Mitoregulin Contributes to Creatine Shuttling and Cardiolipin Protection in Mice Muscle
Abstract
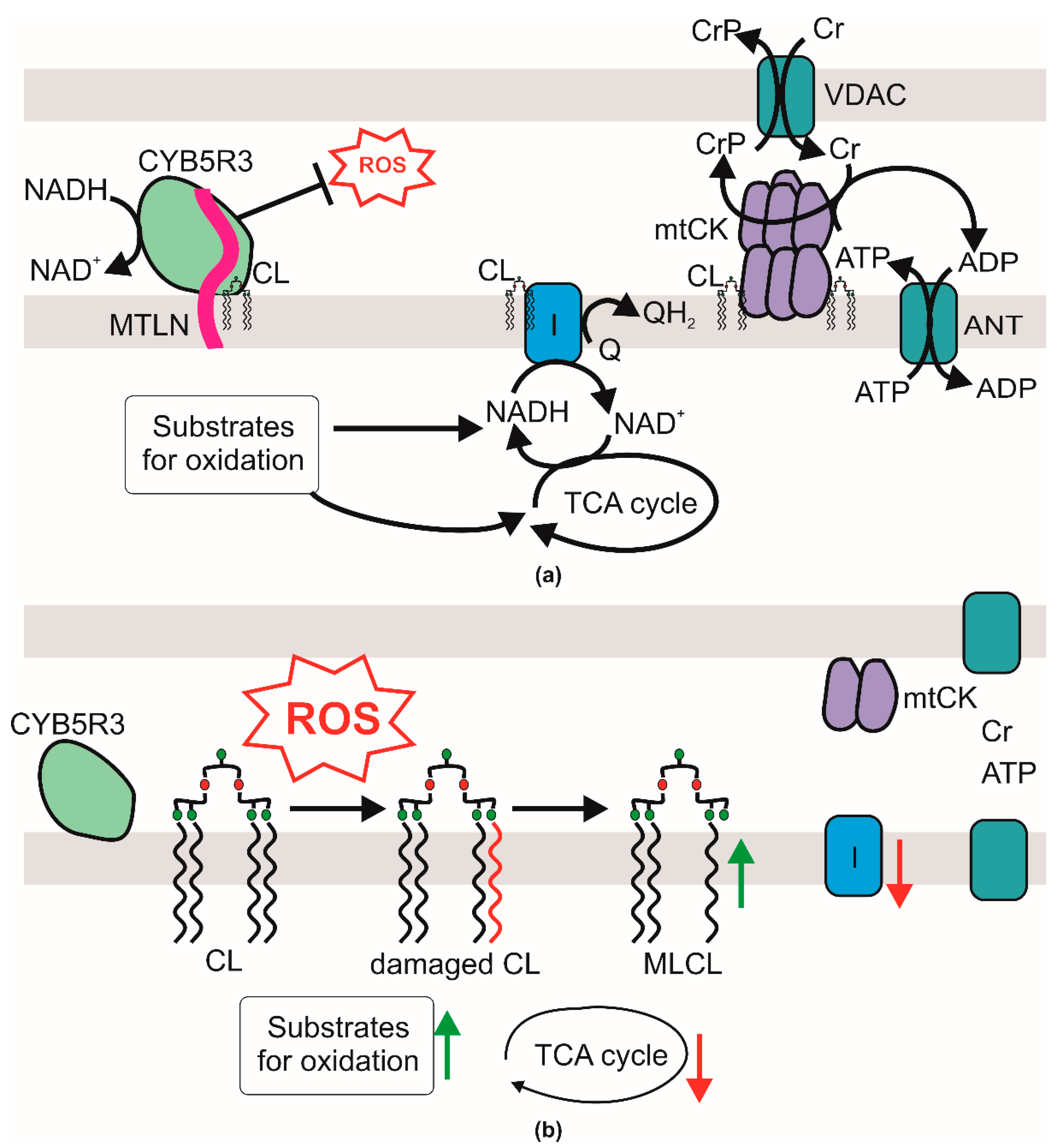
1. Introduction
2. Results
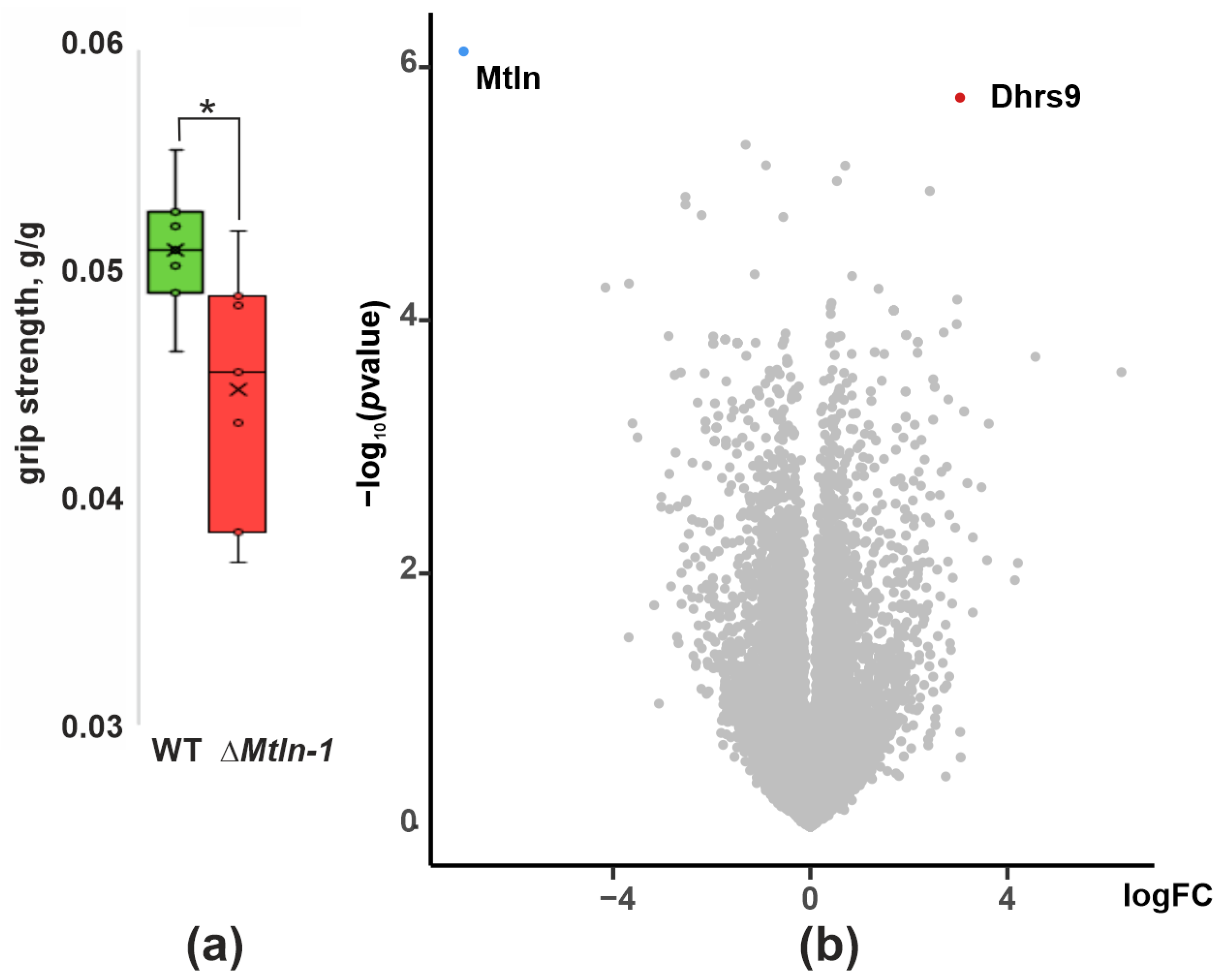
2.1. Muscle Strength of ΔMtln Mice
2.2. Influence of Mtln Gene Inactivation on Muscle Transcriptome
2.3. Histopathological Analysis of ΔMtln Mice Muscles
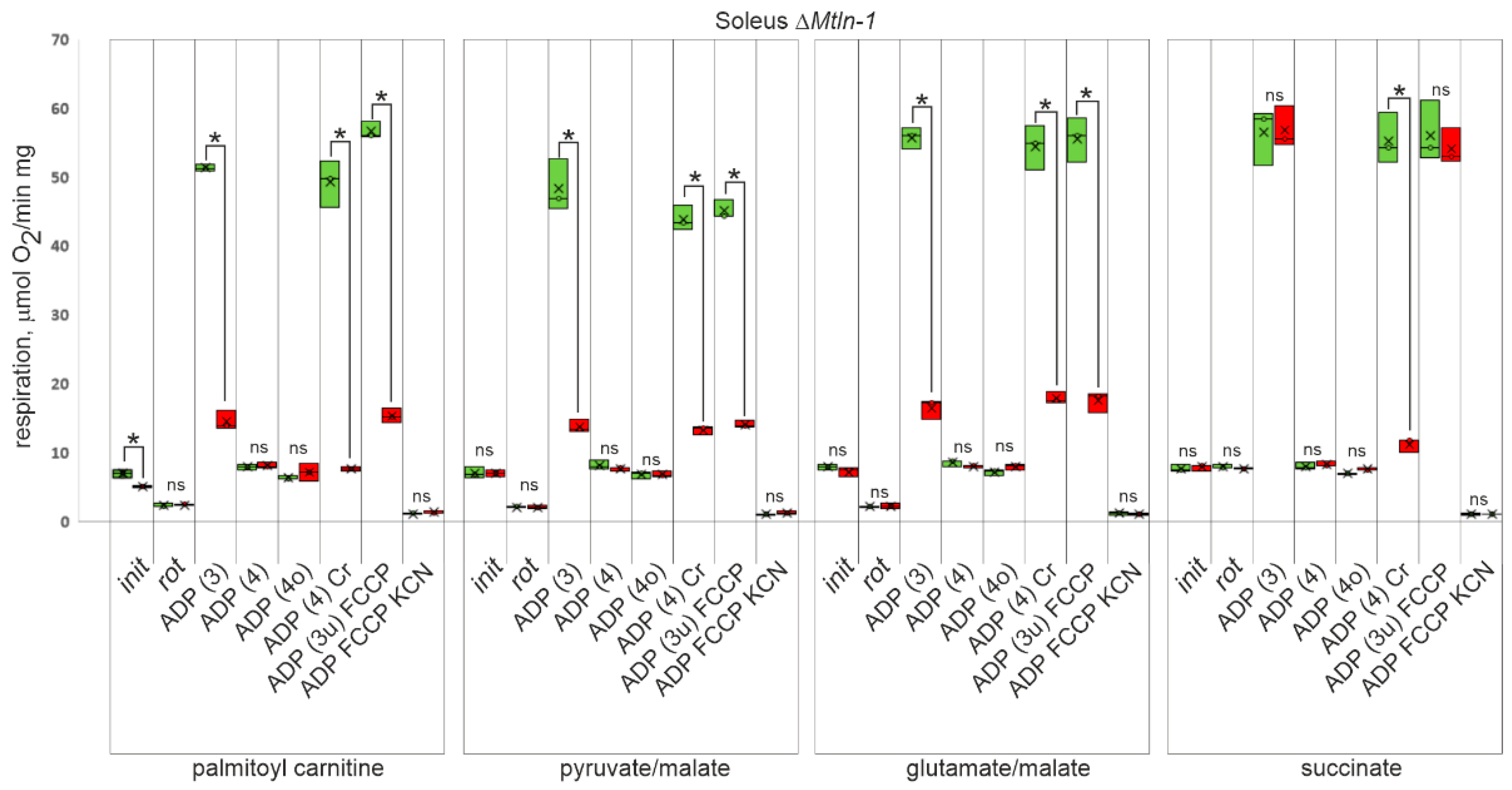
2.4. Respiration of Muscle Mitochondria from ΔMtln Mice
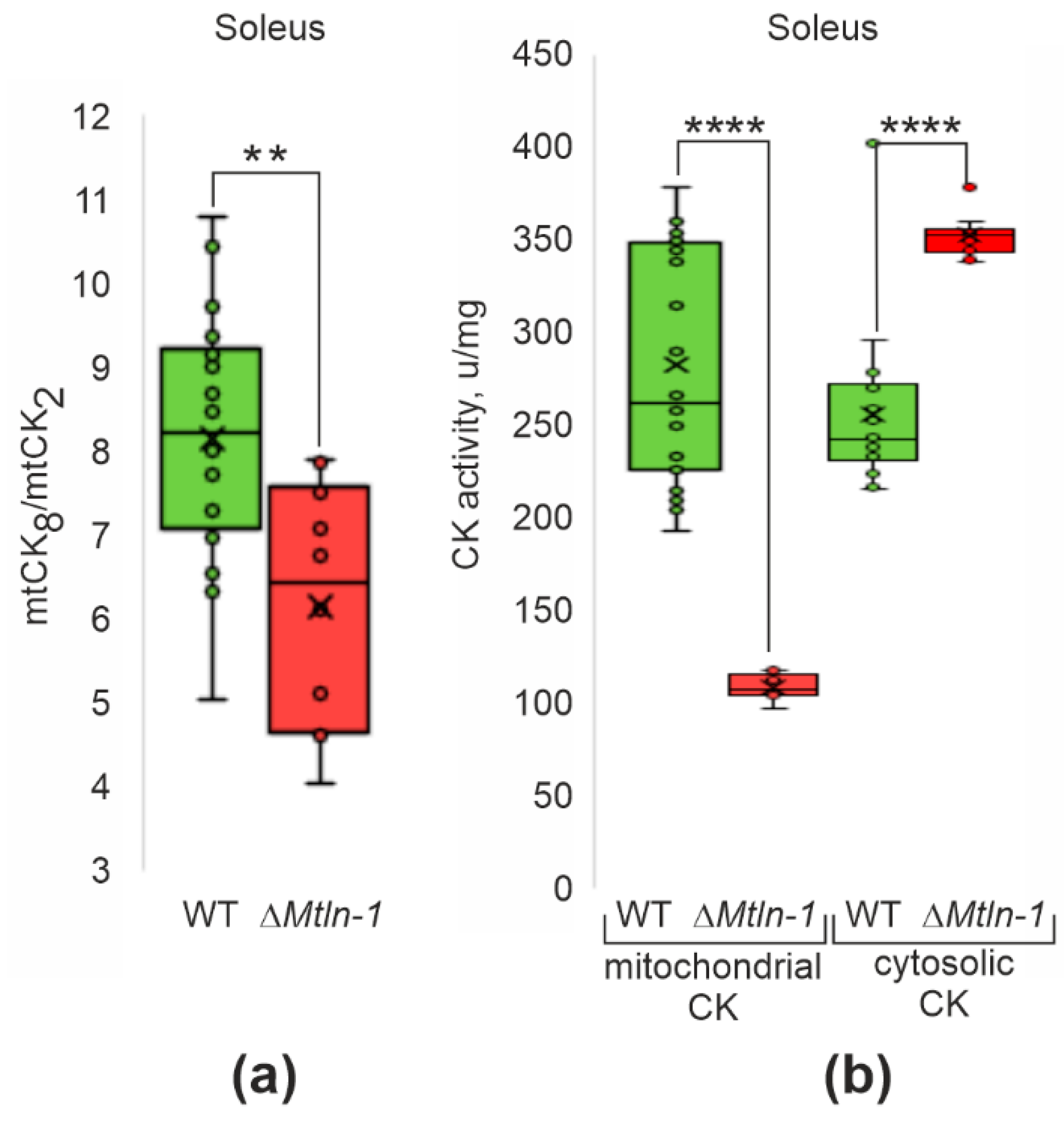
2.5. Influence of Mtln on Creatine Shuttle System Functioning
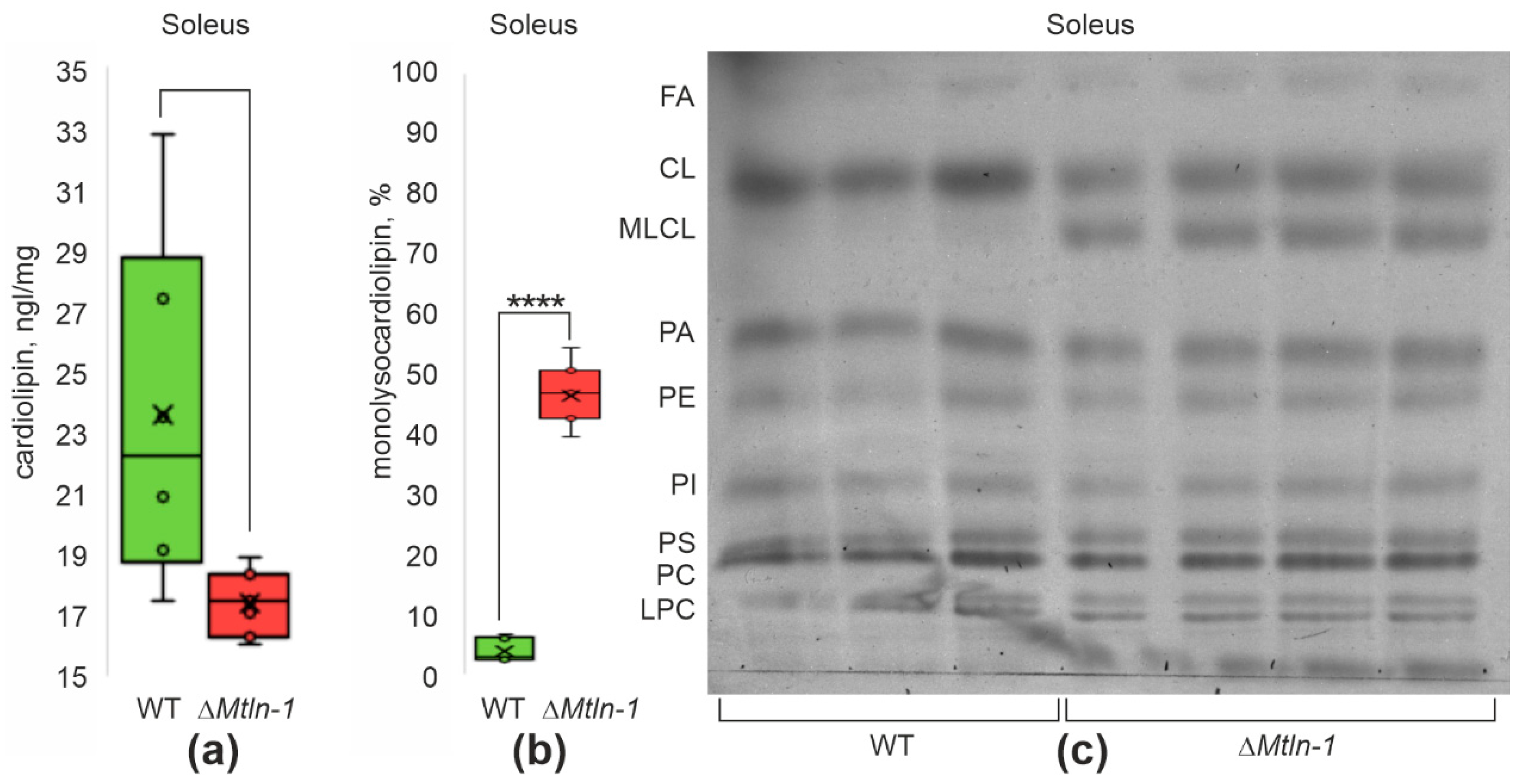
2.6. Influence of Mtln on Cardiolipin Structure and Content
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chugunova, A.; Navalayeu, T.; Dontsova, O.; Sergiev, P. Mining for Small Translated ORFs. J. Proteome Res. 2018, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sergiev, P.V.; Rubtsova, M.P. Little but Loud. The Diversity of Functions of Small Proteins and Peptides—Translational Products of Short Reading Frames. Biochem. Mosc. 2021, 86, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Couso, J.-P.; Patraquim, P. Classification and Function of Small Open Reading Frames. Nat. Rev. Mol. Cell Biol. 2017, 18, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Makarewich, C.A.; Olson, E.N. Mining for Micropeptides. Trends Cell Biol. 2017, 27, 685–696. [Google Scholar] [CrossRef]
- Ruiz-Orera, J.; Albà, M.M. Translation of Small Open Reading Frames: Roles in Regulation and Evolutionary Innovation. Trends Genet. 2019, 35, 186–198. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef]
