza, C.; Gianfrilli, D.; Giannetta, E.; Lemma, A.; Pofi, R.; Barbagallo, F.; Manganaro, L.; Martino, G.; Lombardo, F.; et al. Differential Diagnosis of Nonpalpable Testicular Lesions: Qualitative and Quantitative Contrast-enhanced US of Benign and Malignant Testicular Tumors. Radiology 2014, 273, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Manganaro, L.; Vinci, V.; Pozza, C.; Saldari, M.; Gianfrilli, D.; Pofi, R.; Bernardo, S.; Cantisani, V.; Lenzi, A.; Scialpi, M.; et al. A prospective study on contrast-enhanced magnetic resonance imaging of testicular lesions: Distinctive features of Leydig cell tumours. Eur. Radiol. 2015, 25, 3586–3595. [Google Scholar] [CrossRef] [PubMed]
- Paffenholz, P.; Held, L.; Loosen, S.H.; Pfister, D.; Heidenreich, A. Testis Sparing Surgery for Benign Testicular Masses: Diagnostics and Therapeutic Approaches. J. Urol. 2018, 200, 353–360. [Google Scholar] [CrossRef]
- Tarsitano, M.G.; Bandak, M.; Jørgensen, N.; Skakkebaek, N.E.; Juul, A.; Lenzi, A.; Daugaard, G.; Rajpert-De Meyts, E. Quantification of the Leydig cell compartment in testicular biopsies and association with biochemical Leydig cell dysfunction in testicular cancer survivors. Andrology 2018, 6, 748–755. [Google Scholar] [CrossRef] [PubMed]
- Pozza, C.; Pofi, R.; Tenuta, M.; Tarsitano, M.G.; Sbardella, E.; Fattorini, G.; Cantisani, V.; Lenzi, A.; Isidori, A.M.; Gianfrilli, D. Clinical presentation, management and follow-up of 83 patients with Leydig cell tumors of the testis: A prospective case-cohort study. Hum. Reprod. 2019, 34, 1389–1403. [Google Scholar] [CrossRef]
- Pozza, C.; Kanakis, G.; Carlomagno, F.; Lemma, A.; Pofi, R.; Tenuta, M.; Minnetti, M.; Tarsitano, M.G.; Sesti, F.; Paoli, D.; et al. Testicular ultrasound score: A new proposal for a scoring system to predict testicular function. Andrology 2020, 8, 1051–1063. [Google Scholar] [CrossRef]
- Cardarelli, S.; Miele, A.E.; Campolo, F.; Massimi, M.; Mancini, P.; Biagioni, S.; Naro, F.; Giorgi, M.; Saliola, M. Cellular Redox Metabolism Is Modulated by the Distinct Localization of Cyclic Nucleotide Phosphodiesterase 5A Isoforms. Int. J. Mol. Sci. 2022, 23, 8587. [Google Scholar] [CrossRef]
- Campolo, F.; Pofi, R.; Venneri, M.A.; Isidori, A.M. Priming metabolism with the type 5 phosphodiesterase: The role of cGMP-hydrolyzing enzymes. Curr. Opin. Pharmacol. 2021, 60, 298–305. [Google Scholar] [CrossRef]
- Ravnskjaer, K.; Madiraju, A.; Montminy, M. Role of the cAMP Pathway in Glucose and Lipid Metabolism. Handb. Exp. Pharmacol. 2015, 233, 29–49. [Google Scholar]
- Smith, S.A.; Newby, A.C.; Bond, M. Ending Restenosis: Inhibition of Vascular Smooth Muscle Cell Proliferation by cAMP. Cells 2019, 8, 1447. [Google Scholar] [CrossRef]
- Pizzoni, A.; Zhang, X.; Naim, N.; Altschuler, D.L. Soluble cyclase-mediated nuclear cAMP synthesis is sufficient for cell proliferation. Proc. Natl. Acad. Sci. USA 2023, 120, e2208749120. [Google Scholar] [CrossRef]
- Lerner, A.; Epstein, P.M. Cyclic nucleotide phosphodiesterases as targets for treatment of haematological malignancies. Biochem. J. 2006, 393, 21–41. [Google Scholar] [CrossRef]
