CCL2-Mediated Stromal Interactions Drive Macrophage Polarization to Increase Breast Tumorigenesis
Abstract
1. Introduction
2. Results
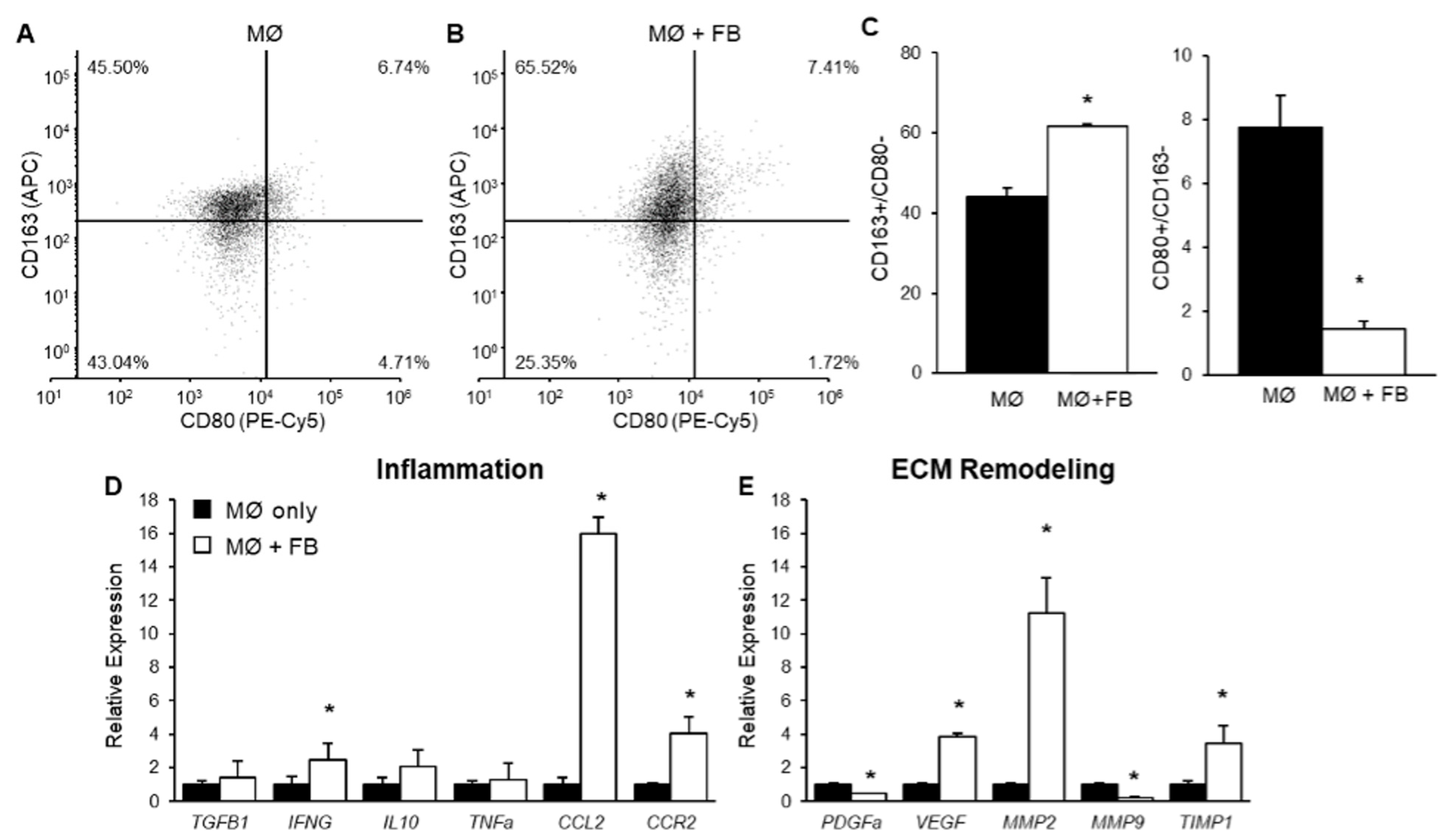
2.1. The Effect of Mammary Fibroblasts on THP-1-Derived Macrophage Phenotype in Co-Culture
2.2. The Effect of Mammary Fibroblasts on THP-1 Derived Macrophage Gene Expression
2.3. The Effect of THP-1 Derived Macrophages on Mammary Fibroblast Gene Expression in Co-Culture
2.4. The Effect of CCL2 on Mammary Fibroblast Collagen Production in Co-Culture
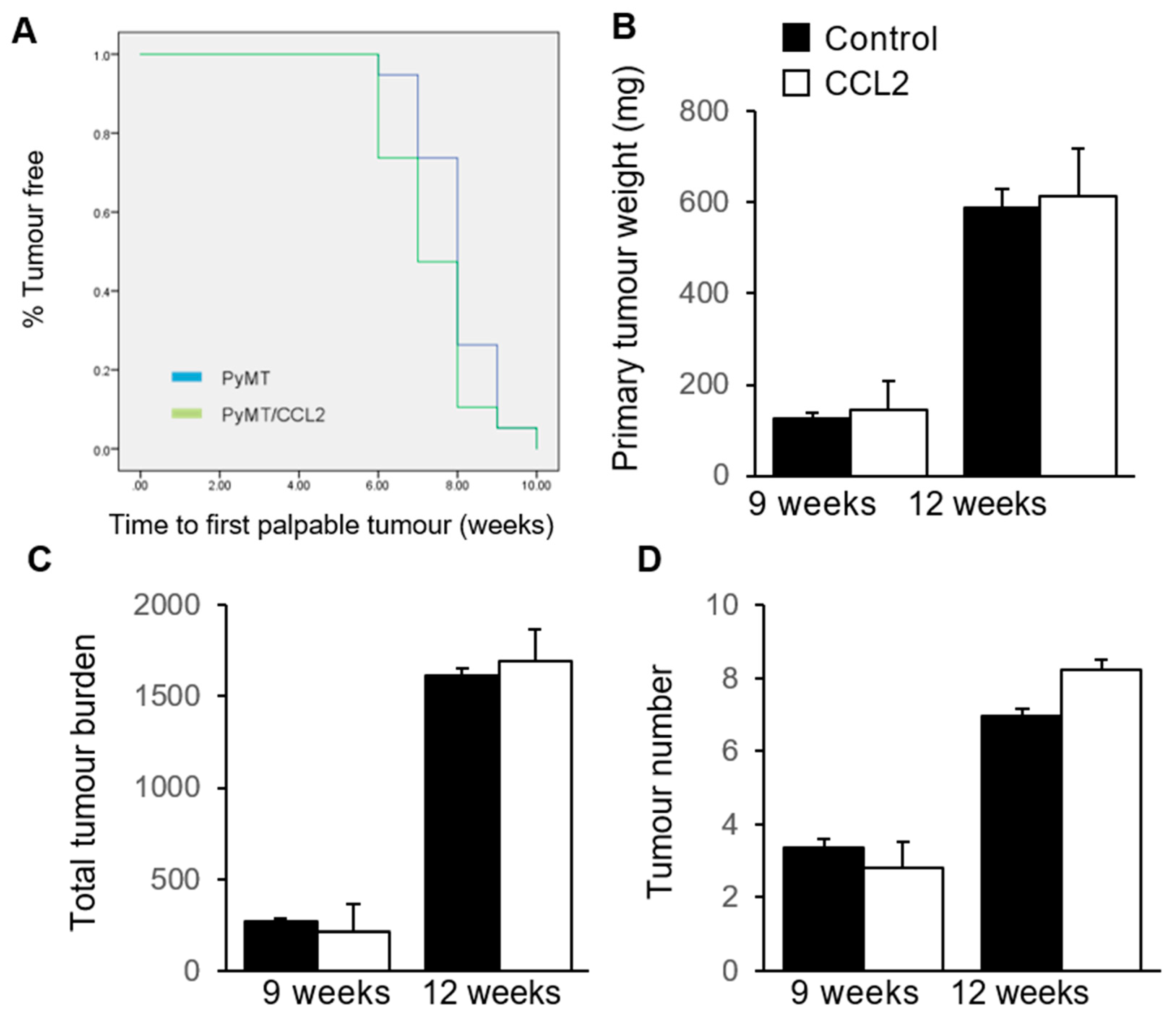
2.5. The Effect of CCL2 on Tumour Development in PyMT Mammary Tumour Models
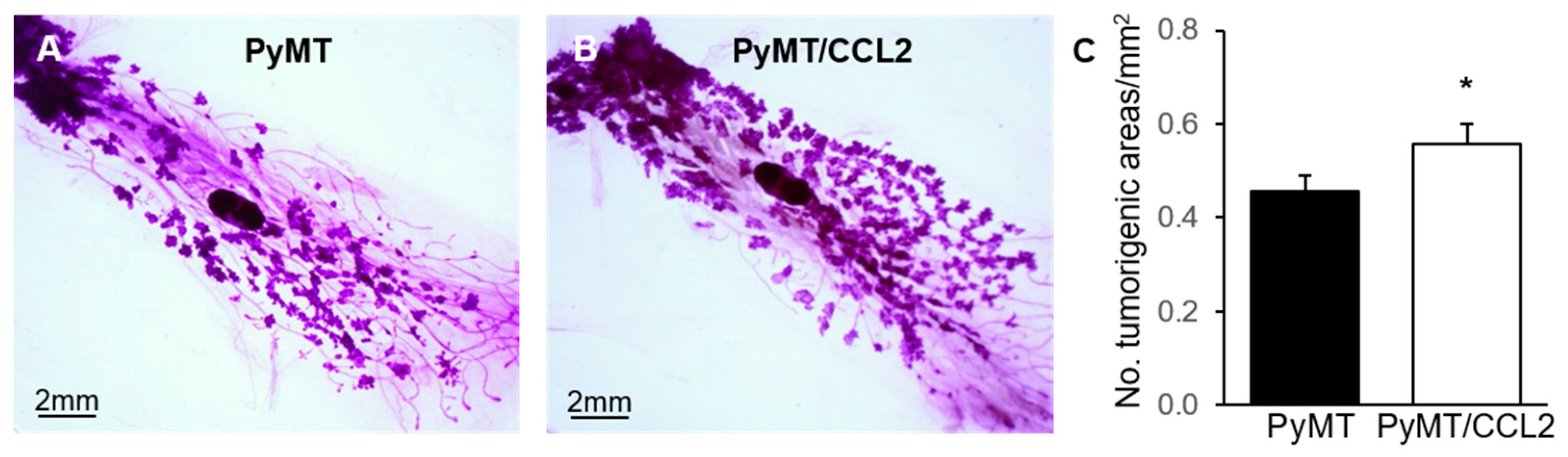
2.6. The Effect of CCL2 on Early Tumourigenesis in 9 Week Old PyMT Mice
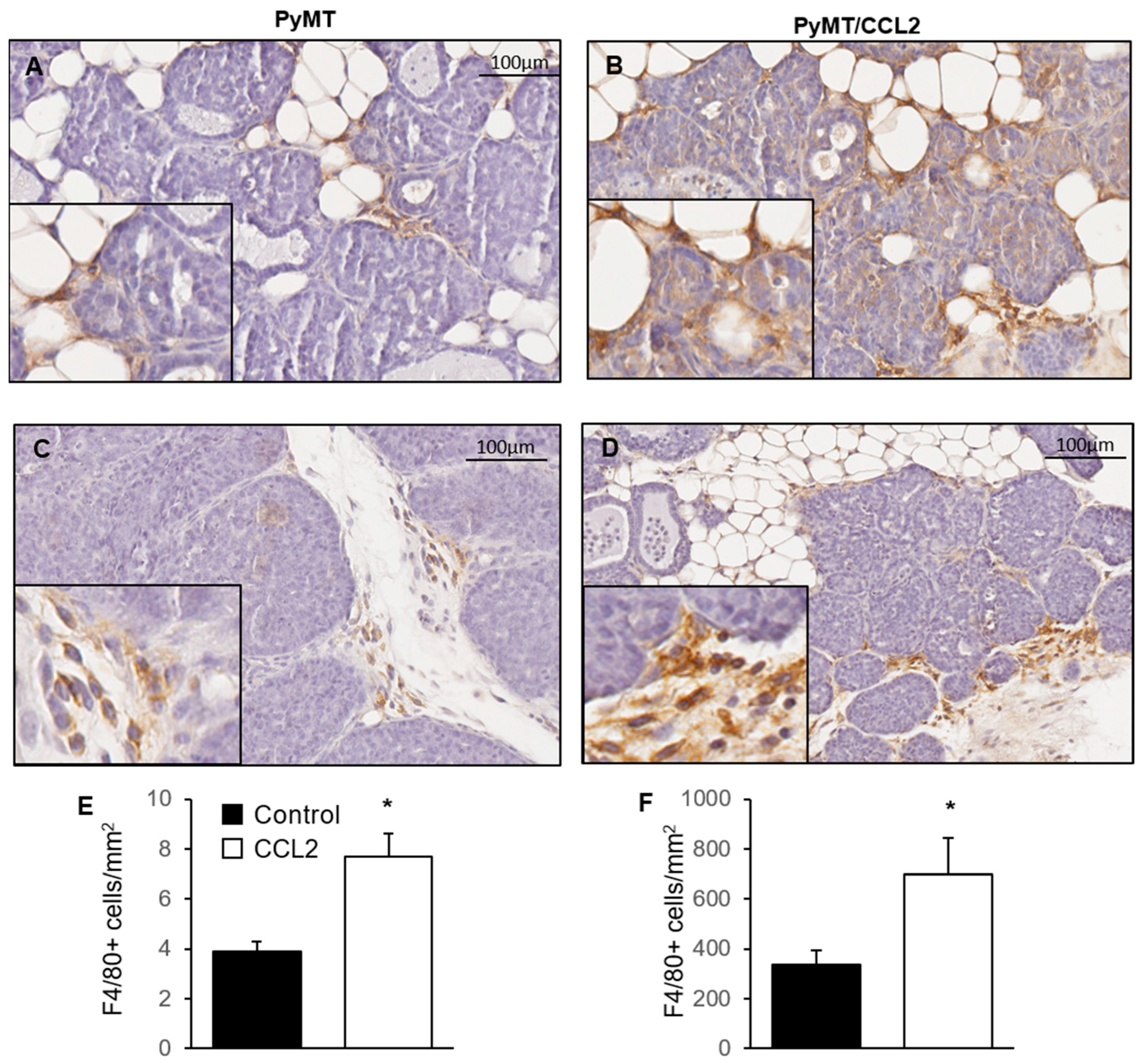
2.7. The Effect of CCL2 on Macrophage Infiltration of Mammary Tumours
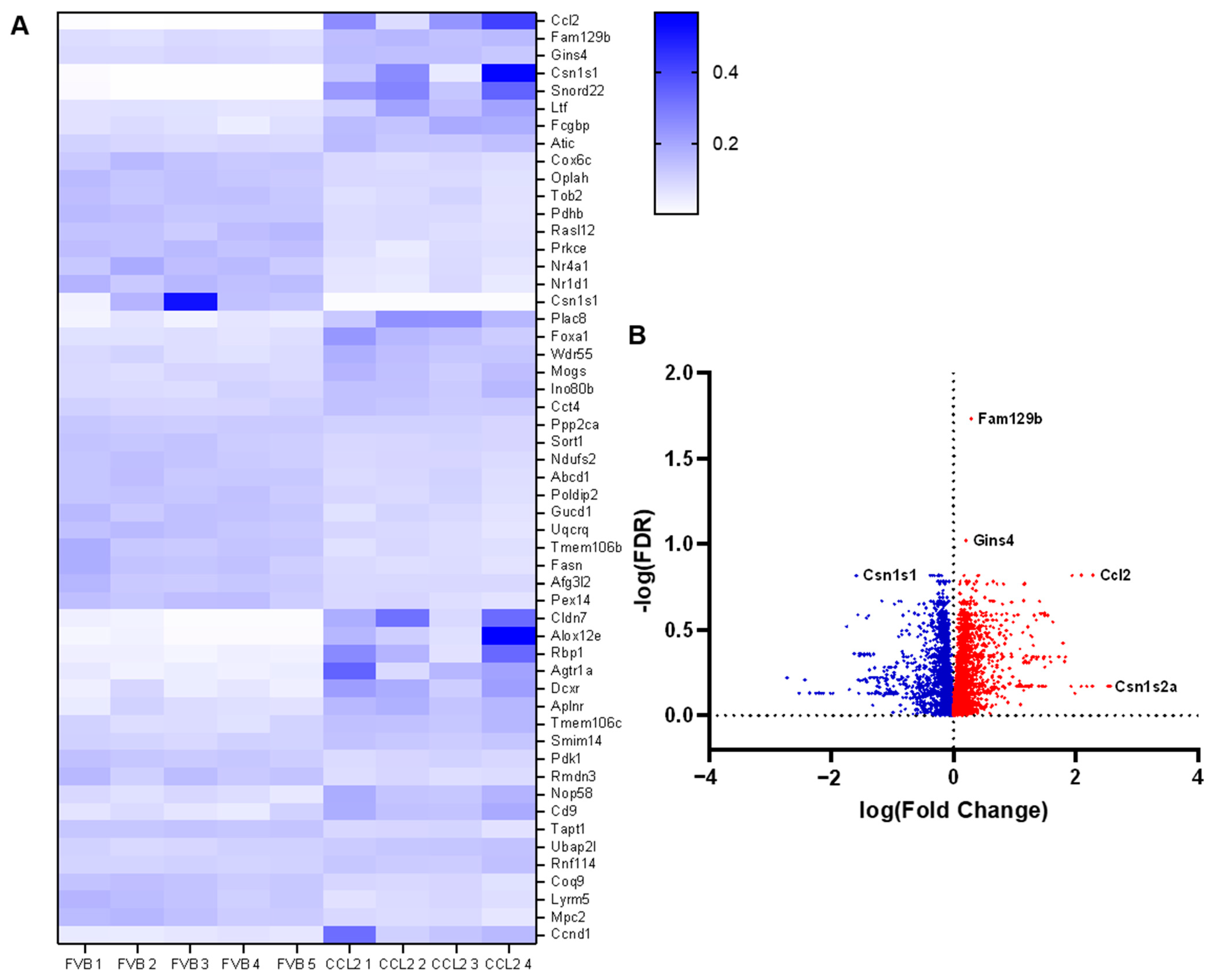
2.8. The Effect of CCL2 on Global Gene Expression in the Mammary Gland
3. Discussion
3.1. The Effect of CCL2 on ECM Regulation and Mammographic Density
3.2. Mammary Fibroblasts Drive Differentiation of Macrophages
3.3. Crosstalk between M2 Polarized Macrophages and Mammary Fibroblasts
3.4. The Role of CCL2 in Tumour Development and Progression
3.5. Constitutive CCL2 Expression Downregulates Fatty Acid Synthesis and Metabolism
3.6. Limitations
4. Materials and Methods
4.1. Isolation and Culture of Human Primary Mammary Fibroblasts
4.2. Culture of THP-1 Monocytes
4.3. Indirect Co-Culture of Human Mammary Fibroblasts and Differentiated THP-1 Cells
4.4. Enzyme-Linked Immunosorbent Assay
4.5. Sirius Red Analysis of Insoluble Collagen
4.6. Flow Cytometry of THP-1 Macrophages
4.7. Mice
4.7.1. Mmtv-Ccl2 Mice
4.7.2. MMTV-PyMT Mice
4.8. Generation of PyMT/CCL2 Cohort
4.9. Tumour Detection by Palpation
4.10. Histology and Immunohistochemistry
4.10.1. Tissue Collection, Embedding and Sectioning of Mouse Tissues
4.10.2. Mammary Gland Whole Mount Preparation
4.10.3. Haematoxylin and Eosin Staining
4.10.4. Immunohistochemistry to Detect F4/80
4.11. Image Capture and Quantification
4.11.1. Haematoxylin and Eosin Stained Mouse Tissues
4.11.2. Whole Mounted Mammary Glands
4.11.3. F4/80 Quantification in Mouse Tumours
4.11.4. Genotyping Mice
4.11.5. RNA Extraction from Frozen Tissues
4.11.6. Next Generation Sequencing of Mmtv-Ccl2 Mammary Glands
4.12. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deshmane, S.L.; Kremlev, S.; Amini, S.; Sawaya, B.E. Monocyte chemoattractant protein-1 (mcp-1): An overview. J. Interferon Cytokine Res. 2009, 29, 313–326. [Google Scholar] [CrossRef]
- Beall, C.J.; Mahajan, S.; Kuhn, D.E.; Kolattukudy, P.E. Site-directed mutagenesis of monocyte chemoattractant protein-1 identifies two regions of the polypeptide essential for biological activity. Biochem. J. 1996, 313 Pt 2, 633–640. [Google Scholar] [CrossRef]
- Cushing, S.D.; Berliner, J.A.; Valente, A.J.; Territo, M.C.; Navab, M.; Parhami, F.; Gerrity, R.; Schwartz, C.J.; Fogelman, A.M. Minimally modified low density lipoprotein induces monocyte chemotactic protein 1 in human endothelial cells and smooth muscle cells. Proc. Natl. Acad. Sci. USA 1990, 87, 5134–5138. [Google Scholar] [CrossRef] [PubMed]
