G-QINDER Tool: Bioinformatically Predicted Formation of Different Four-Stranded DNA Motifs from (GT)n and (GA)n Repeats
Abstract
1. Introduction
2. Results
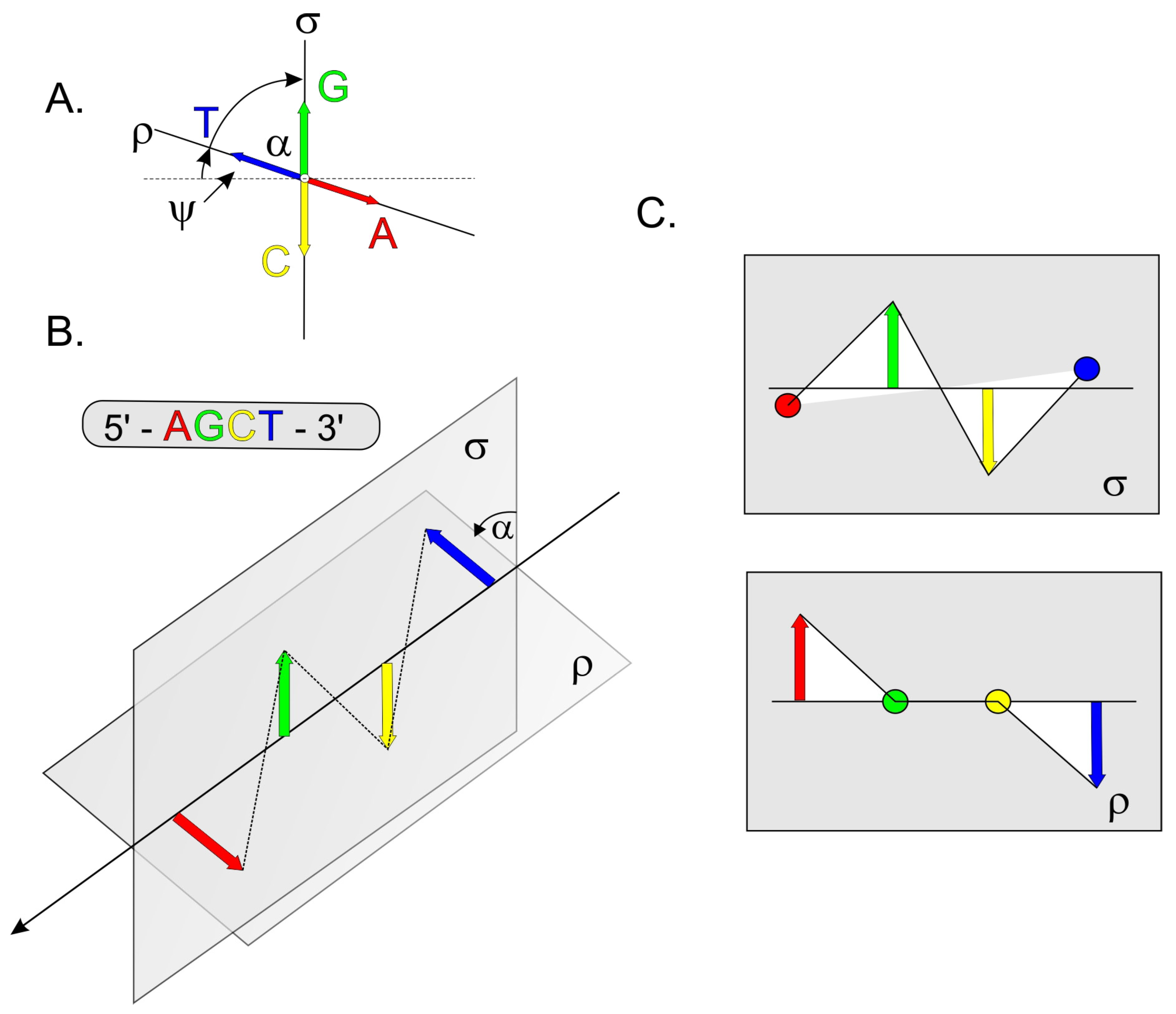
2.1. Analysis of Selected DNA Sequences Using the G-QINDER Tool
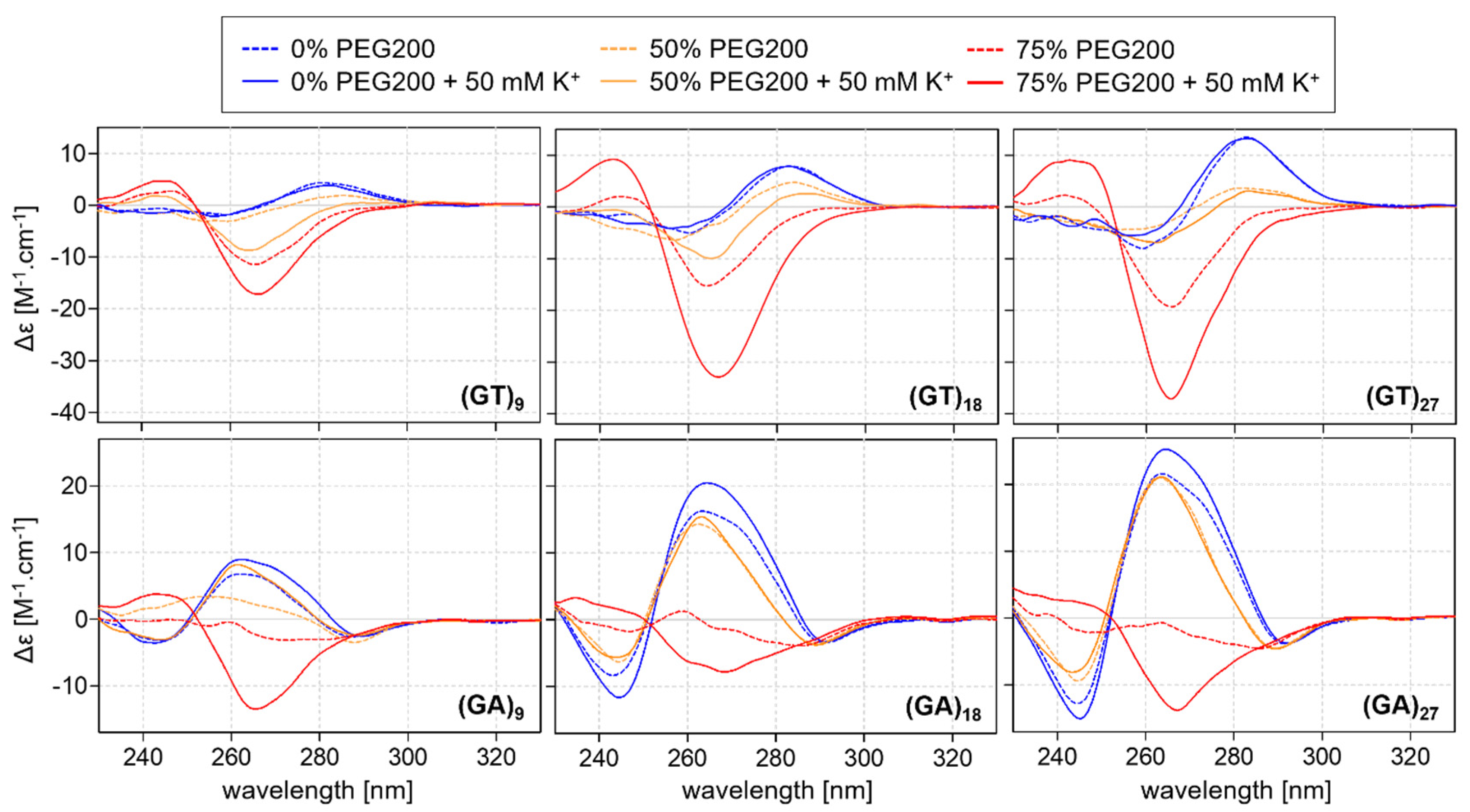
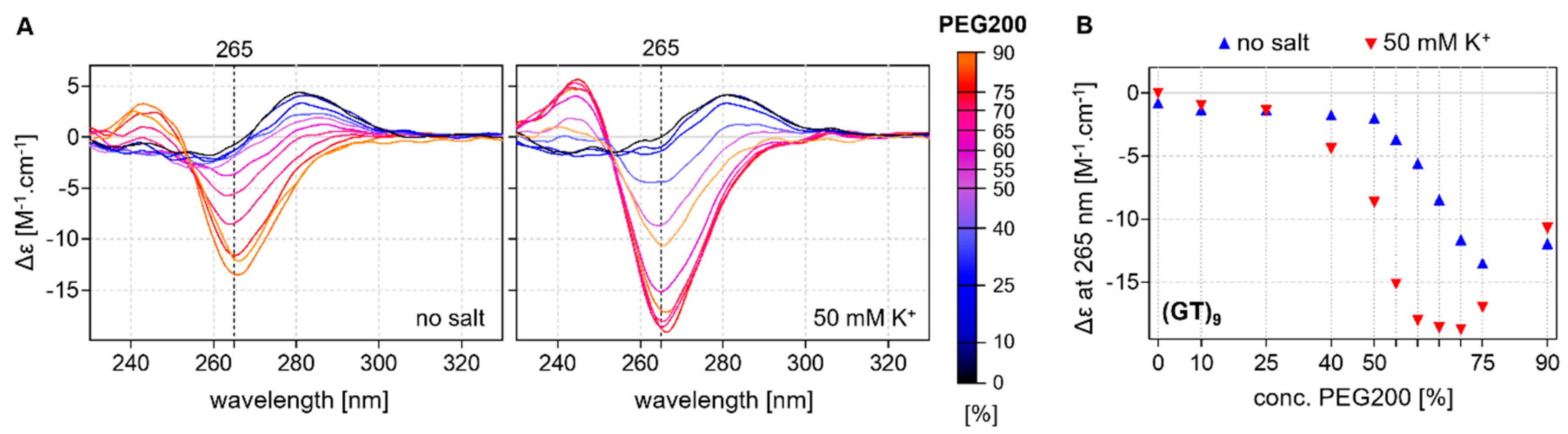
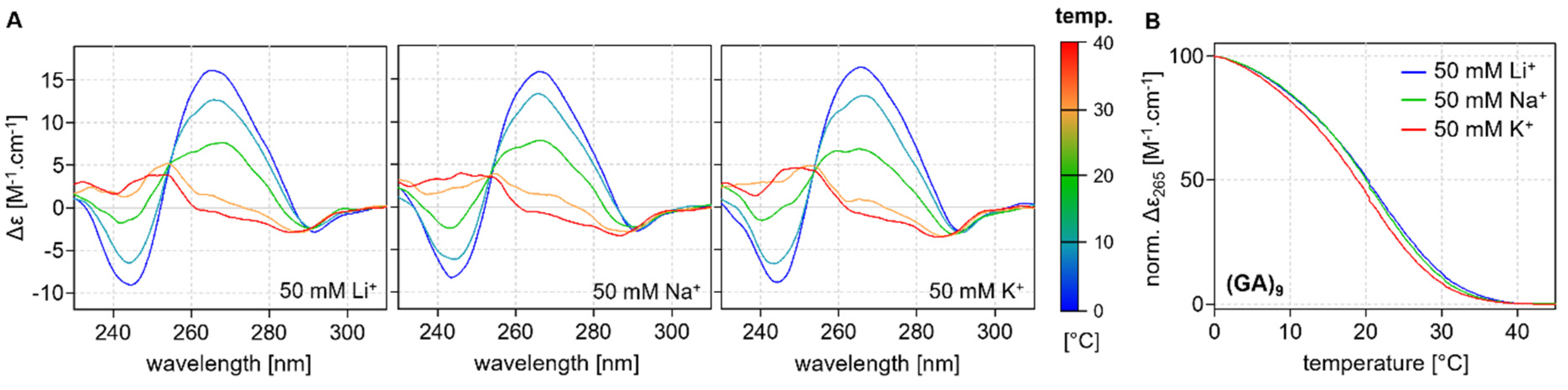
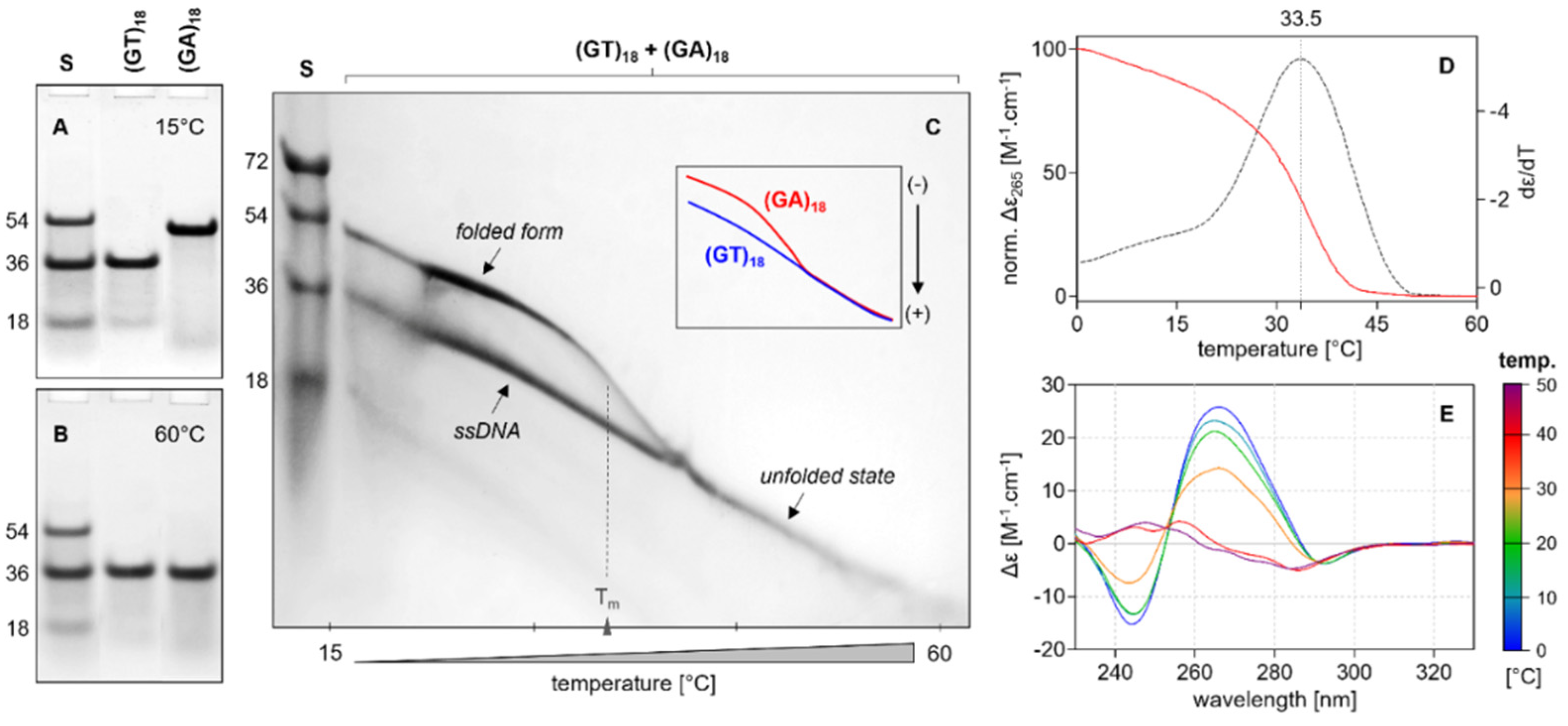
2.2. CD Analysis of d(GT)n and d(GA)n Sequences
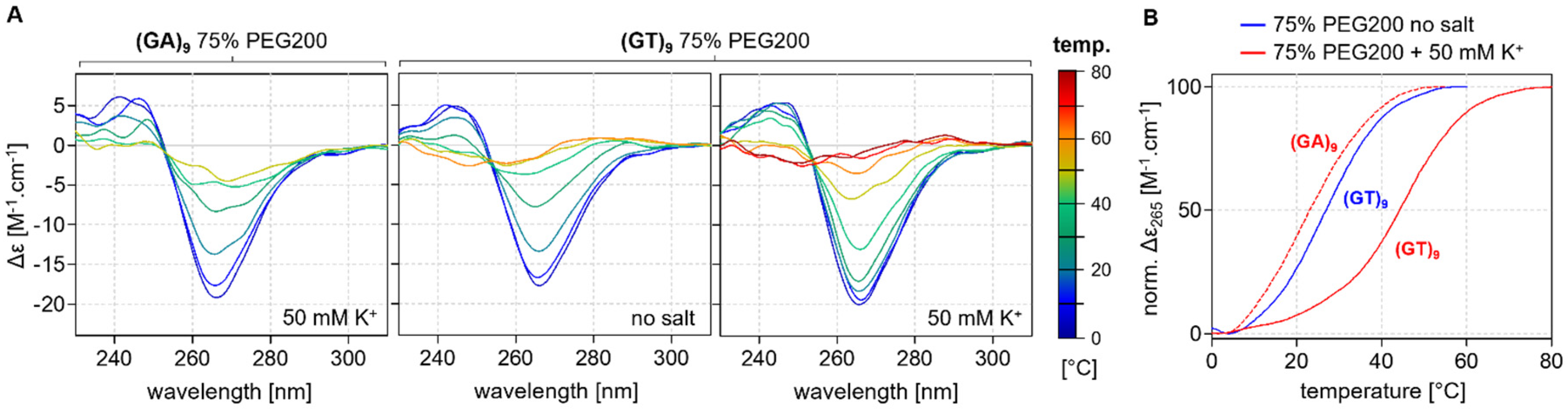
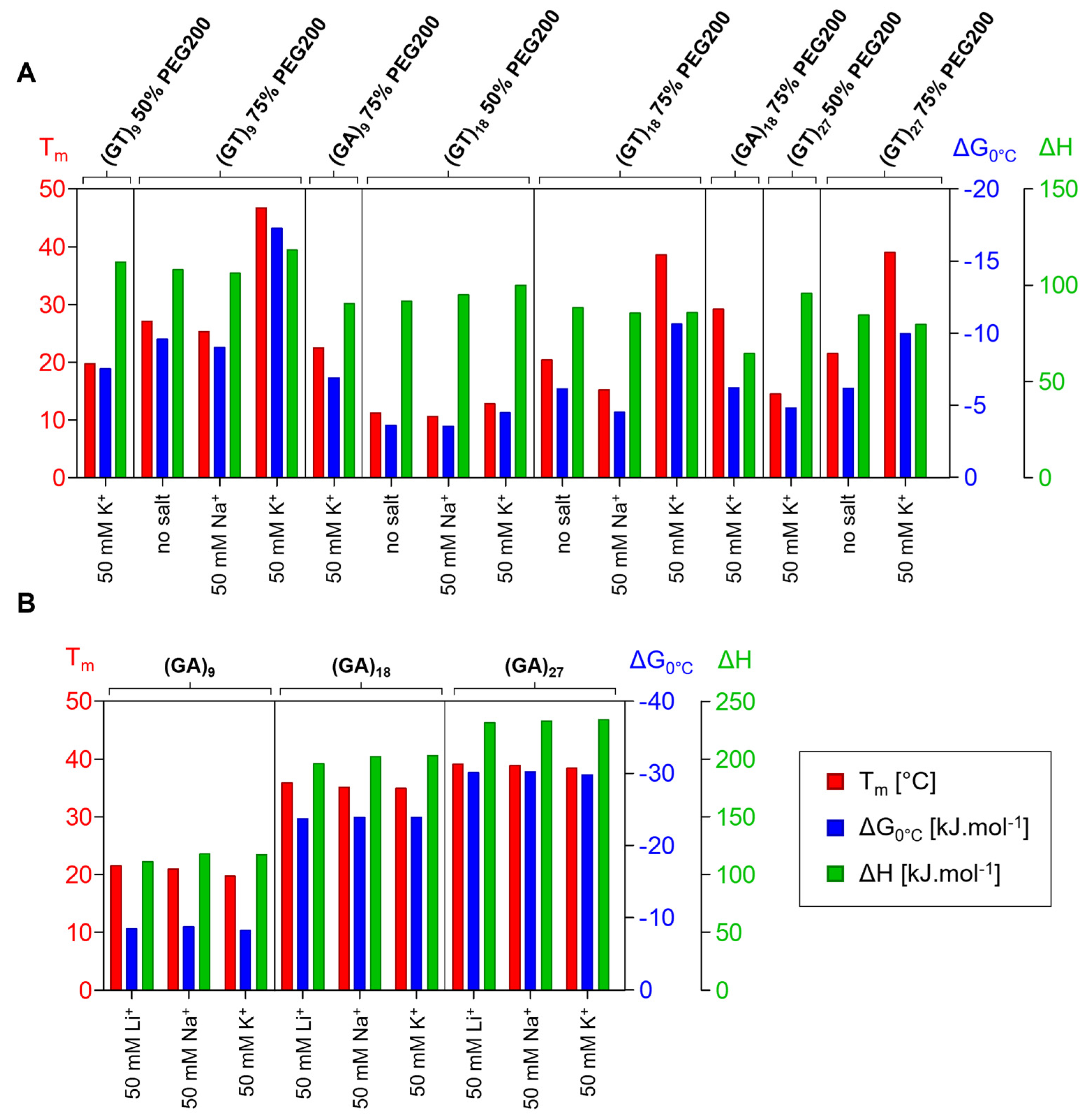
2.3. Thermodynamic Stability
2.4. Electrophoretic Analysis
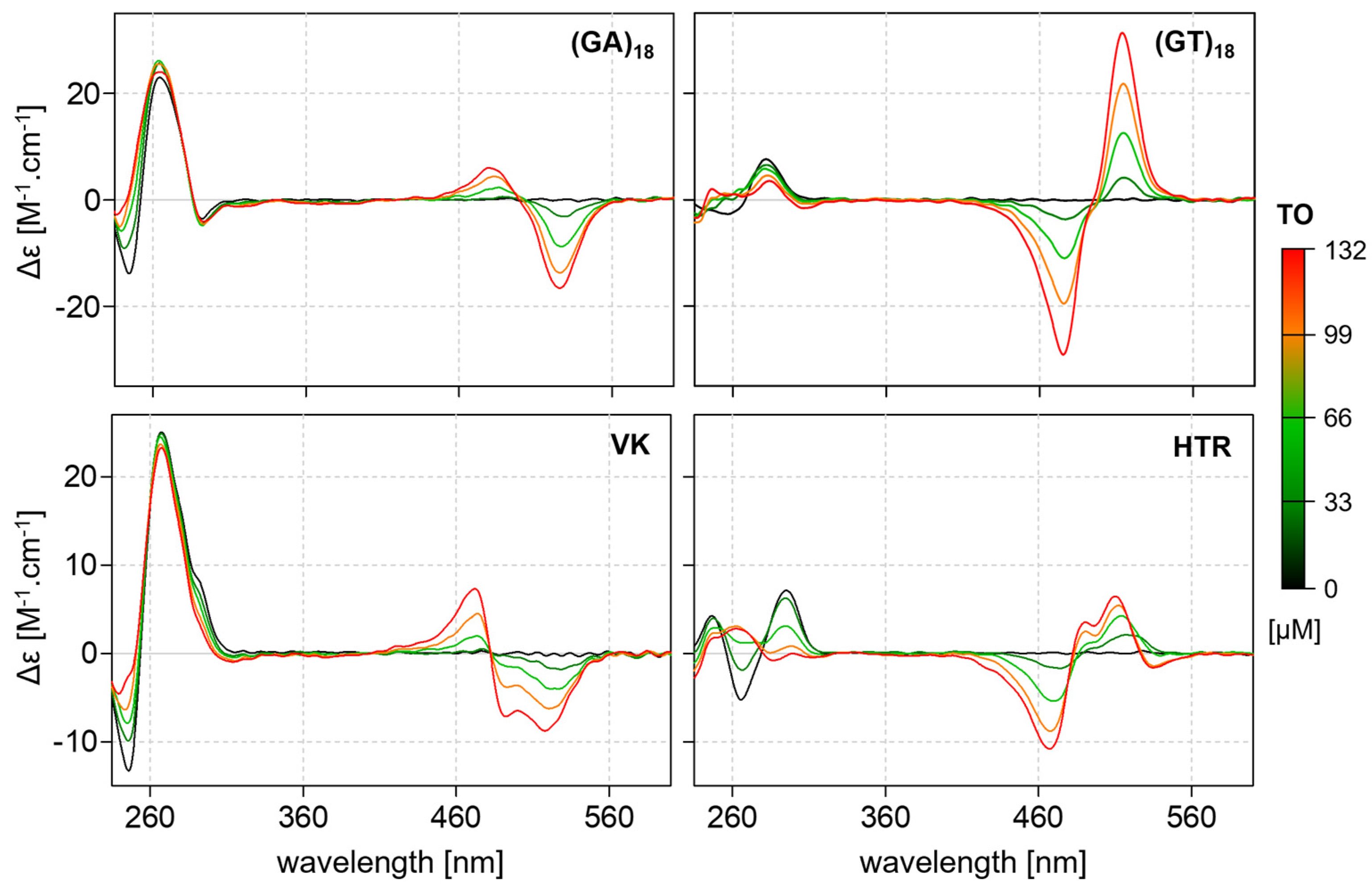
2.5. Interactions between Noncanonical Motifs and Thiazole Orange
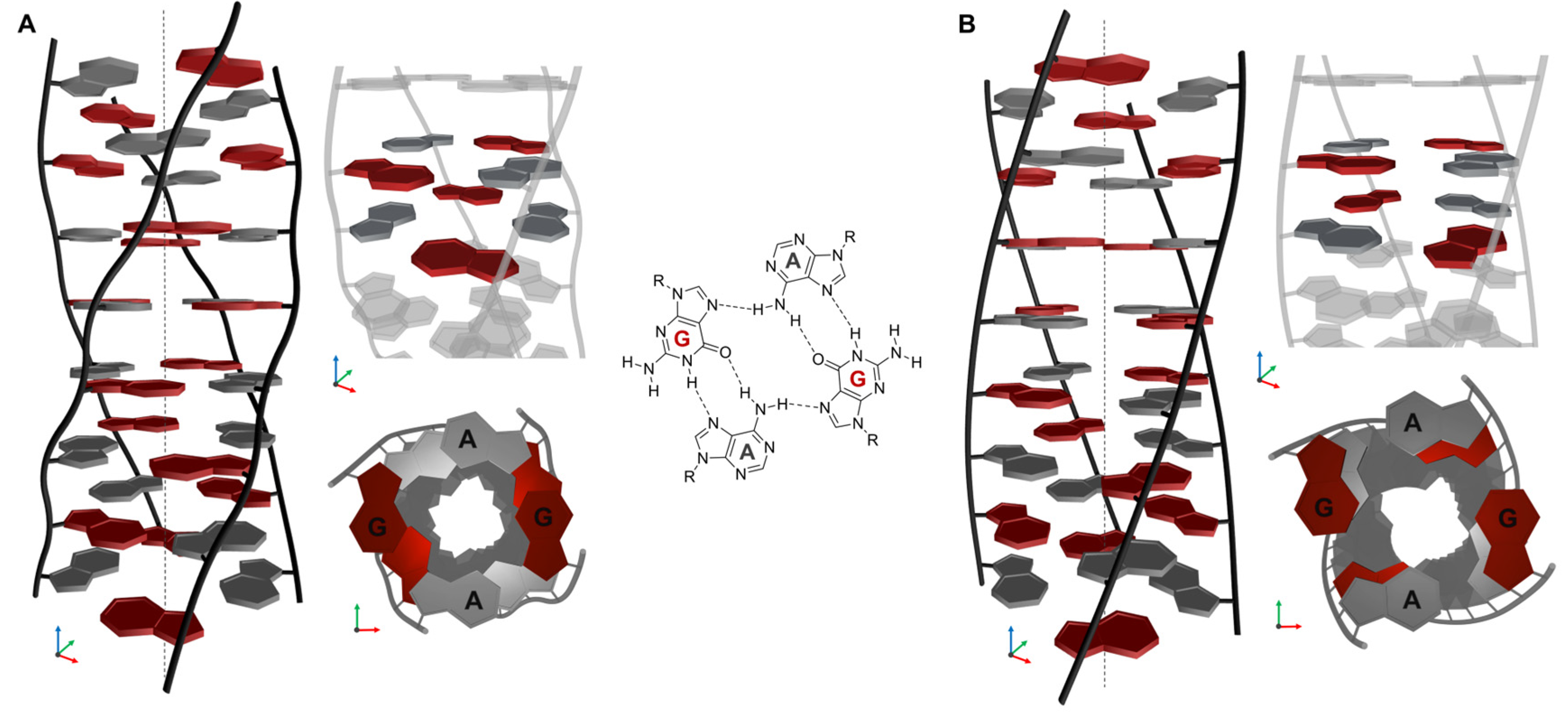
2.6. Structural Model of (AG)n
3. Materials and Methods
3.1. Circular Dichroism Spectroscopy
CD Melting Curves
3.2. Electrophoresis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bacolla, A.; Wojciechowska, M.; Kosmider, B.; Larson, J.E.; Wells, R.D. The Involvement of Non-B DNA Structures in Gross Chromosomal Rearrangements. DNA Repair 2006, 5, 1161–1170. [Google Scholar] [CrossRef]
- Choi, J.; Majima, T. Conformational Changes of Non-B DNA. Chem. Soc. Rev. 2011, 40, 5893. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Bacolla, A.; Wang, G.; Vasquez, K.M. Non-B DNA Structure-Induced Genetic Instability and Evolution. Cell. Mol. Life Sci. 2009, 67, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Kuryavyi, V.; Patel, D.J. DNA Architecture: From G to Z. Curr. Opin. Struc. Biol. 2006, 16, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, Topology and Structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal Structure of Parallel Quadruplexes from Human Telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Dai, J.; Carver, M.; Punchihewa, C.; Jones, R.A.; Yang, D. Structure of the Hybrid-2 Type Intramolecular Human Telomeric G-Quadruplex in K+ Solution: Insights into Structure Polymorphism of the Human Telomeric Sequence. Nucleic Acids Res. 2007, 35, 4927–4940. [Google Scholar] [CrossRef]
- Dai, J.; Punchihewa, C.; Ambrus, A.; Chen, D.; Jones, R.A.; Yang, D. Structure of the Intramolecular Human Telomeric G-Quadruplex in Potassium Solution: A Novel Adenine Triple Formation. Nucleic Acids Res. 2007, 35, 2440–2450. [Google Scholar] [CrossRef]
- Lim, K.W.; Amrane, S.; Bouaziz, S.; Xu, W.; Mu, Y.; Patel, D.J.; Luu, K.N.; Phan, A.T. Structure of the Human Telomere in K+ Solution: A Stable Basket-Type G-Quadruplex with Only Two G-Tetrad Layers. J. Am. Chem. Soc. 2009, 131, 4301–4309. [Google Scholar] [CrossRef]
- Víglaský, V.; Tlučková, K.; Bauer, Ľ. The First Derivative of a Function of Circular Dichroism Spectra: Biophysical Study of Human Telomeric G-Quadruplex. Eur. Biophys. J. 2010, 40, 29–37. [Google Scholar] [CrossRef]
- Miyoshi, D.; Fujimoto, T.; Sugimoto, N. Molecular Crowding and Hydration Regulating of G-Quadruplex Formation. Top. Curr. Chem. 2012, 330, 87–110. [Google Scholar] [CrossRef]
- Miyoshi, D.; Sugimoto, N. Molecular Crowding Effects on Structure and Stability of DNA. Biochimie 2008, 90, 1040–1051. [Google Scholar] [CrossRef]