- Kranias, E.G.; Hajjar, R.J. Modulation of Cardiac Contractility by the Phopholamban/SERCA2a Regulatome. Circ. Res. 2012, 110, 1646–1660. [Google Scholar] [CrossRef]
- Tupling, A.R.; Bombardier, E.; Gupta, S.C.; Hussain, D.; Vigna, C.; Bloemberg, D.; Quadrilatero, J.; Trivieri, M.G.; Babu, G.J.; Backx, P.H.; et al. Enhanced Ca2+ Transport and Muscle Relaxation in Skeletal Muscle from Sarcolipin-Null Mice. Am. J. Physiol. Cell Physiol. 2011, 301, C841–C849. [Google Scholar] [CrossRef]
- Chugunova, A.; Loseva, E.; Mazin, P.; Mitina, A.; Navalayeu, T.; Bilan, D.; Vishnyakova, P.; Marey, M.; Golovina, A.; Serebryakova, M.; et al. LINC00116 Codes for a Mitochondrial Peptide Linking Respiration and Lipid Metabolism. Proc. Natl. Acad. Sci. USA 2019, 116, 4940–4945. [Google Scholar] [CrossRef]
- Stein, C.S.; Jadiya, P.; Zhang, X.; McLendon, J.M.; Abouassaly, G.M.; Witmer, N.H.; Anderson, E.J.; Elrod, J.W.; Boudreau, R.L. Mitoregulin: A LncRNA-Encoded Microprotein That Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep. 2018, 23, 3710–3720.e8. [Google Scholar] [CrossRef]
- Makarewich, C.A.; Baskin, K.K.; Munir, A.Z.; Bezprozvannaya, S.; Sharma, G.; Khemtong, C.; Shah, A.M.; McAnally, J.R.; Malloy, C.R.; Szweda, L.I.; et al. MOXI Is a Mitochondrial Micropeptide That Enhances Fatty Acid β-Oxidation. Cell Rep. 2018, 23, 3701–3709. [Google Scholar] [CrossRef]
- Friesen, M.; Warren, C.R.; Yu, H.; Toyohara, T.; Ding, Q.; Florido, M.H.C.; Sayre, C.; Pope, B.D.; Goff, L.A.; Rinn, J.L.; et al. Mitoregulin Controls β-Oxidation in Human and Mouse Adipocytes. Stem Cell Rep. 2020, 14, 590–602. [Google Scholar] [CrossRef]