- Standiford, T.J.; Kunkel, S.L.; Phan, S.H.; Rollins, B.J.; Strieter, R.M. Alveolar macrophage-derived cytokines induce monocyte chemoattractant protein-1 expression from human pulmonary type ii-like epithelial cells. J. Biol. Chem. 1991, 266, 9912–9918. [Google Scholar] [CrossRef]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. Ccl2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, H.; Sangai, T.; Ishii, G.; Ikehara, A.; Nagashima, T.; Miyazaki, M.; Ochiai, A. Stromal mcp-1 in mammary tumors induces tumor-associated macrophage infiltration and contributes to tumor progression. Int. J. Cancer 2009, 125, 1276–1284. [Google Scholar] [CrossRef]
- Soria, G.; Ben-Baruch, A. The inflammatory chemokines ccl2 and ccl5 in breast cancer. Cancer Lett. 2008, 267, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Ueno, T.; Toi, M.; Saji, H.; Muta, M.; Bando, H.; Kuroi, K.; Koike, M.; Inadera, H.; Matsushima, K. Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin. Cancer Res. 2000, 6, 3282–3289. [Google Scholar]
- Archer, M.; Dasari, P.; Evdokiou, A.; Ingman, W.V. Biological mechanisms and therapeutic opportunities in mammographic density and breast cancer risk. Cancers 2021, 13, 5391. [Google Scholar] [CrossRef]
- Huo, C.W.; Chew, G.L.; Britt, K.L.; Ingman, W.V.; Henderson, M.A.; Hopper, J.L.; Thompson, E.W. Mammographic density-a review on the current understanding of its association with breast cancer. Breast Cancer Res. Treat. 2014, 144, 479–502. [Google Scholar] [CrossRef]
- Sun, X.; Glynn, D.J.; Hodson, L.J.; Huo, C.; Britt, K.; Thompson, E.W.; Woolford, L.; Evdokiou, A.; Pollard, J.W.; Robertson, S.A.; et al. Ccl2-driven inflammation increases mammary gland stromal density and cancer susceptibility in a transgenic mouse model. Breast Cancer Res. 2017, 19, 4. [Google Scholar] [CrossRef] [PubMed]
- Huo, C.W.; Chew, G.; Hill, P.; Huang, D.; Ingman, W.; Hodson, L.; Brown, K.A.; Magenau, A.; Allam, A.H.; McGhee, E.; et al. High mammographic density is associated with an increase in stromal collagen and immune cells within the mammary epithelium. Breast Cancer Res. 2015, 17, 79. [Google Scholar] [CrossRef] [PubMed]
- Ghadge, A.G.; Dasari, P.; Stone, J.; Thompson, E.W.; Robker, R.L.; Ingman, W.V. Pubertal mammary gland development is a key determinant of adult mammographic density. Semin. Cell Dev. Biol. 2021, 114, 143–158. [Google Scholar] [CrossRef]
- Archer, M.; Dasari, P.; Walsh, D.; Britt, K.L.; Evdokiou, A.; Ingman, W.V. Immune regulation of mammary fibroblasts and the impact of mammographic density. J. Clin. Med. 2022, 11, 799. [Google Scholar] [CrossRef]
- Huo, C.W.; Hill, P.; Chew, G.; Neeson, P.J.; Halse, H.; Williams, E.D.; Henderson, M.A.; Thompson, E.W.; Britt, K.L. High mammographic density in women is associated with protumor inflammation. Breast Cancer Res. 2018, 20, 92. [Google Scholar] [CrossRef]