- Krafčíková, P.; Demkovičová, E.; Víglaský, V. Ebola Virus Derived G-Quadruplexes: Thiazole Orange Interaction. BBA-Gen. Subj. 2017, 1861, 1321–1328. [Google Scholar] [CrossRef] [PubMed]
- Phan, A.T.; Patel, D.J. Two-Repeat Human Telomeric d(TAGGGTTAGGGT) Sequence Forms Interconverting Parallel and Antiparallel G-Quadruplexes in Solution: Distinct Topologies, Thermodynamic Properties, and Folding/Unfolding Kinetics. J. Am. Chem. Soc. 2003, 125, 15021–15027. [Google Scholar] [CrossRef]
- Ying, L.; Green, J.J.; Li, H.; Klenerman, D.; Balasubramanian, S. Studies on the Structure and Dynamics of the Human Telomeric G Quadruplex by Single-Molecule Fluorescence Resonance Energy Transfer. Proc. Natl. Acad. Sci. USA 2003, 100, 14629–14634. [Google Scholar] [CrossRef]
- Li, J. Not so Crystal Clear: The Structure of the Human Telomere G-Quadruplex in Solution Differs from That Present in a Crystal. Nucleic Acids Res. 2005, 33, 4649–4659. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Okumus, B.; Kim, D.S.; Ha, T. Extreme Conformational Diversity in Human Telomeric DNA. Proc. Natl. Acad. Sci. USA 2005, 102, 18938–18943. [Google Scholar] [CrossRef]
- Víglaský, V. Hidden Information Revealed Using the Orthogonal System of Nucleic Acids. Int. J. Mol. Sci. 2022, 23, 1804. [Google Scholar] [CrossRef] [PubMed]
- Bedrat, A.; Lacroix, L.; Mergny, J.-L. Re-Evaluation of G-Quadruplex Propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L. Prevalence of Quadruplexes in the Human Genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef]