- Vafai, S.B.; Mootha, V.K. Mitochondrial Disorders as Windows into an Ancient Organelle. Nature 2012, 491, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Keshavan, N.; Rahman, S. Natural History of Mitochondrial Disorders: A Systematic Review. Essays Biochem. 2018, 62, 423–442. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-F.; Xiao, M.-H.; Chen, H.-X.; Meng, Y.; Zhao, N.; Yang, L.; Tang, H.; Wang, J.-L.; Liu, X.; Zhu, Y.; et al. A Novel Mitochondrial Micropeptide MPM Enhances Mitochondrial Respiratory Activity and Promotes Myogenic Differentiation. Cell Death Dis. 2019, 10, 528. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fan, J.; Han, L.; Qi, H.; Wang, Y.; Wang, H.; Chen, S.; Du, L.; Li, S.; Zhang, Y.; et al. The Micropeptide LEMP Plays an Evolutionarily Conserved Role in Myogenesis. Cell Death Dis. 2020, 11, 357. [Google Scholar] [CrossRef] [PubMed]
- Averina, O.A.; Permyakov, O.A.; Emelianova, M.A.; Grigoryeva, O.O.; Gulyaev, M.V.; Pavlova, O.S.; Mariasina, S.S.; Frolova, O.Y.; Kurkina, M.V.; Baydakova, G.V.; et al. Mitochondrial Peptide Mtln Contributes to Oxidative Metabolism in Mice. Biochimie 2023, 204, 136–139. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the Unification of Biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium; Carbon, S.; Douglass, E.; Good, B.M.; Unni, D.R.; Harris, N.L.; Mungall, C.J.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; et al. The Gene Ontology Resource: Enriching a GOld Mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Afzal, N.; Lederer, W.J.; Jafri, M.S.; Mannella, C.A. Effect of Crista Morphology on Mitochondrial ATP Output: A Computational Study. Curr. Res. Physiol. 