- Dasari, P.; Sharkey, D.J.; Noordin, E.; Glynn, D.J.; Hodson, L.J.; Chin, P.Y.; Evdokiou, A.; Robertson, S.A.; Ingman, W.V. Hormonal regulation of the cytokine microenvironment in the mammary gland. J. Reprod. Immunol. 2014, 106, 58–66. [Google Scholar] [CrossRef]
- Hodson, L.J.; Chua, A.C.; Evdokiou, A.; Robertson, S.A.; Ingman, W.V. Macrophage phenotype in the mammary gland fluctuates over the course of the estrous cycle and is regulated by ovarian steroid hormones. Biol. Reprod. 2013, 89, 65. [Google Scholar] [CrossRef]
- Need, E.F.; Atashgaran, V.; Ingman, W.V.; Dasari, P. Hormonal regulation of the immune microenvironment in the mammary gland. J. Mammary Gland Biol. Neoplasia 2014, 19, 229–239. [Google Scholar] [CrossRef]
- Chua, A.C.; Hodson, L.J.; Moldenhauer, L.M.; Robertson, S.A.; Ingman, W.V. Dual roles for macrophages in ovarian cycle-associated development and remodelling of the mammary gland epithelium. Development 2010, 137, 4229–4238. [Google Scholar] [CrossRef] [PubMed]
- Ingman, W.V.; Wyckoff, J.; Gouon-Evans, V.; Condeelis, J.; Pollard, J.W. Macrophages promote collagen fibrillogenesis around terminal end buds of the developing mammary gland. Dev. Dyn. 2006, 235, 3222–3229. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, S.; Yasuda, K.; Suzuki, K.; Tahara, K.; Higashi, H.; Era, S. Macrophage infiltration and its prognostic implications in breast cancer: The relationship with vegf expression and microvessel density. Oncol. Rep. 2005, 14, 425–431. [Google Scholar] [CrossRef]
- Lewis, C.E.; Pollard, J.W. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006, 66, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Bernhardt, S.M.; Glynn, D.J.; Hodson, L.J.; Woolford, L.; Evdokiou, A.; Yan, C.; Du, H.; Robertson, S.A.; Ingman, W.V. Attenuated tgfb signalling in macrophages decreases susceptibility to dmba-induced mammary cancer in mice. Breast Cancer Res. 2021, 23, 39. [Google Scholar] [CrossRef] [PubMed]
- Bates, J.P.; Derakhshandeh, R.; Jones, L.; Webb, T.J. Mechanisms of immune evasion in breast cancer. BMC Cancer 2018, 18, 556. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ingman, W.V. Cytokine networks that mediate epithelial cell-macrophage crosstalk in the mammary gland: Implications for development and cancer. J. Mammary Gland Biol. Neoplasia 2014, 19, 191–201. [Google Scholar] [CrossRef]
- Mosser, D.M.; Hamidzadeh, K.; Goncalves, R. Macrophages and the maintenance of homeostasis. Cell Mol. Immunol. 2021, 18, 579–587. [Google Scholar] [CrossRef]
- Li, J.; Yang, J.; Xing, R.; Wang, Y. A novel inflammation-related signature for predicting prognosis and characterizing the tumor microenvironment in colorectal cancer. Aging 2023, 15. [Google Scholar] [CrossRef]
- Mantovani, A.; Biswas, S.K.; Galdiero, M.R.; Sica, A.; Locati, M. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013, 229, 176–185. [Google Scholar] [CrossRef]
- Braga, T.T.; Agudelo, J.S.; Camara, N.O. Macrophages during the fibrotic process: M2 as friend and foe. Front. Immunol. 2015, 6, 602. [Google Scholar] [CrossRef]