- Barbagallo, F.; Xu, B.; Reddy, G.R.; West, T.; Wang, Q.; Fu, Q.; Li, M.; Shi, Q.; Ginsburg, K.S.; Ferrier, W.; et al. Genetically Encoded Biosensors Reveal PKA Hyperphosphorylation on the Myofilaments in Rabbit Heart Failure. Circ. Res. 2016, 119, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Pinto, E.M.; Faucz, F.R.; Paza, L.Z.; Wu, G.; Fernandes, E.S.; Bertherat, J.; Stratakis, C.A.; Lalli, E.; Ribeiro, R.C.; Rodriguez-Galindo, C.; et al. Germline Variants in Phosphodiesterase Genes and Genetic Predisposition to Pediatric Adrenocortical Tumors. Cancers 2020, 12, 506. [Google Scholar] [CrossRef] [PubMed]
- Bolger, G.B. The cAMP-signaling cancers: Clinically-divergent disorders with a common central pathway. Front. Endocrinol. 2022, 13, 1024423. [Google Scholar] [CrossRef]
- Zhang, L.; Murray, F.; Zahno, A.; Kanter, J.R.; Chou, D.; Suda, R.; Fenlon, M.; Rassenti, L.; Cottam, H.; Kipps, T.J.; et al. Cyclic nucleotide phosphodiesterase profiling reveals increased expression of phosphodiesterase 7B in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2008, 105, 19532–19537. [Google Scholar] [CrossRef]
- Cesarini, V.; Martini, M.; Vitiani, L.R.; Gravina, G.L.; Di Agostino, S.; Graziani, G.; D’Alessandris, Q.G.; Pallini, R.; Larocca, L.M.; Rossi, P.; et al. Type 5 phosphodiesterase regulates glioblastoma multiforme aggressiveness and clinical outcome. Oncotarget 2017, 8, 13223–13239. [Google Scholar] [CrossRef]
- Goldhoff, P.; Warrington, N.M.; Limbrick, D.D.; Hope, A.; Woerner, B.M.; Jackson, E.; Perry, A.; Piwnica-Worms, D.; Rubin, J.B. Targeted Inhibition of Cyclic AMP Phosphodiesterase-4 Promotes Brain Tumor Regression. Clin. Cancer Res. 2008, 14, 7717–7725. [Google Scholar] [CrossRef]
- Chen, Y.; Qi, C.; Xia, L.; Li, G. Identification of novel genetic etiology and key molecular pathways for seminoma via network-based studies. Int. J. Oncol. 2017, 51, 1280–1290. [Google Scholar] [CrossRef]
- Rouillard, A.D.; Gundersen, G.W.; Fernandez, N.F.; Wang, Z.; Monteiro, C.D.; McDermott, M.G.; Ma’ayan, A. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database 2016, 2016, baw100. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, S.; Kitapci, T.H.; Blumenfeld, H.; Besaratinia, A. Secondhand smoke affects reproductive functions by altering the mouse testis transcriptome, and leads to select intron retention in Pde1a. Environ. Int. 2022, 161, 107086. [Google Scholar] [CrossRef] [PubMed]
- Horvath, A.; Korde, L.; Greene, M.H.; Libe, R.; Osorio, P.; Faucz, F.R.; Raffin-Sanson, M.L.; Tsang, K.M.; Drori-Herishanu, L.; Patronas, Y.; et al. Functional Phosphodiesterase 11A Mutations May Modify the Risk of Familial and Bilateral Testicular Germ Cell Tumors. Cancer Res. 2009, 69, 5301–5306. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, M.F.; Horvath, A.; Bornstein, E.R.; Almeida, M.Q.; Xekouki, P.; Faucz, F.R.; Gourgari, E.; Nadella, K.; Remmers, E.F.; Quezado, M.; et al. Cyclic AMP and c-KIT Signaling in Familial Testicular Germ Cell Tumor Predisposition. J. Clin. Endocrinol. Metab. 2013, 98, E1393–E1400. [Google Scholar] [CrossRef] [PubMed]