2021, 4, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; Hepple, R.T.; Burelle, Y. Mitochondrial Functional Specialization in Glycolytic and Oxidative Muscle Fibers: Tailoring the Organelle for Optimal Function. Am. J. Physiol. Cell Physiol. 2012, 302, C629–C641. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Khuchua, Z.; Boero, J.; Mark Payne, R.; Strauss, A.W. Oxidative Myocytes of Heart and Skeletal Muscle Express Abundant Sarcomeric Mitochondrial Creatine Kinase. Histochem. J. 1999, 31, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Schlattner, U.; Wallimann, T. Octamers of Mitochondrial Creatine Kinase Isoenzymes Differ in Stability and Membrane Binding. J. Biol. Chem. 2000, 275, 17314–17320. [Google Scholar] [CrossRef] [PubMed]
- Jussupow, A.; Di Luca, A.; Kaila, V.R.I. How Cardiolipin Modulates the Dynamics of Respiratory Complex I. Sci. Adv. 2019, 5, eaav1850. [Google Scholar] [CrossRef]
- Paradies, G.; Paradies, V.; Ruggiero, F.M.; Petrosillo, G. Role of Cardiolipin in Mitochondrial Function and Dynamics in Health and Disease: Molecular and Pharmacological Aspects. Cells 2019, 8, 728. [Google Scholar] [CrossRef]
- Schlattner, U.; Kay, L.; Tokarska-Schlattner, M. Mitochondrial Proteolipid Complexes of Creatine Kinase. In Membrane Protein Complexes: Structure and Function; Harris, J.R., Boekema, E.J., Eds.; Subcellular Biochemistry; Springer: Singapore, 2018; Volume 87, pp. 365–408. ISBN 978-981-10-7756-2. [Google Scholar]