- Todd, A.K. Highly Prevalent Putative Quadruplex Sequence Motifs in Human DNA. Nucleic Acids Res. 2005, 33, 2901–2907. [Google Scholar] [CrossRef] [PubMed]
- Kudlicki, A.S. G-Quadruplexes Involving Both Strands of Genomic DNA Are Highly Abundant and Colocalize with Functional Sites in the Human Genome. PLoS ONE 2016, 11, e0146174. [Google Scholar] [CrossRef]
- Varizhuk, A.; Ischenko, D.; Tsvetkov, V.; Novikov, R.; Kulemin, N.; Kaluzhny, D.; Vlasenok, M.; Naumov, V.; Smirnov, I.; Pozmogova, G. The Expanding Repertoire of G4 DNA Structures. Biochimie 2017, 135, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS Mapper: A Web-Based Server for Predicting G-Quadruplexes in Nucleotide Sequences. Nucleic Acids Res. 2006, 34, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Hon, J.; Martínek, T.; Zendulka, J.; Lexa, M. Pqsfinder: An Exhaustive and Imperfection-Tolerant Search Tool for Potential Quadruplex-Forming Sequences in R. Bioinformatics 2017, 33, 3373–3379. [Google Scholar] [CrossRef]
- Eddy, J.; Maizels, N. Gene Function Correlates with Potential for G4 DNA Formation in the Human Genome. Nucleic Acids Res. 2006, 34, 3887–3896. [Google Scholar] [CrossRef]
- Beaudoin, J.-D.; Jodoin, R.; Perreault, J.-P. New Scoring System to Identify RNA G-Quadruplex Folding. Nucleic Acids Res. 2013, 42, 1209–1223. [Google Scholar] [CrossRef]
- Garant, J.-M.; Perreault, J.-P.; Scott, M.S. Motif Independent Identification of Potential RNA G-Quadruplexes by G4RNA Screener. Bioinformatics 2017, 33, 3532–3537. [Google Scholar] [CrossRef]
