Association between Expression of Connective Tissue Genes and Prostate Cancer Growth and Progression
Abstract
1. Introduction
2. Results
2.1. Clinicopathological Characteristics of Patients
2.2. Decipher Risk Category and Postoperative Features (ECE, LN Invasion, ISUP and eBCR)
2.3. Gene Expression Correlation with Pejorative Outcomes
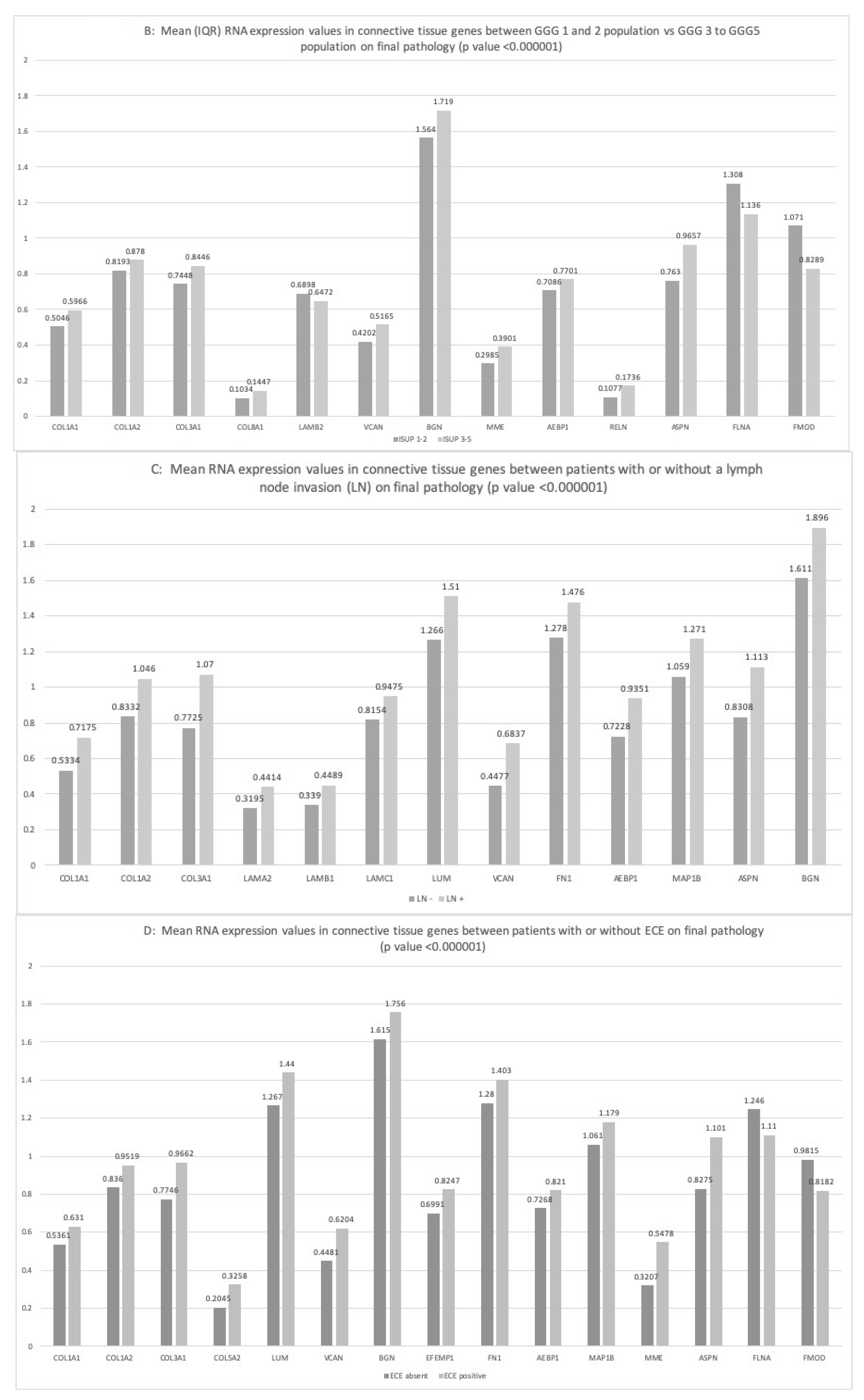
2.3.1. ISUP Score
2.3.2. Lymph Node Invasion
2.3.3. Extra-Capsular-Extension
2.3.4. Pathway Analysis of Our Connective Tissue Genes Selection
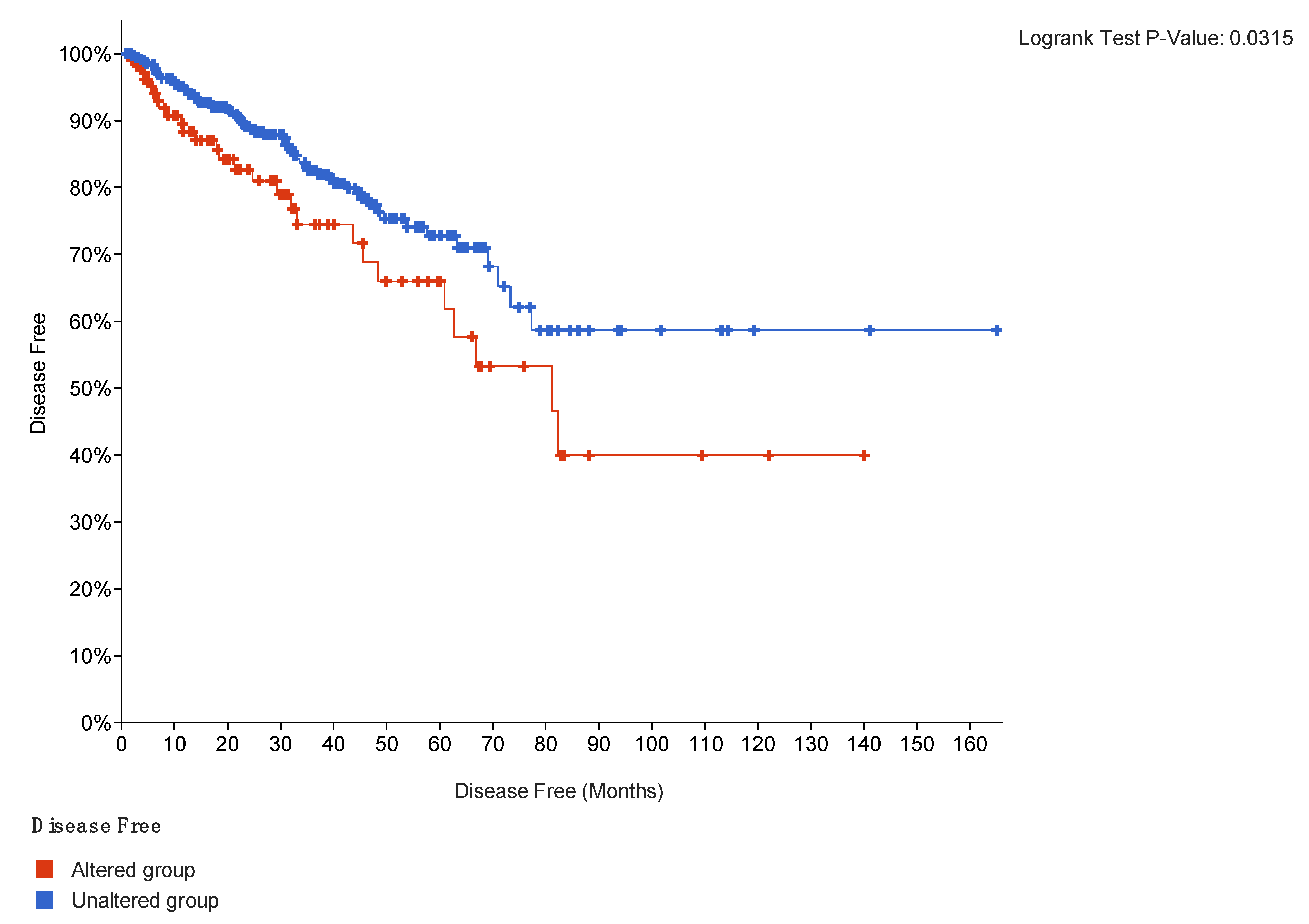
2.4. Outcomes Correlation
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. RNA Expression Analysis
4.3. Gene Selection
4.4. Correlation with Clinical Outcomes
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PSA | Prostate Specific Antigen |
| RNA | Ribonucleic Acid |
| BCR/eBCR | Biochemical Recurrence/early Biochemical Recurrence |
| TCGA | The Cancer Genome Atlas |
| ECE | Extra-capsular extension |
| ECM | Extra Cellular Matrix |
| DNA | Deoxyribonucleic Acid |
| MAPK | Mitogen Activated Protein Kinase |
| OS | Overall Survival |
| PFS | Progression-free Survival |
| ISUP | International Society of Urological Pathology |
| SCAN | Single Channel Array Normalization |
| LNI | Lymph Node Invasion |
| GGG | Gleason Group Grade |
| COL | Collagen |
| VCAN | Versican |
| VIM | vimentin |
| BGN | Biglycan |
| FN1 | Fibronectin |
| FLNA | Filamin A |
| LAM | Laminin |
| AEBP1 | AE binding protein |
| MME | Membrane Metalloendopeptidase |
| FMOD | Fibromodulin |
| LUM | Lumican |
| IQR | InterQuartile Range |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Schatten, H. Brief Overview of Prostate Cancer Statistics, Grading, Diagnosis and Treatment Strategies. In Cell & Molecular Biology of Prostate Cancer; Advances in Experimental Medicine and Biology; Springer: Cham, Swizerland, 2018; Volume 1095, pp. 1–14. [Google Scholar]
- Alford, A.V.; Brito, J.M.; Yadav, K.K.; Yadav, S.S.; Tewari, A.K.; Renzulli, J. The Use of Biomarkers in Prostate Cancer Screening and Treatment. Rev. Urol. 2017, 19, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Narayan, V.; Jiang, S.; Warlick, C.A.M. Early Stage Cancer in Older Adults: Prostate—Avoiding Overtreatment and Undertreatment. Cancer J. 2017, 23, 238–241. [Google Scholar] [CrossRef]
- Ayala, G.; Tuxhorn, J.A.; Wheeler, T.M.; Frolov, A.; Scardino, P.T.; Ohori, M.; Wheeler, M.; Spitler, J.; Rowley, D.R. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin. Cancer Res. 2003, 9, 4792–4801. [Google Scholar] [PubMed]