- Ploeger, D.T.; Hosper, N.A.; Schipper, M.; Koerts, J.A.; de Rond, S.; Bank, R.A. Cell plasticity in wound healing: Paracrine factors of m1/ m2 polarized macrophages influence the phenotypical state of dermal fibroblasts. Cell Commun. Signal. 2013, 11, 29. [Google Scholar] [CrossRef]
- Song, E.; Ouyang, N.; Horbelt, M.; Antus, B.; Wang, M.; Exton, M.S. Influence of alternatively and classically activated macrophages on fibrogenic activities of human fibroblasts. Cell Immunol. 2000, 204, 19–28. [Google Scholar] [CrossRef]
- Zhu, Z.; Ding, J.; Ma, Z.; Iwashina, T.; Tredget, E.E. Alternatively activated macrophages derived from thp-1 cells promote the fibrogenic activities of human dermal fibroblasts. Wound Repair Regen. 2017, 25, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Gok Yavuz, B.; Gunaydin, G.; Gedik, M.E.; Kosemehmetoglu, K.; Karakoc, D.; Ozgur, F.; Guc, D. Cancer associated fibroblasts sculpt tumour microenvironment by recruiting monocytes and inducing immunosuppressive pd-1+ tams. Sci. Rep. 2019, 9, 3172. [Google Scholar] [CrossRef] [PubMed]
- Ronca, R.; Van Ginderachter, J.A.; Turtoi, A. Paracrine interactions of cancer-associated fibroblasts, macrophages and endothelial cells: Tumor allies and foes. Curr. Opin. Oncol. 2018, 30, 45–53. [Google Scholar] [CrossRef]
- Zhang, A.; Qian, Y.; Ye, Z.; Chen, H.; Xie, H.; Zhou, L.; Shen, Y.; Zheng, S. Cancer-associated fibroblasts promote m2 polarization of macrophages in pancreatic ductal adenocarcinoma. Cancer Med. 2017, 6, 463–470. [Google Scholar] [CrossRef]
- LeBleu, V.S.; Kalluri, R. A peek into cancer-associated fibroblasts: Origins, functions and translational impact. Dis. Model. Mech. 2018, 11, dmm029447. [Google Scholar] [CrossRef]
- Ireland, L.V.; Mielgo, A. Macrophages and fibroblasts, key players in cancer chemoresistance. Front. Cell Dev. Biol. 2018, 6, 131. [Google Scholar] [CrossRef] [PubMed]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T.; Sahebkar, A. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Porta, C.; Riboldi, E.; Ippolito, A.; Sica, A. Molecular and epigenetic basis of macrophage polarized activation. Semin. Immunol. 2015, 27, 237–248. [Google Scholar] [CrossRef]
- Boutilier, A.J.; Elsawa, S.F. Macrophage polarization states in the tumor microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.Y.; Jones, J.G.; Li, P.; Zhu, L.; Whitney, K.D.; Muller, W.J.; Pollard, J.W. Progression to malignancy in the polyoma middle t oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003, 163, 2113–2126. [Google Scholar] [CrossRef]
- Rose, C.E., Jr.; Sung, S.S.; Fu, S.M. Significant involvement of ccl2 (mcp-1) in inflammatory disorders of the lung. Microcirculation 2003, 10, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Antoniades, C.G.; Quaglia, A.; Taams, L.S.; Mitry, R.R.; Hussain, M.; Abeles, R.; Possamai, L.A.; Bruce, M.; McPhail, M.; Starling, C.; et al. Source and characterization of hepatic macrophages in acetaminophen-induced acute liver failure in humans. Hepatology 2012, 56, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Antoniades, H.N.; Neville-Golden, J.; Galanopoulos, T.; Kradin, R.L.; Valente, A.J.; Graves, D.T. Expression of monocyte chemoattractant protein 1 mrna in human idiopathic pulmonary fibrosis. Proc. Natl. Acad. Sci. USA 1992, 89, 5371–5375. [Google Scholar] [CrossRef] [PubMed]