- Sahakyan, A.B.; Chambers, V.S.; Marsico, G.; Santner, T.; Di Antonio, M.; Balasubramanian, S. Machine Learning Model for Sequence-Driven DNA G-Quadruplex Formation. Sci. Rep. 2017, 7, 14535. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.; Qin, J.; zu Siederdissen, C.H.; Tanzer, A.; Amman, F.; Hofacker, I.L.; Stadler, P.F. 2D Meets 4G: G-Quadruplexes in RNA Secondary Structure Prediction. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 10, 832–844. [Google Scholar] [CrossRef]
- Di Salvo, M.; Pinatel, E.; Talà, A.; Fondi, M.; Peano, C.; Alifano, P. G4PromFinder: An Algorithm for Predicting Transcription Promoters in GC-Rich Bacterial Genomes Based on AT-Rich Elements and G-Quadruplex Motifs. BMC Bioinform. 2018, 19, 36. [Google Scholar] [CrossRef]
- Bagshaw, A.T.M. Functional Mechanisms of Microsatellite DNA in Eukaryotic Genomes. Genome Biol. Evol. 2017, 9, 2428–2443. [Google Scholar] [CrossRef] [PubMed]
- Hefferon, T.W.; Groman, J.D.; Yurk, C.E.; Cutting, G.R. A Variable Dinucleotide Repeat in the CFTR Gene Contributes to Phenotype Diversity by Forming RNA Secondary Structures That Alter Splicing. Proc. Natl. Acad. Sci. USA 2004, 101, 3504–3509. [Google Scholar] [CrossRef] [PubMed]
- Pagani, F.; Buratti, E.; Stuani, C.; Romano, M.; Zuccato, E.; Niksic, M.; Giglio, L.; Faraguna, D.; Baralle, F.E. Splicing Factors Induce Cystic Fibrosis Transmembrane Regulator Exon 9 Skipping through a Nonevolutionary Conserved Intronic Element. J. Biol. Chem. 2000, 275, 21041–21047. [Google Scholar] [CrossRef]