- Pathak, A.; Stewart, D.R.; Faucz, F.R.; Xekouki, P.; Bass, S.; Vogt, A.; Zhang, X.; Boland, J.; Yeager, M.; Loud, J.T.; et al. Rare inactivating PDE11A variants associated with testicular germ cell tumors. Endocr. Relat. Cancer 2015, 22, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Faja, F.; Finocchi, F.; Carlini, T.; Rizzo, F.; Pallotti, F.; Spaziani, M.; Balercia, G.; Lenzi, A.; Paoli, D.; Lombardo, F. PDE11A gene polymorphism in testicular cancer: Sperm parameters and hormonal profile. J. Endocrinol. Investig. 2021, 44, 2273–2284. [Google Scholar] [CrossRef]
- Libé, R.; Horvath, A.; Vezzosi, D.; Fratticci, A.; Coste, J.; Perlemoine, K.; Ragazzon, B.; Guillaud-Bataille, M.; Groussin, L.; Clauser, E.; et al. Frequent Phosphodiesterase 11A Gene (PDE11A) Defects in Patients with Carney Complex (CNC) Caused by PRKAR1A Mutations: PDE11A May Contribute to Adrenal and Testicular Tumors in CNC as a Modifier of the Phenotype. J. Clin. Endocrinol. Metab. 2011, 96, E208–E214. [Google Scholar] [CrossRef] [PubMed]
- Baillie, G.S.; Scott, J.D.; Houslay, M.D. Compartmentalisation of phosphodiesterases and protein kinase A: Opposites attract. FEBS Lett. 2005, 579, 3264–3270. [Google Scholar] [CrossRef]
- Grimaldi, P.; Capolunghi, F.; Geremia, R.; Rossi, P. Cyclic Adenosine Monophosphate (cAMP) Stimulation of the Kit Ligand Promoter in Sertoli Cells Requires an Sp1-Binding Region, a Canonical TATA Box, and a cAMP-Induced Factor Binding to an Immediately Downstream GC-Rich Element1. Biol. Reprod. 2003, 69, 1979–1988. [Google Scholar] [CrossRef]
- Landmark, B.F.; Oyen, O.; Skalhegg, B.S.; Fauske, B.; Jahnsen, T.; Hansson, V. Cellular location and age-dependent changes of the regulatory subunits of cAMP-dependent protein kinase in rat testis. Reproduction 1993, 99, 323–334. [Google Scholar] [CrossRef]
- Davidoff, M.S.; Middendorff, R.; Mayer, B.; DeVente, J.; Koesling, D.; Holstein, A.F. Nitric oxide/cGMP pathway components in the Leydig cells of the human testis. Cell Tissue Res. 1996, 287, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Middendorff, R.; Davidoff, M.S.; Behrends, S.; Mewe, M.; Miethens, A.; Müller, D. Multiple roles of the messenger molecule cGMP in testicular function. Andrologia 2000, 32, 55–59. [Google Scholar] [PubMed]
- Walker, W.H. Testosterone signaling and the regulation of spermatogenesis. Spermatogenesis 2011, 1, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Barbagallo, F.; Condorelli, R.A.; Mongioì, L.M.; Cannarella, R.; Aversa, A.; Calogero, A.E.; La Vignera, S. Effects of Bisphenols on Testicular Steroidogenesis. Front. Endocrinol. 2020, 11, 373. [Google Scholar] [CrossRef]