- Kalderen, C.; Stadler, C.; Forsgren, M.; Kvastad, L.; Johansson, E.; Sydow-Backman, M.; Svensson Gelius, S. Ccl2 mediates anti-fibrotic effects in human fibroblasts independently of ccr2. Int. Immunopharmacol. 2014, 20, 66–73. [Google Scholar] [CrossRef]
- Mitchell, C.; Couton, D.; Couty, J.P.; Anson, M.; Crain, A.M.; Bizet, V.; Renia, L.; Pol, S.; Mallet, V.; Gilgenkrantz, H. Dual role of ccr2 in the constitution and the resolution of liver fibrosis in mice. Am. J. Pathol. 2009, 174, 1766–1775. [Google Scholar] [CrossRef]
- Distler, O.; Pap, T.; Kowal-Bielecka, O.; Meyringer, R.; Guiducci, S.; Landthaler, M.; Scholmerich, J.; Michel, B.A.; Gay, R.E.; Matucci-Cerinic, M.; et al. Overexpression of monocyte chemoattractant protein 1 in systemic sclerosis: Role of platelet-derived growth factor and effects on monocyte chemotaxis and collagen synthesis. Arthritis Rheum. 2001, 44, 2665–2678. [Google Scholar] [CrossRef]
- Locati, M.; Mantovani, A.; Sica, A. Macrophage activation and polarization as an adaptive component of innate immunity. Adv. Immunol. 2013, 120, 163–184. [Google Scholar]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized m2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Roszer, T. Understanding the mysterious m2 macrophage through activation markers and effector mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef]
- Ferrer, R.A.; Saalbach, A.; Grunwedel, M.; Lohmann, N.; Forstreuter, I.; Saupe, S.; Wandel, E.; Simon, J.C.; Franz, S. Dermal fibroblasts promote alternative macrophage activation improving impaired wound healing. J. Investig. Dermatol. 2017, 137, 941–950. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Jetten, N.; Verbruggen, S.; Gijbels, M.J.; Post, M.J.; De Winther, M.P.; Donners, M.M. Anti-inflammatory m2, but not pro-inflammatory m1 macrophages promote angiogenesis in vivo. Angiogenesis 2014, 17, 109–118. [Google Scholar] [CrossRef]
- Murray, P.J. Macrophage polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- Tang, X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 2013, 332, 3–10. [Google Scholar] [CrossRef]
- Mukhtar, R.A.; Nseyo, O.; Campbell, M.J.; Esserman, L.J. Tumor-associated macrophages in breast cancer as potential biomarkers for new treatments and diagnostics. Expert Rev. Mol. Diagn. 2011, 11, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Medrek, C.; Ponten, F.; Jirstrom, K.; Leandersson, K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012, 12, 306. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Li, Q.; Shi, J.; Wei, J.; Li, P.; Chang, C.H.; Shultz, L.D.; Ren, G. Lung fibroblasts facilitate pre-metastatic niche formation by remodeling the local immune microenvironment. Immunity 2022, 55, 1483–1500.e9. [Google Scholar] [CrossRef]