- Hui, J.; Hung, L.-H.; Heiner, M.; Schreiner, S.; Neumüller, N.; Reither, G.; Haas, S.A.; Bindereif, A. Intronic CA-Repeat and CA-Rich Elements: A New Class of Regulators of Mammalian Alternative Splicing. EMBO J. 2005, 24, 1988–1998. [Google Scholar] [CrossRef]
- Jansson-Fritzberg, L.I.; Sousa, C.I.; Smallegan, M.J.; Song, J.J.; Gooding, A.R.; Kasinath, V.; Rinn, J.L.; Cech, T.R. DNMT1 Inhibition by pUG-Fold Quadruplex RNA. RNA 2022, 29, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Roschdi, S.; Yan, J.; Nomura, Y.; Escobar, C.A.; Petersen, R.J.; Bingman, C.A.; Tonelli, M.; Vivek, R.; Montemayor, E.J.; Wickens, M.; et al. An Atypical RNA Quadruplex Marks RNAs as Vectors for Gene Silencing. Nat. Sctruct. Mol. Biol. 2022, 29, 1113–1121. [Google Scholar] [CrossRef]
- Tlučková, K.; Marušič, M.; Tóthová, P.; Bauer, L.; Šket, P.; Plavec, J.; Viglasky, V. Human Papillomavirus G-Quadruplexes. Biochemistry 2013, 52, 7207–7216. [Google Scholar] [CrossRef]
- Kocman, V.; Plavec, J. A Tetrahelical DNA Fold Adopted by Tandem Repeats of Alternating GGG and GCG Tracts. Nat. Commun. 2014, 15, 5831. [Google Scholar] [CrossRef]
- Kocman, V.; Plavec, J. Tetrahelical Structural Family Adopted by AGCGA-Rich Regulatory DNA Regions. Nat. Commun. 2017, 17, 15355. [Google Scholar] [CrossRef]
- Das, P.; Ngo, K.H.; Winnerdy, F.R.; Maity, A.; Bakalar, B.; Mechulam, Y.; Schmitt, E.; Phan, A.T. Bulges in Left-Handed G-Quadruplexes. Nucleic Acids Res. 2021, 49, 1724–1736. [Google Scholar] [CrossRef] [PubMed]