- Dudek, J. Role of Cardiolipin in Mitochondrial Signaling Pathways. Front. Cell Dev. Biol. 2017, 5, 90. [Google Scholar] [CrossRef]
- Stroud, D.A.; Surgenor, E.E.; Formosa, L.E.; Reljic, B.; Frazier, A.E.; Dibley, M.G.; Osellame, L.D.; Stait, T.; Beilharz, T.H.; Thorburn, D.R.; et al. Accessory Subunits Are Integral for Assembly and Function of Human Mitochondrial Complex I. Nature 2016, 538, 123–126. [Google Scholar] [CrossRef]
- Samluk, L.; Urbanska, M.; Kisielewska, K.; Mohanraj, K.; Kim, M.-J.; Machnicka, K.; Liszewska, E.; Jaworski, J.; Chacinska, A. Cytosolic Translational Responses Differ under Conditions of Severe Short-Term and Long-Term Mitochondrial Stress. MBoC 2019, 30, 1864–1877. [Google Scholar] [CrossRef]
- Keyes, S.R.; Cinti, D.L. Biochemical Properties of Cytochrome B5-Dependent Microsomal Fatty Acid Elongation and Identification of Products. J. Biol. Chem. 1980, 255, 11357–11364. [Google Scholar] [CrossRef]
- Oshino, N.; Imai, Y.; Sato, R. A Function of Cytochrome B5 in Fatty Acid Desaturation by Rat Liver Microsomes. J. Biochem. 1971, 69, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, M.; Tamura, M.; Yoshida, S.; Yubisui, T. Palmitoyl-CoA Elongation in Brain Microsomes: Dependence on Cytochrome b5 and NADH-Cytochrome b5 Reductase. J. Neurochem. 1985, 45, 1390–1395. [Google Scholar] [CrossRef]
- Martin-Montalvo, A.; Sun, Y.; Diaz-Ruiz, A.; Ali, A.; Gutierrez, V.; Palacios, H.H.; Curtis, J.; Siendones, E.; Ariza, J.; Abulwerdi, G.A.; et al. Cytochrome B5 Reductase and the Control of Lipid Metabolism and Healthspan. NPJ Aging Mech. Dis. 2016, 2, 16006. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.A.; Mejia, E.M.; Mitchell, R.W.; Choy, P.C.; Sparagna, G.C.; Hatch, G.M. Human Trifunctional Protein Alpha Links Cardiolipin Remodeling to Beta-Oxidation. PLoS ONE 2012, 7, e48628. [Google Scholar] [CrossRef]