- Don, J.; Stelzer, G. The expanding family of CREB/CREM transcription factors that are involved with spermatogenesis. Mol. Cell. Endocrinol. 2002, 187, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Dufau, M.L.; Mendelson, C.R.; Catt, K.J. A highly sensitive in vitro bioassay for luteinizing hormone and chorionic gonadotropin: Testosterone production by dispersed leydig cells. J. Clin. Endocrinol. Metab. 1974, 39, 610–613. [Google Scholar] [CrossRef]
- Andric, S.A.; Janjic, M.M.; Stojkov, N.J.; Kostic, T.S. Testosterone-Induced Modulation of Nitric Oxide-cGMP Signaling Pathway and Androgenesis in the Rat Leydig Cells1. Biol. Reprod. 2010, 83, 434–442. [Google Scholar] [CrossRef]
- Walker, W.H.; Habener, J.F. Role of transcription factors CREB and CREM in cAMP-regulated transcription during spermatogenesis. Trends Endocrinol. Metab. 1996, 7, 133–138. [Google Scholar] [CrossRef]
- Behr, R.; Weinbauer, G.F. cAMP response element modulator (CREM): An essential factor for spermatogenesis in primates? Int. J. Androl. 2001, 24, 126–135. [Google Scholar] [CrossRef]
- Blendy, J.A.; Kaestner, K.H.; Weinbauer, G.F.; Nieschlag, E.; Schütz, G. Severe impairment of permatogenesis in mice lacking the CREM gene. Nature 1996, 380, 162–165. [Google Scholar] [CrossRef]
- Scobey, M.J.; Bertera, S.; Somers, J.P.; Watkins, S.C.; Zeleznik, A.J.; Walker, W.H. Delivery of a Cyclic Adenosine 3′,5′-Monophosphate Response Element-Binding Protein (CREB) Mutant to Seminiferous Tubules Results in Impaired Spermatogenesis 1. Endocrinology 2001, 142, 948–954. [Google Scholar] [CrossRef]
- Buffone, M.G.; Wertheimer, E.V.; Visconti, P.E.; Krapf, D. Central role of soluble adenylyl cyclase and cAMP in sperm physiology. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 2610–2620. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Grow, E.J.; Yi, C.; Mlcochova, H.; Maher, G.J.; Lindskog, C.; Murphy, P.J.; Wike, C.L.; Carrell, D.T.; Goriely, A.; et al. Chromatin and Single-Cell RNA-Seq Profiling Reveal Dynamic Signaling and Metabolic Transitions during Human Spermatogonial Stem Cell Development. Cell Stem Cell 2017, 21, 533–546.e6. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Nie, X.; Giebler, M.; Mlcochova, H.; Wang, Y.; Grow, E.J.; Kim, R.; Tharmalingam, M.; Matilionyte, G.; Lindskog, C.; et al. The Dynamic Transcriptional Cell Atlas of Testis Development during Human Puberty. Cell Stem Cell 2020, 26, 262–276.e4. [Google Scholar] [CrossRef] [PubMed]
- Nestler, T.; Dalvi, P.; Haidl, F.; Wittersheim, M.; von Brandenstein, M.; Paffenholz, P.; Wagener-Ryczek, S.; Pfister, D.; Koitzsch, U.; Hellmich, M.; et al. Transcriptome analysis reveals upregulation of immune response pathways at the invasive tumour front of metastatic seminoma germ cell tumours. Br. J. Cancer 2022, 126, 937–947. [Google Scholar] [CrossRef]
- Mo, L.; Yu, Z.; Lv, Y.; Cheng, J.; Yan, H.; Lu, W.; Su, C.; Ling, Q.; Mo, Z. Single-Cell RNA Sequencing of Metastatic Testicular Seminoma Reveals the Cellular and Molecular Characteristics of Metastatic Cell Lineage. Front. Oncol. 2022, 12, 871489. [Google Scholar] [CrossRef]
- Kotula-Balak, M.; Duliban, M.; Gurgul, A.; Krakowska, I.; Grzmil, P.; Bilinska, B.; Wolski, J.K. Transcriptome analysis of human Leydig cell tumours reveals potential mechanisms underlying its development. Andrologia 2021, 53, e14222. [Google Scholar] [CrossRef]