- Bauer, L.; Tlučková, K.; Tóthová, P.; Viglaský, V. G-Quadruplex Motifs Arranged in Tandem Occurring in Telomeric Repeats and the Insulin-Linked Polymorphic Region. Biochemistry 2011, 50, 7484–7492. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Evans, D.H.; Morgan, A.R. Polypurine DNAs and RNAs Form Secondary Structures Which May Be Tetra-Stranded. Nucleic Acids Res. 1980, 8, 4305–4320. [Google Scholar] [CrossRef] [PubMed]
- Rippe, K.; Fritsch, V.; Westhof, E.; Jovin, T.M. Alternating d(G-A) Sequences Form a Parallel-Stranded DNA Homoduplex. EMBO J. 1992, 11, 3777–3786. [Google Scholar] [CrossRef]
- Huertas, D.; Bellsolell, L.; Casasnovas, J.M.; Coll, M.; Azorín, F. Alternating d(GA)n DNA Sequences Form Antiparallel Stranded Homoduplexes Stabilized by the Formation of G.A Base Pairs. EMBO J. 1993, 12, 4029–4038. [Google Scholar] [CrossRef]
- Kejnovská, I.; Kypr, J.; Vorlíčková, M. Circular Dichroism Spectroscopy of Conformers of (Guanine + Adenine) Repeat Strands of DNA. Chirality 2003, 15, 584–592. [Google Scholar] [CrossRef]
- Poniková, S.; Tlučková, K.; Antalík, M.; Víglaský, V.; Hianik, T. The Circular Dichroism and Differential Scanning Calorimetry Study of the Properties of DNA Aptamer Dimers. Biophys. Chem. 2011, 155, 29–35. [Google Scholar] [CrossRef]
- Sokoloski, J.E.; Godfrey, S.A.; Dombrowski, S.E.; Bevilacqua, P.C. Prevalence of Syn Nucleobases in the Active Sites of Functional RNAs. RNA 2011, 17, 1775–1787. [Google Scholar] [CrossRef]
| Name | DNA Sequence | nts | Qs | |
|---|---|---|---|---|
| Ψ = 15° | 30° | |||
| (TG)6 | GTGTGTGTGTGT | 12 | 1.73 | 2.06 |
| (TG)9 | GTGTGTGTGTGTGTGTGT | 18 | 1.78 | 2.12 |
| (TG)18 | GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGT | 36 | 1.84 | 2.18 |