- Yan, W.; Jenkins, C.M.; Han, X.; Mancuso, D.J.; Sims, H.F.; Yang, K.; Gross, R.W. The Highly Selective Production of 2-Arachidonoyl Lysophosphatidylcholine Catalyzed by Purified Calcium-Independent Phospholipase A2gamma: Identification of a Novel Enzymatic Mediator for the Generation of a Key Branch Point Intermediate in Eicosanoid Signaling. J. Biol. Chem. 2005, 280, 26669–26679. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-Y.; Moon, S.H.; Jenkins, C.M.; Li, M.; Sims, H.F.; Guan, S.; Gross, R.W. The Phospholipase IPLA2γ Is a Major Mediator Releasing Oxidized Aliphatic Chains from Cardiolipin, Integrating Mitochondrial Bioenergetics and Signaling. J. Biol. Chem. 2017, 292, 10672–10684. [Google Scholar] [CrossRef]
- Zhong, H.; Lu, J.; Xia, L.; Zhu, M.; Yin, H. Formation of Electrophilic Oxidation Products from Mitochondrial Cardiolipin in Vitro and in Vivo in the Context of Apoptosis and Atherosclerosis. Redox Biol. 2014, 2, 878–883. [Google Scholar] [CrossRef] [PubMed]
- Wahjudi, P.N.; Yee, J.K.; Martinez, S.R.; Zhang, J.; Teitell, M.; Nikolaenko, L.; Swerdloff, R.; Wang, C.; Lee, W.N.P. Turnover of Nonessential Fatty Acids in Cardiolipin from the Rat Heart. J. Lipid Res. 2011, 52, 2226–2233. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Mayette, J.; Rapoport, S.I.; Bazinet, R.P. Selective Remodeling of Cardiolipin Fatty Acids in the Aged Rat Heart. Lipids Health Dis. 2006, 5, 2. [Google Scholar] [CrossRef]