- Li, M.; Knight, D.A.; Li, A.S.; Smyth, M.J.; Stewart, T.J. A role for ccl2 in both tumor progression and immunosurveillance. Oncoimmunology 2013, 2, e25474. [Google Scholar] [CrossRef]
- Nesbit, M.; Schaider, H.; Miller, T.H.; Herlyn, M. Low-level monocyte chemoattractant protein-1 stimulation of monocytes leads to tumor formation in nontumorigenic melanoma cells. J. Immunol. 2001, 166, 6483–6490. [Google Scholar] [CrossRef]
- Nakasone, Y.; Fujimoto, M.; Matsushita, T.; Hamaguchi, Y.; Huu, D.L.; Yanaba, M.; Sato, S.; Takehara, K.; Hasegawa, M. Host-derived mcp-1 and mip-1alpha regulate protective anti-tumor immunity to localized and metastatic b16 melanoma. Am. J. Pathol. 2012, 180, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Lei, Z.; Zhao, J.; Gong, W.; Liu, J.; Chen, Z.; Liu, Y.; Li, D.; Yuan, Y.; Zhang, G.M.; et al. Ccl2/ccr2 pathway mediates recruitment of myeloid suppressor cells to cancers. Cancer Lett. 2007, 252, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Allavena, P.; Sica, A.; Garlanda, C.; Mantovani, A. The yin-yang of tumor-associated macrophages in neoplastic progression and immune surveillance. Immunol. Rev. 2008, 222, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Cedile, O.; Wlodarczyk, A.; Owens, T. Ccl2 recruits t cells into the brain in a ccr2-independent manner. APMIS 2017, 125, 945–956. [Google Scholar] [CrossRef]
- Flaishon, L.; Becker-Herman, S.; Hart, G.; Levo, Y.; Kuziel, W.A.; Shachar, I. Expression of the chemokine receptor ccr2 on immature b cells negatively regulates their cytoskeletal rearrangement and migration. Blood 2004, 104, 933–941. [Google Scholar] [CrossRef]
- Lanca, T.; Costa, M.F.; Goncalves-Sousa, N.; Rei, M.; Grosso, A.R.; Penido, C.; Silva-Santos, B. Protective role of the inflammatory ccr2/ccl2 chemokine pathway through recruitment of type 1 cytotoxic gammadelta t lymphocytes to tumor beds. J. Immunol. 2013, 190, 6673–6680. [Google Scholar] [CrossRef]
- Penido, C.; Costa, M.F.; Souza, M.C.; Costa, K.A.; Candea, A.L.; Benjamim, C.F.; Henriques, M. Involvement of cc chemokines in gammadelta t lymphocyte trafficking during allergic inflammation: The role of ccl2/ccr2 pathway. Int. Immunol. 2008, 20, 129–139. [Google Scholar] [CrossRef]
- Zysk, A.; DeNichilo, M.O.; Panagopoulos, V.; Zinonos, I.; Liapis, V.; Hay, S.; Ingman, W.; Ponomarev, V.; Atkins, G.; Findlay, D.; et al. Adoptive transfer of ex vivo expanded vγ9vδ2 t cells in combination with zoledronic acid inhibits cancer growth and limits osteolysis in a murine model of osteolytic breast cancer. Cancer Lett. 2017, 386, 141–150. [Google Scholar] [CrossRef]
- Ahmad, R.; Al-Roub, A.; Kochumon, S.; Akther, N.; Thomas, R.; Kumari, M.; Koshy, M.S.; Tiss, A.; Hannun, Y.A.; Tuomilehto, J.; et al. The synergy between palmitate and tnf-α for ccl2 production is dependent on the trif/irf3 pathway: Implications for metabolic inflammation. J. Immunol. 2018, 200, 3599–3611. [Google Scholar] [CrossRef]