- Den, R.B.; Santiago-Jimenez, M.; Alter, J.; Schliekelman, M.; Wagner, J.R.; Ii, J.F.R.; Lee, D.I.; Brito, C.G.; Monahan, K.; Gburek, B.; et al. Decipher correlation patterns post prostatectomy: Initial experience from 2342 prospective patients. Prostate Cancer Prostatic Dis. 2016, 19, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Schauer, I.G.; Rowley, D.R. The functional role of reactive stroma in benign prostatic hyperplasia. Differentiation 2011, 82, 200–210. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, Q.; Li, A.; Huang, W.; Cai, Z.; Chen, W. Extracellular matrix protein 1 (ECM1) is associated with carcinogenesis potential of human bladder cancer. OncoTargets Ther. 2019, 12, 1423–1432. [Google Scholar] [CrossRef]
- Yang, F.; Tuxhorn, J.A.; Ressler, S.J.; McAlhany, S.J.; Dang, T.D.; Rowley, D.R. Stromal expression of connective tissue growth factor promotes angiogenesis and prostate cancer tumorigenesis. Cancer Res. 2005, 65, 8887–8895. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Knittel, J.G.; Yan, L.; Rueden, C.T.; White, J.G.; Keely, P.J. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008, 6, 11. [Google Scholar] [CrossRef]
- Tlsty, T.D.; Hein, P.W. Know thy neighbor: Stromal cells can contribute oncogenic signals. Curr. Opin. Genet. Dev. 2001, 11, 54–59. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, H.; Wang, J.; Zhou, L.; Liu, S. Collagen stiffness promoted non-muscle-invasive bladder cancer progression to muscle-invasive bladder cancer. OncoTargets Ther. 2019, 12, 3441–3457. [Google Scholar] [CrossRef] [PubMed]
- Ligorio, M.; Sil, S.; Malagon-Lopez, J.; Nieman, L.T.; Misale, S.; Di Pilato, M.; Ebright, R.Y.; Karabacak, M.N.; Kulkarni, A.S.; Liu, A.; et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. Cell 2019, 178, 160–175.e27. [Google Scholar] [CrossRef]
- Shuttleworth, R.; Trucu, D. Multiscale Modelling of Fibres Dynamics and Cell Adhesion within Moving Boundary Cancer Invasion. Bull. Math. Biol. 2019, 81, 2176–2219. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Livas, T.; Kyprianou, N. Anoikis and EMT: Lethal “Liaisons” during Cancer Progression. Crit. Rev. Oncog. 2016, 21, 155–168. [Google Scholar] [CrossRef]
