Discovering the Mechanisms of Oleodaphnone as a Potential HIV Latency-Reversing Agent by Transcriptome Profiling
Abstract
1. Introduction
2. Results
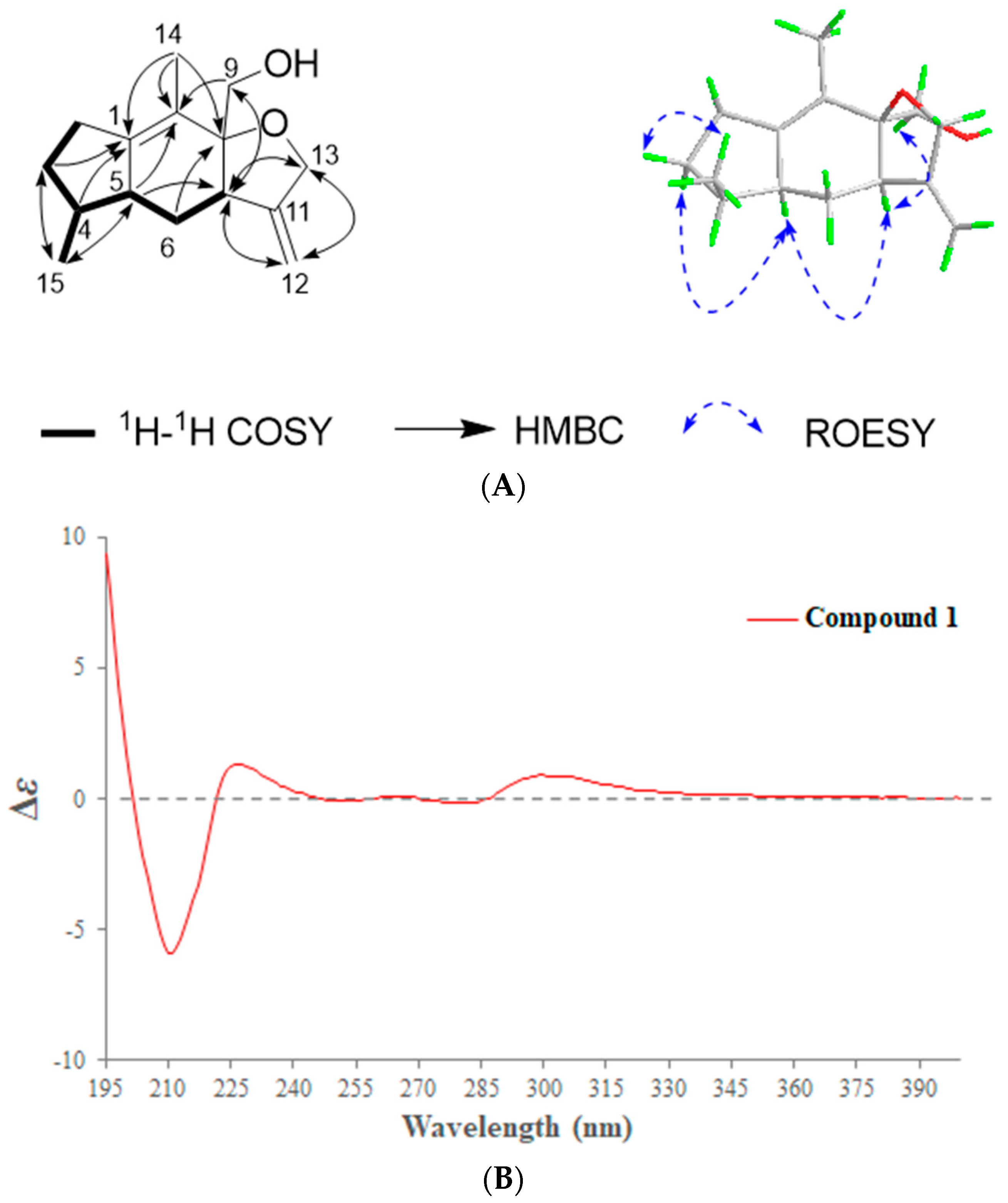
2.1. Phytochemical Investigation of the Roots of Wikstroemia chamaedaphne
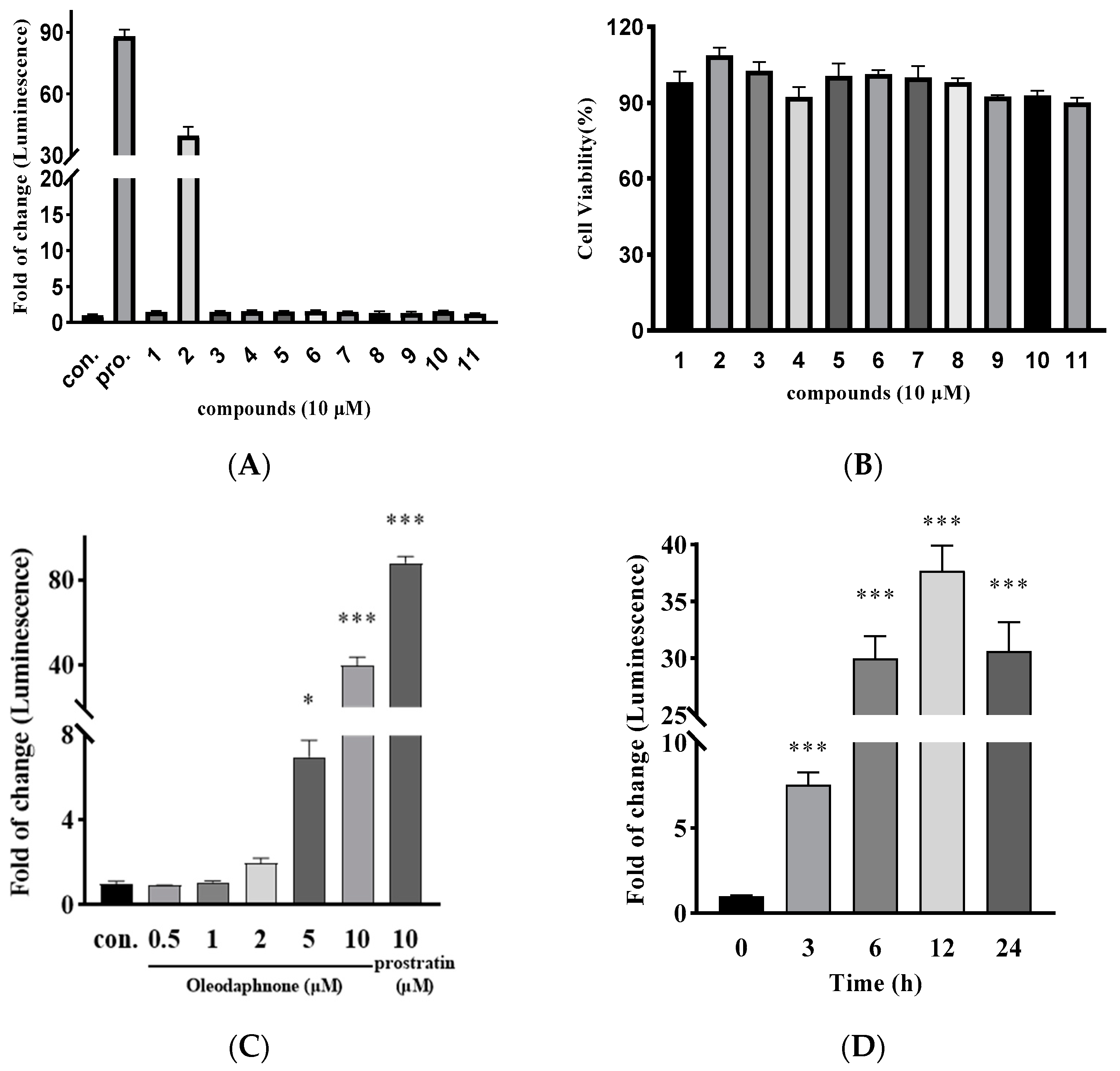