- Li, L.; Dong, J.; Yan, L.; Yong, J.; Liu, X.; Hu, Y.; Fan, X.; Wu, X.; Guo, H.; Wang, X.; et al. Single-Cell RNA-Seq Analysis Maps Development of Human Germline Cells and Gonadal Niche Interactions. Cell Stem Cell 2017, 20, 858–873.e4. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Y.; Gao, Y.; Lin, Z.; Yang, S.; Wang, T.; Wang, Q.; Xie, N.; Hua, R.; Liu, M.; et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018, 28, 879–896. [Google Scholar] [CrossRef]
- Green, C.D.; Ma, Q.; Manske, G.L.; Shami, A.N.; Zheng, X.; Marini, S.; Moritz, L.; Sultan, C.; Gurczynski, S.J.; Moore, B.B.; et al. A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev. Cell 2018, 46, 651–667.e10. [Google Scholar] [CrossRef]
- Hermann, B.P.; Cheng, K.; Singh, A.; Roa-De La Cruz, L.; Mutoji, K.N.; Chen, I.-C.; Gildersleeve, H.; Lehle, J.D.; Mayo, M.; Westernströer, B.; et al. The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep. 2018, 25, 1650–1667.e8. [Google Scholar] [CrossRef]
- Lukassen, S.; Bosch, E.; Ekici, A.B.; Winterpacht, A. Single-cell RNA sequencing of adult mouse testes. Sci. Data 2018, 5, 180192. [Google Scholar] [CrossRef]
- Wang, M.; Liu, X.; Chang, G.; Chen, Y.; An, G.; Yan, L.; Gao, S.; Xu, Y.; Cui, Y.; Dong, J.; et al. Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell Stem Cell 2018, 23, 599–614.e4. [Google Scholar] [CrossRef]
- Grive, K.J.; Hu, Y.; Shu, E.; Grimson, A.; Elemento, O.; Grenier, J.K.; Cohen, P.E. Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing. PLOS Genet. 2019, 15, e1007810. [Google Scholar] [CrossRef]
- Shami, A.N.; Zheng, X.; Munyoki, S.K.; Ma, Q.; Manske, G.L.; Green, C.D.; Sukhwani, M.; Orwig, K.E.; Li, J.Z.; Hammoud, S.S. Single-Cell RNA Sequencing of Human, Macaque, and Mouse Testes Uncovers Conserved and Divergent Features of Mammalian Spermatogenesis. Dev. Cell 2020, 54, 529–547.e12. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ning, B.; Shi, T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front. Genet. 2019, 10, 317. [Google Scholar] [CrossRef]
- Naro, C.; Cesari, E.; Sette, C. Splicing regulation in brain and testis: Common themes for highly specialized organs. Cell Cycle 2021, 20, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kong, Q.; Wang, J.; Jiang, Y.; Hua, H. Complex roles of cAMP–PKA–CREB signaling in cancer. Exp. Hematol. Oncol. 2020, 9, 32. [Google Scholar] [CrossRef] [PubMed]
- Rothenbuhler, A.; Horvath, A.; Libé, R.; Faucz, F.R.; Fratticci, A.; Raffin Sanson, M.L.; Vezzosi, D.; Azevedo, M.; Levy, I.; Almeida, M.Q.; et al. Identification of novel genetic variants in phosphodiesterase 8B (PDE8B), a cAMP-specific phosphodiesterase highly expressed in the adrenal cortex, in a cohort of patients with adrenal tumours. Clin. Endocrinol. 2012, 77, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Mei, L.; Fan, X.; Tang, C.; Ji, X.; Hu, X.; Shi, W.; Qian, Y.; Hussain, M.; Wu, J.; et al. Phosphodiesterase 5/protein kinase G signal governs stemness of prostate cancer stem cells through Hippo pathway. Cancer Lett. 2016, 378, 38–50. [Google Scholar] [CrossRef]