- Zhu, X.; Assoian, R.K. Integrin-dependent activation of MAP kinase: A link to shape-dependent cell proliferation. Mol. Biol. Cell 1995, 6, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Schwarze, S.; Kyprianou, N. Anoikis disruption of focal adhesion-Akt signaling impairs renal cell carcinoma. Eur. Urol. 2011, 59, 734–744. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- Dalela, D.; Löppenberg, B.; Sood, A.; Sammon, J.; Abdollah, F. Contemporary Role of the Decipher® Test in Prostate Cancer Management: Current Practice and Future Perspectives. Rev. Urol. 2016, 18, 1–9. [Google Scholar]
- Shore, N.; Concepcion, R.; Saltzstein, D.; Lucia, M.S.; van Breda, A.; Welbourn, W.; Lewine, N.; Gustavsen, G.; Pothier, K.; Brawer, M.K. Clinical utility of a biopsy-based cell cycle gene expression assay in localized prostate cancer. Curr. Med. Res. Opin. 2013, 30, 547–553. [Google Scholar] [CrossRef]
- Nakagawa, T.; Kollmeyer, T.M.; Morlan, B.W.; Anderson, S.K.; Bergstralh, E.J.; Davis, B.; Asmann, Y.W.; Klee, G.G.; Ballman, K.; Jenkins, R.B. A tissue biomarker panel predicting systemic progression after PSA recurrence post-definitive prostate cancer therapy. PLoS ONE 2008, 3, e2318. [Google Scholar] [CrossRef]
- Tomas, D.; Spajić, B.; Milošević, M.; Demirović, A.; Marušić, Z.; Krušlin, B. Intensity of stromal changes predicts biochemical recurrence-free survival in prostatic carcinoma. Scand. J. Urol. Nephrol. 2010, 44, 284–290. [Google Scholar] [CrossRef] [PubMed]
- De Vivar, A.D.; Sayeeduddin, M.; Rowley, D.; Cubilla, A.; Miles, B.; Kadmon, D.; Ayala, G. Histologic features of stromogenic carcinoma of the prostate (carcinomas with reactive stroma grade 3). Hum. Pathol. 2017, 63, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Dakhova, O.; Ozen, M.; Creighton, C.J.; Li, R.; Ayala, G.; Rowley, D.; Ittmann, M. Global gene expression analysis of reactive stroma in prostate cancer. Clin. Cancer Res. 2009, 15, 3979–3989. [Google Scholar] [CrossRef]
- Miles, B.; Ittmann, M.; Wheeler, T.; Sayeeduddin, M.; Cubilla, A.; Rowley, D.; Bu, P.; Ding, Y.; Gao, Y.; Lee, M.; et al. Moving Beyond Gleason Scoring. Arch. Pathol. Lab. Med. 2019, 143, 565–570. [Google Scholar] [CrossRef]
- Gordetsky, J.; Epstein, J. Grading of prostatic adenocarcinoma: Current state and prognostic implications. Diagn. Pathol. 2016, 11, 25. [Google Scholar] [CrossRef]
- Pierorazio, P.M.; Walsh, P.C.; Partin, A.W.; Epstein, J.I. Prognostic Gleason grade grouping: Data based on the modified Gleason scoring system. BJU Int. 2013, 111, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Burdick, M.J.; Reddy, C.A.; Ulchaker, J.; Angermeier, K.; Altman, A.; Chehade, N.; Mahadevan, A.; Kupelian, P.A.; Klein, E.A.; Ciezki, J.P. Comparison of biochemical relapse-free survival between primary Gleason score 3 and primary