- Sullivan, E.M.; Pennington, E.R.; Sparagna, G.C.; Torres, M.J.; Neufer, P.D.; Harris, M.; Washington, J.; Anderson, E.J.; Zeczycki, T.N.; Brown, D.A.; et al. Docosahexaenoic Acid Lowers Cardiac Mitochondrial Enzyme Activity by Replacing Linoleic Acid in the Phospholipidome. J. Biol. Chem. 2018, 293, 466–483. [Google Scholar] [CrossRef]
- Wallimann, T.; Tokarska-Schlattner, M.; Schlattner, U. The Creatine Kinase System and Pleiotropic Effects of Creatine. Amino Acids 2011, 40, 1271–1296. [Google Scholar] [CrossRef]
- Ye, C.; Shen, Z.; Greenberg, M.L. Cardiolipin Remodeling: A Regulatory Hub for Modulating Cardiolipin Metabolism and Function. J. Bioenerg. Biomembr. 2016, 48, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Averina, O.A.; Vysokikh, M.Y.; Permyakov, O.A.; Sergiev, P.V. Simple Recommendations for Improving Efficiency in Generating Genome-Edited Mice. Acta Nat. 2020, 12, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Rogers, D.C.; Peters, J.; Martin, J.E.; Ball, S.; Nicholson, S.J.; Witherden, A.S.; Hafezparast, M.; Latcham, J.; Robinson, T.L.; Quilter, C.A.; et al. SHIRPA, a Protocol for Behavioral Assessment: Validation for Longitudinal Study of Neurological Dysfunction in Mice. Neurosci. Lett. 2001, 306, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Vyssokikh, M.Y.; Holtze, S.; Averina, O.A.; Lyamzaev, K.G.; Panteleeva, A.A.; Marey, M.V.; Zinovkin, R.A.; Severin, F.F.; Skulachev, M.V.; Fasel, N.; et al. Mild Depolarization of the Inner Mitochondrial Membrane Is a Crucial Component of an Anti-Aging Program. Proc. Natl. Acad. Sci. USA 2020, 117, 6491–6501. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Averina, O.A.; Permyakov, O.A.; Emelianova, M.A.; Grigoryeva, O.O.; Lovat, M.L.; Egorova, A.E.; Grinchenko, A.V.; Kumeiko, V.V.; Marey, M.V.; Manskikh, V.N.; et al. Mitoregulin Contributes to Creatine Shuttling and Cardiolipin Protection in Mice Muscle. Int. J. Mol. Sci. 2023, 24, 7589. https://doi.org/10.3390/ijms24087589
Averina OA, Permyakov OA, Emelianova MA, Grigoryeva OO, Lovat ML, Egorova AE, Grinchenko AV, Kumeiko VV, Marey MV, Manskikh VN, et al. Mitoregulin Contributes to Creatine Shuttling and Cardiolipin Protection in Mice Muscle. International Journal of Molecular Sciences. 2023; 24(8):7589. https://doi.org/10.3390/ijms24087589
Chicago/Turabian StyleAverina, Olga A., Oleg A. Permyakov, Mariia A. Emelianova, Olga O. Grigoryeva, Maxim L. Lovat, Anna E. Egorova, Andrei V. Grinchenko, Vadim V. Kumeiko, Maria V. Marey, Vasily N. Manskikh, and et al. 2023. "Mitoregulin Contributes to Creatine Shuttling and Cardiolipin Protection in Mice Muscle" International Journal of Molecular Sciences 24, no. 8: 7589. https://doi.org/10.3390/ijms24087589
APA StyleAverina, O. A., Permyakov, O. A., Emelianova, M. A., Grigoryeva, O. O., Lovat, M. L., Egorova, A. E., Grinchenko, A. V., Kumeiko, V. V., Marey, M. V., Manskikh, V. N., Dontsova, O. A., Vysokikh, M. Y., & Sergiev, P. V. (2023). Mitoregulin Contributes to Creatine Shuttling and Cardiolipin Protection in Mice Muscle. International Journal of Molecular Sciences, 24(8), 7589. https://doi.org/10.3390/ijms24087589