- Haukeland, J.W.; Damas, J.K.; Konopski, Z.; Loberg, E.M.; Haaland, T.; Goverud, I.; Torjesen, P.A.; Birkeland, K.; Bjoro, K.; Aukrust, P. Systemic inflammation in nonalcoholic fatty liver disease is characterized by elevated levels of ccl2. J. Hepatol. 2006, 44, 1167–1174. [Google Scholar] [CrossRef]
- Roh, Y.S.; Seki, E. Chemokines and chemokine receptors in the development of nafld. Adv. Exp. Med. Biol. 2018, 1061, 45–53. [Google Scholar] [PubMed]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef]
- Kinlaw, W.B.; Baures, P.W.; Lupien, L.E.; Davis, W.L.; Kuemmerle, N.B. Fatty acids and breast cancer: Make them on site or have them delivered. J. Cell Physiol. 2016, 231, 2128–2141. [Google Scholar] [CrossRef] [PubMed]
- Monaco, M.E. Fatty acid metabolism in breast cancer subtypes. Oncotarget 2017, 8, 29487–29500. [Google Scholar] [CrossRef]
- Hilvo, M.; Matej Orešiè, A. Regulation of lipid metabolism in breast cancer provides diagnostic and therapeutic opportunities. Clin. Lipidol. 2012, 7, 177–188. [Google Scholar] [CrossRef]
- Milgraum, L.Z.; Witters, L.A.; Pasternack, G.R.; Kuhajda, F.P. Enzymes of the fatty acid synthesis pathway are highly expressed in in situ breast carcinoma. Clin. Cancer Res. 1997, 3, 2115–2120. [Google Scholar]
- Alo, P.L.; Visca, P.; Marci, A.; Mangoni, A.; Botti, C.; Di Tondo, U. Expression of fatty acid synthase (fas) as a predictor of recurrence in stage i breast carcinoma patients. Cancer 1996, 77, 474–482. [Google Scholar] [CrossRef]
- Alo, P.L.; Visca, P.; Trombetta, G.; Mangoni, A.; Lenti, L.; Monaco, S.; Botti, C.; Serpieri, D.E.; Di Tondo, U. Fatty acid synthase (fas) predictive strength in poorly differentiated early breast carcinomas. Tumori 1999, 85, 35–40. [Google Scholar] [CrossRef]
- Yang, Y.; Morin, P.J.; Han, W.F.; Chen, T.; Bornman, D.M.; Gabrielson, E.W.; Pizer, E.S. Regulation of fatty acid synthase expression in breast cancer by sterol regulatory element binding protein-1c. Exp. Cell Res. 2003, 282, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Suburu, J.; Shi, L.; Wu, J.; Wang, S.; Samuel, M.; Thomas, M.J.; Kock, N.D.; Yang, G.; Kridel, S.; Chen, Y.Q. Fatty acid synthase is required for mammary gland development and milk production during lactation. Am. J. Physiol. Endocrinol. Metab. 2014, 306, E1132–E1143. [Google Scholar] [CrossRef]
- Guy, C.T.; Cardiff, R.D.; Muller, W.J. Induction of mammary tumors by expression of polyomavirus middle t oncogene: A transgenic mouse model for metastatic disease. Mol. Cell Biol. 1992, 12, 954–961. [Google Scholar] [PubMed]
- Davie, S.A.; Maglione, J.E.; Manner, C.K.; Young, D.; Cardiff, R.D.; MacLeod, C.L.; Ellies, L.G. Effects of fvb/nj and c57bl/6j strain backgrounds on mammary tumor phenotype in inducible nitric oxide synthase deficient mice. Transgenic Res. 2007, 16, 193–201. [Google Scholar] [CrossRef] [PubMed]
- National Health and Medical Research Council. Australian Code for the Care and Use of Animals for Scientific Purposes, 8th ed.; National Health and Medical Research Council: Canberra, Australia, 2013; ISBN 1864965975.