Gleason score 4 for biopsy Gleason score 7 prostate cancer. Int. J. Radiat. Oncol. Biol. Phys. 2009, 73, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- LeBert, D.; Squirrell, J.M.; Freisinger, C.; Rindy, J.; Golenberg, N.; Frecentese, G.; Gibson, A.; Eliceiri, K.W.; Huttenlocher, A. Damage-induced reactive oxygen species regulate vimentin and dynamic collagen-based projections to mediate wound repair. eLife 2018, 16, 7. [Google Scholar] [CrossRef]
- Lai, C.-J.; Lin, C.-Y.; Liao, W.-Y.; Hour, T.-C.; Wang, H.-D.; Chuu, C.-P. CD44 Promotes Migration and Invasion of Docetaxel-Resistant Prostate Cancer Cells Likely via Induction of Hippo-Yap Signaling. Cells 2019, 8, 295. [Google Scholar] [CrossRef]
- Konac, E.; Kiliccioglu, I.; Sogutdelen, E.; Dikmen, A.U.; Albayrak, G.; Bilen, C.Y. Do the expressions of epithelial-mesenchymal transition proteins, periostin, integrin-α4 and fibronectin correlate with clinico-pathological features and prognosis of metastatic castration-resistant prostate cancer? Exp. Biol. Med. 2017, 242, 1795–1801. [Google Scholar] [CrossRef]
- Suh, Y.J.; Hall, M.S.; Huang, Y.L.; Moon, S.Y.; Song, W.; Ma, M.; Bonassar, L.J.; Segall, J.E.; Wu, M. Glycation of collagen matrices promotes breast tumor cell invasion. Integr. Biol. 2019, 11, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Li, C.; Feng, K.; Palmer, S.; Appleton, P.L.; Lang, S.; McGloin, D.; Huang, Z.; Nabi, G. Second harmonic generation (SHG) imaging of cancer heterogeneity in ultrasound guided biopsies of prostate in men suspected with prostate cancer. J. Biophotonics 2016, 10, 911–918. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.Y.; Koh, D.H.; Goh, H.J.; Kim, M.S.; Lee, J.S.; Jang, W.S.; Choi, Y.D. Clinical significance and predictors of oncologic outcome after radical prostatectomy for invisible prostate cancer on multiparametric MRI. BMC Cancer 2018, 18, 1057. [Google Scholar] [CrossRef] [PubMed]
- Ruppender, N.; Larson, S.; Lakely, B.; Kollath, L.; Brown, L.; Coleman, I.; Coleman, R.; Nguyen, H.; Nelson, P.S.; Corey, E.; et al. Cellular Adhesion Promotes Prostate Cancer Cells Escape from Dormancy. PLoS ONE 2015, 10, e0130565. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhong, J.; Zhao, Z.; Sheng, J.; Wang, J.; Liu, J.; Cui, K.; Chang, J.; Zhao, H.; Wong, S. Epithelial derived CTGF promotes breast tumor progression via inducing EMT and collagen I fibers deposition. Oncotarget 2015, 6, 25320–25338. [Google Scholar] [CrossRef]