| (TG)27 | GTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGT | 54 | 1.85 | 2.20 |
| (GA)6 | GAGAGAGAGAGA | 12 | 1.02 | 0.69 |
| (GA)9 | GAGAGAGAGAGAGAGAGA | 18 | 1.05 | 0.71 |
| (GA)18 | GAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGA | 36 | 1.08 | 0.73 |
| (GA)27 | GAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGAGA | 54 | 1.09 | 0.74 |
| VK | GGGAGCGAGGGAGCGAGGGAGCGAGGGAGCG | 31 | 1.28 | 1.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trizna, L.; Osif, B.; Víglaský, V. G-QINDER Tool: Bioinformatically Predicted Formation of Different Four-Stranded DNA Motifs from (GT)n and (GA)n Repeats. Int. J. Mol. Sci. 2023, 24, 7565. https://doi.org/10.3390/ijms24087565
Trizna L, Osif B, Víglaský V. G-QINDER Tool: Bioinformatically Predicted Formation of Different Four-Stranded DNA Motifs from (GT)n and (GA)n Repeats. International Journal of Molecular Sciences. 2023; 24(8):7565. https://doi.org/10.3390/ijms24087565
Chicago/Turabian StyleTrizna, Lukáš, Branislav Osif, and Viktor Víglaský. 2023. "G-QINDER Tool: Bioinformatically Predicted Formation of Different Four-Stranded DNA Motifs from (GT)n and (GA)n Repeats" International Journal of Molecular Sciences 24, no. 8: 7565. https://doi.org/10.3390/ijms24087565
APA StyleTrizna, L., Osif, B., & Víglaský, V. (2023). G-QINDER Tool: Bioinformatically Predicted Formation of Different Four-Stranded DNA Motifs from (GT)n and (GA)n Repeats. International Journal of Molecular Sciences, 24(8), 7565. https://doi.org/10.3390/ijms24087565