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]


| KEGG Pathway | Gene Count | p Value |
|---|---|---|
| Proteoglycans in Cancer | 62 | 1.39 × 10−8 |
| DNA Replication | 16 | 5.57 × 10−5 |
| Cell Cycle | 36 | 7.36 × 10−5 |
| PI3K-Akt Signalling Pathway | 76 | 4.23 × 10−4 |
| Pathways in Cancer | 84 | 4.40 × 10−4 |
| Leukocyte Migration | 33 | 5.35 × 10−4 |
| mRNA Surveillance | 27 | 0.0012 |
| Nucleotide Excision Repair | 15 | 0.0033 |
| ECM Receptor Interaction | 23 | 0.0077 |
| Mismatch Repair | 9 | 0.0102 |
| Oestrogen Signalling Pathway | 24 | 0.0143 |
| P53 Signalling Pathway | 18 | 0.0155 |
| Ras Signalling Pathway | 47 | 0.0162 |
| cAMP Signalling Pathway | 41 | 0.0201 |
| Prolactin Signalling Pathway | 18 | 0.0345 |
| TGFB Signalling Pathway | 20 | 0.0389 |
| Transcriptional Misregulation in Cancer | 34 | 0.0425 |
| VEGF Signalling Pathway | 15 | 0.051 |
| KEGG Pathway | Gene Count | p Value |
|---|---|---|
| Metabolic Pathways | 386 | 5.82 × 10−42 |
| Non-Alcoholic Fatty Liver Disease | 88 | 1.71 × 10−29 |
| Citric Acid Cycle | 28 | 4.54 × 10−17 |
| Fatty Acid Metabolism | 34 | 1.26 × 10−14 |
| PPAR Signalling Pathway | 40 | 1.63 × 10−11 |
| Fatty Acid Degradation | 28 | 6.55 × 10−10 |
| Insulin Signalling Pathway | 54 | 6.58 × 10−10 |
| AMPK Signalling Pathway | 50 | 1.38 × 10−9 |
| Glycolysis/Gluconeogenesis | 30 | 1.21 × 10−7 |
| Adipokine Signalling Pathway | 30 | 1.13 × 10−6 |
| Fatty Acid Elongation | 16 | 2.05 × 10−6 |
| Regulation of Lipolysis in Adipocytes | 23 | 4.84 × 10−5 |
| Biosynthesis of Unsaturated Fatty Acids | 14 | 1.25 × 10−4 |
| T Cell Receptor Signalling Pathway | 29 | 0.004 |
| B Cell Receptor Signalling Pathway | 20 | 0.016 |
| Fatty Acid Biosynthesis | 7 | 0.018 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Archer, M.; Bernhardt, S.M.; Hodson, L.J.; Woolford, L.; Van der Hoek, M.; Dasari, P.; Evdokiou, A.; Ingman, W.V. CCL2-Mediated Stromal Interactions Drive Macrophage Polarization to Increase Breast Tumorigenesis. Int. J. Mol. Sci. 2023, 24, 7385. https://doi.org/10.3390/ijms24087385
Archer M, Bernhardt SM, Hodson LJ, Woolford L, Van der Hoek M, Dasari P, Evdokiou A, Ingman WV. CCL2-Mediated Stromal Interactions Drive Macrophage Polarization to Increase Breast Tumorigenesis. International Journal of Molecular Sciences. 2023; 24(8):7385. https://doi.org/10.3390/ijms24087385
Chicago/Turabian StyleArcher, Maddison, Sarah M. Bernhardt, Leigh J. Hodson, Lucy Woolford, Mark Van der Hoek, Pallave Dasari, Andreas Evdokiou, and Wendy V. Ingman. 2023. "CCL2-Mediated Stromal Interactions Drive Macrophage Polarization to Increase Breast Tumorigenesis" International Journal of Molecular Sciences 24, no. 8: 7385. https://doi.org/10.3390/ijms24087385
APA StyleArcher, M., Bernhardt, S. M., Hodson, L. J., Woolford, L., Van der Hoek, M., Dasari, P., Evdokiou, A., & Ingman, W. V. (2023). CCL2-Mediated Stromal Interactions Drive Macrophage Polarization to Increase Breast Tumorigenesis. International Journal of Molecular Sciences, 24(8), 7385. https://doi.org/10.3390/ijms24087385