- Chen, C.; Cao, F.-H.; Li, Z.-G.; Zhang, L.-G.; Liu, J.; Chen, N.; Yao, A.-L.; Kang, S.-S.; Gao, W.-X.; Han, H.; et al. Bioinformatics analysis of differentially expressed proteins in prostate cancer based on proteomics data. OncoTargets Ther. 2016, 9, 1545–1557. [Google Scholar] [CrossRef]
- Jia, Z.; Rahmatpanah, F.B.; Chen, X.; Lernhardt, W.; Wang, Y.; Xia, X.Q.; Sawyers, A.; Sutton, M.; McClelland, M.; Mercola, D. Expression changes in the stroma of prostate cancer predict subsequent relapse. PLoS ONE 2012, 7, e41371. [Google Scholar] [CrossRef]
- Cheong, M.-L.; Lai, T.-H.; Wu, W.-B. Connective tissue growth factor mediates transforming growth factor β-induced collagen expression in human endometrial stromal cells. PLoS ONE 2019, 14, e0210765. [Google Scholar] [CrossRef]
- Zhang, S.; Li, B.; Tang, W.; Ni, L.; Ma, H.; Lu, M.; Meng, Q. Effects of connective tissue growth factor on prostate cancer bone metastasis and osteoblast differentiation. Oncol. Lett. 2018, 16, 2305–2311. [Google Scholar] [CrossRef]
- Moran-Jones, K.; Gloss, B.S.; Murali, R.; Chang, D.K.; Colvin, E.K.; Jones, M.D.; Yuen, S.; Howell, V.M.; Brown, L.M.; Wong, C.W.; et al. Connective tissue growth factor as a novel therapeutic target in high grade serous ovarian cancer. Oncotarget 2015, 6, 44551–44562. [Google Scholar] [CrossRef]
- Lockstone, H.E. Exon array data analysis using Affymetrix power tools and R statistical software. Brief. Bioinform. 2011, 12, 634–644. [Google Scholar] [CrossRef] [PubMed]

| Covariates | Odds Ratio | 95% Minimal Confidence Limit | 95% Maximal Confidence Limit | p Value |
|---|---|---|---|---|
| ECE | 4.154 | 0.475 | 36.3 | 0.1978 |
| LN invasion | 0.865 | 0.061 | 12.256 | 0.9149 |
| ISUP 3 | 2.138 | 1.392 | 3.283 | 0.229 |
| ISUP 4 and 5 | 9.535 | 3.649 | 24.917 | 0.0001 |
| early BCR | 2.115 | 1.108 | 4.038 | 0.0231 |
| Queried Genes | COL1A1 | COL1A2 | COL3A1 | LUM | VCAN | FN1 | AEBP1 | MAP1B | ASPN | COMP | MME |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic alteration repartition | 3% | 4% | 4% | 5% | 3% | 5% | 5% | 5% | 4% | 4% | 4% |
| Genes | Co-Occurrence with | Number of Co-Occurrences | p Value |
|---|---|---|---|
| COL1A1 | COL1A2 | 32 | <0.01 |
| COL1A3 | 32 | ||
| AEBP1 | 21 | ||
| ASPN | 20 | ||
| COMP | 13 | ||
| LUM | 11 | ||
| COL1A2 | COL3A1 | 32 | <0.01 |
| AEBP1 | 22 | ||
| ASPN | 21 | ||
| VCAN | 15 | ||
| LUM | 13 | ||
| COMP | 13 | ||
| COL3A1 | ASPN | 20 | <0.01 |
| AEBP1 | 19 | ||
| VCAN | 15 | ||
| COMP | 13 | ||
| LUM | 10 | ||
| MAP1B | 6 | ||
| LUM | AEBP1 | 8 | 0.019 |
| VCAN | 7 | ||
| ASPN | 7 | ||