- Susmi, T.F.; Rahman, A.; Khan, M.M.R.; Yasmin, F.; Islam, M.S.; Nasif, O.; Alharbi, S.A.; Batiha, G.E.-S.; Hossain, M.U. Prognostic and clinicopathological insights of phosphodiesterase 9A gene as novel biomarker in human colorectal cancer. BMC Cancer 2021, 21, 577. [Google Scholar] [CrossRef]
- Nazir, M.; Senkowski, W.; Nyberg, F.; Blom, K.; Edqvist, P.-H.; Jarvius, M.; Andersson, C.; Gustafsson, M.G.; Nygren, P.; Larsson, R.; et al. Targeting tumor cells based on Phosphodiesterase 3A expression. Exp. Cell Res. 2017, 361, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhou, J.; Zhao, Z.; Zhu, Y.; Xing, J.; An, J.; Guo, X. Low Expression of Phosphodiesterase 2 (PDE2A) Promotes the Progression by Regulating Mitochondrial Morphology and ATP Content and Predicts Poor Prognosis in Hepatocellular Carcinoma. Cells 2022, 12, 68. [Google Scholar] [CrossRef] [PubMed]
- Catalano, S.; Campana, A.; Giordano, C.; Győrffy, B.; Tarallo, R.; Rinaldi, A.; Bruno, G.; Ferraro, A.; Romeo, F.; Lanzino, M.; et al. Expression and Function of Phosphodiesterase Type 5 in Human Breast Cancer Cell Lines and Tissues: Implications for Targeted Therapy. Clin. Cancer Res. 2016, 22, 2271–2282. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Lindsey, A.; Li, N.; Lee, K.; Ramirez-Alcantara, V.; Canzoneri, J.C.; Fajardo, A.; da Silva, L.M.; Thomas, M.; Piazza, J.T.; et al. Phosphodiesterase 10A is overexpressed in lung tumor cells and inhibitors selectively suppress growth by blocking β-catenin and MAPK signaling. Oncotarget 2017, 8, 69264–69280. [Google Scholar] [CrossRef]
- Bolger, G.B. The PDE-Opathies: Diverse Phenotypes Produced by a Functionally Related Multigene Family. Trends Genet. 2021, 37, 669–681. [Google Scholar] [CrossRef]
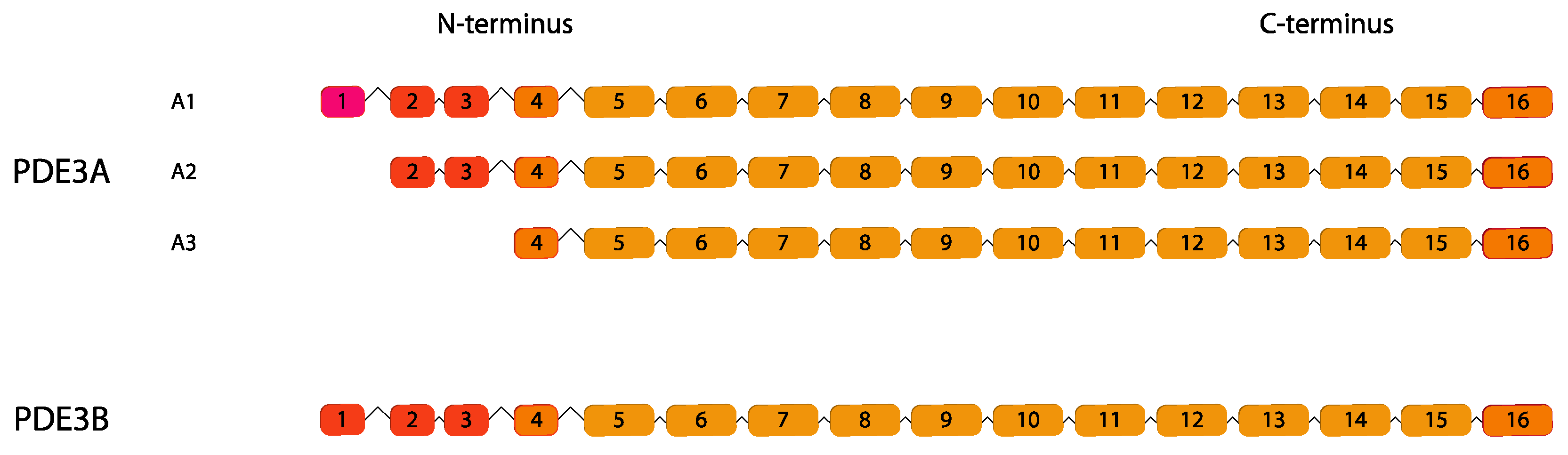

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campolo, F.; Assenza, M.R.; Venneri, M.A.; Barbagallo, F. Once upon a Testis: The Tale of Cyclic Nucleotide Phosphodiesterase in Testicular Cancers. Int. J. Mol. Sci. 2023, 24, 7617. https://doi.org/10.3390/ijms24087617
Campolo F, Assenza MR, Venneri MA, Barbagallo F. Once upon a Testis: The Tale of Cyclic Nucleotide Phosphodiesterase in Testicular Cancers. International Journal of Molecular Sciences. 2023; 24(8):7617. https://doi.org/10.3390/ijms24087617
Chicago/Turabian StyleCampolo, Federica, Maria Rita Assenza, Mary Anna Venneri, and Federica Barbagallo. 2023. "Once upon a Testis: The Tale of Cyclic Nucleotide Phosphodiesterase in Testicular Cancers" International Journal of Molecular Sciences 24, no. 8: 7617. https://doi.org/10.3390/ijms24087617
APA StyleCampolo, F., Assenza, M. R., Venneri, M. A., & Barbagallo, F. (2023). Once upon a Testis: The Tale of Cyclic Nucleotide Phosphodiesterase in Testicular Cancers. International Journal of Molecular Sciences, 24(8), 7617. https://doi.org/10.3390/ijms24087617