| VCAN | COMP | 11 | <0.01 |
| AEBP1 | 11 | ||
| ASPN | 10 |
| Gene Name | Protein | Function |
|---|---|---|
| COL1A1, COL1A2 | Collagen type 1 | Most abundant collagen, interstitial matrix component. |
| COL3A1 | Collagen type 3 | Fibrillar collagen group, mostly found in vascular and microvascular system. |
| VIM | Vimentin | Intermediate filament expressed in mesenchymal cells. |
| VCAN | Versican | Proteoglycan involved in cell adhesion. |
| BGN | Biglycan | Proteoglycan involved in collagen fibril assembly. |
| ASPN | Asporin | Extra-cellular matrix. |
| FN1 | Fibronectin | Extra-cellular matrix glycoprotein, binding to integrins. |
| LAMA4, LAMB1, LAMC1 | Laminin | Major component of the basal lamina (basement membrane). |
| AEBP1 | AE binding protein | Role in adipogenesis and smooth muscle differentiation. |
| FLNA | Filamin | Actin filaments link to membrane glycoproteins. |
| MME | Membrane Metalloendopeptidase | Type 2 transmembrane glycoprotein. |
| FMOD | Fibromodulin | Fibrillogenesis inhibition of collagen type 1 and 2. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Treacy, P.-J.; Martini, A.; Falagario, U.G.; Ratnani, P.; Wajswol, E.; Beksac, A.T.; Wiklund, P.; Nair, S.; Kyprianou, N.; Durand, M.; et al. Association between Expression of Connective Tissue Genes and Prostate Cancer Growth and Progression. Int. J. Mol. Sci. 2023, 24, 7520. https://doi.org/10.3390/ijms24087520
Treacy P-J, Martini A, Falagario UG, Ratnani P, Wajswol E, Beksac AT, Wiklund P, Nair S, Kyprianou N, Durand M, et al. Association between Expression of Connective Tissue Genes and Prostate Cancer Growth and Progression. International Journal of Molecular Sciences. 2023; 24(8):7520. https://doi.org/10.3390/ijms24087520
Chicago/Turabian StyleTreacy, Patrick-Julien, Alberto Martini, Ugo Giovanni Falagario, Parita Ratnani, Ethan Wajswol, Alp Tuna Beksac, Peter Wiklund, Sujit Nair, Natasha Kyprianou, Matthieu Durand, and et al. 2023. "Association between Expression of Connective Tissue Genes and Prostate Cancer Growth and Progression" International Journal of Molecular Sciences 24, no. 8: 7520. https://doi.org/10.3390/ijms24087520
APA StyleTreacy, P.-J., Martini, A., Falagario, U. G., Ratnani, P., Wajswol, E., Beksac, A. T., Wiklund, P., Nair, S., Kyprianou, N., Durand, M., & Tewari, A. K. (2023). Association between Expression of Connective Tissue Genes and Prostate Cancer Growth and Progression. International Journal of Molecular Sciences, 24(8), 7520. https://doi.org/10.3390/ijms24087520