2.2. Oleodaphnone Is an Efficient HIV LRA
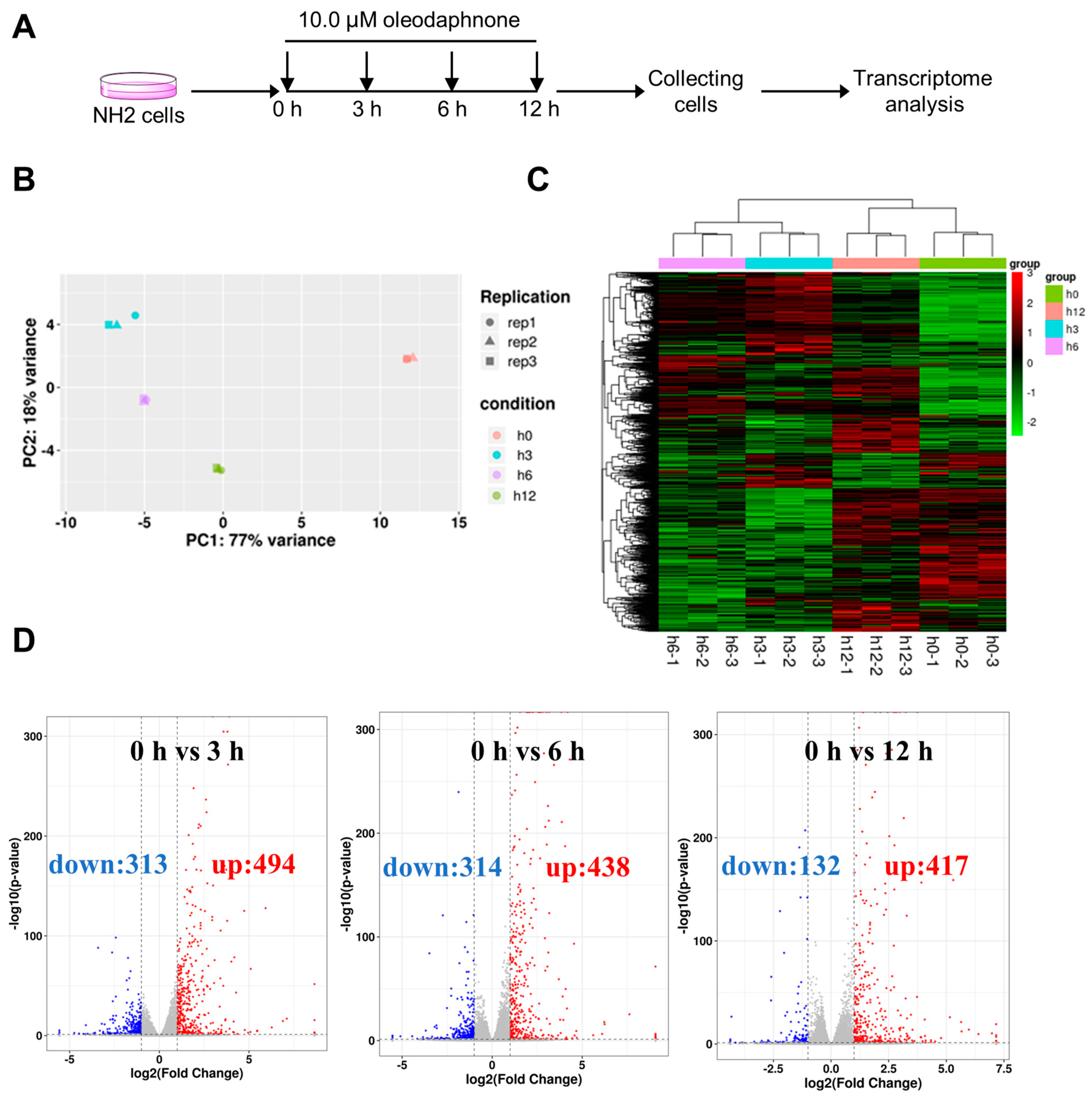
2.3. Oleodaphnone Reprograms the Transcriptome of NH2 Cells
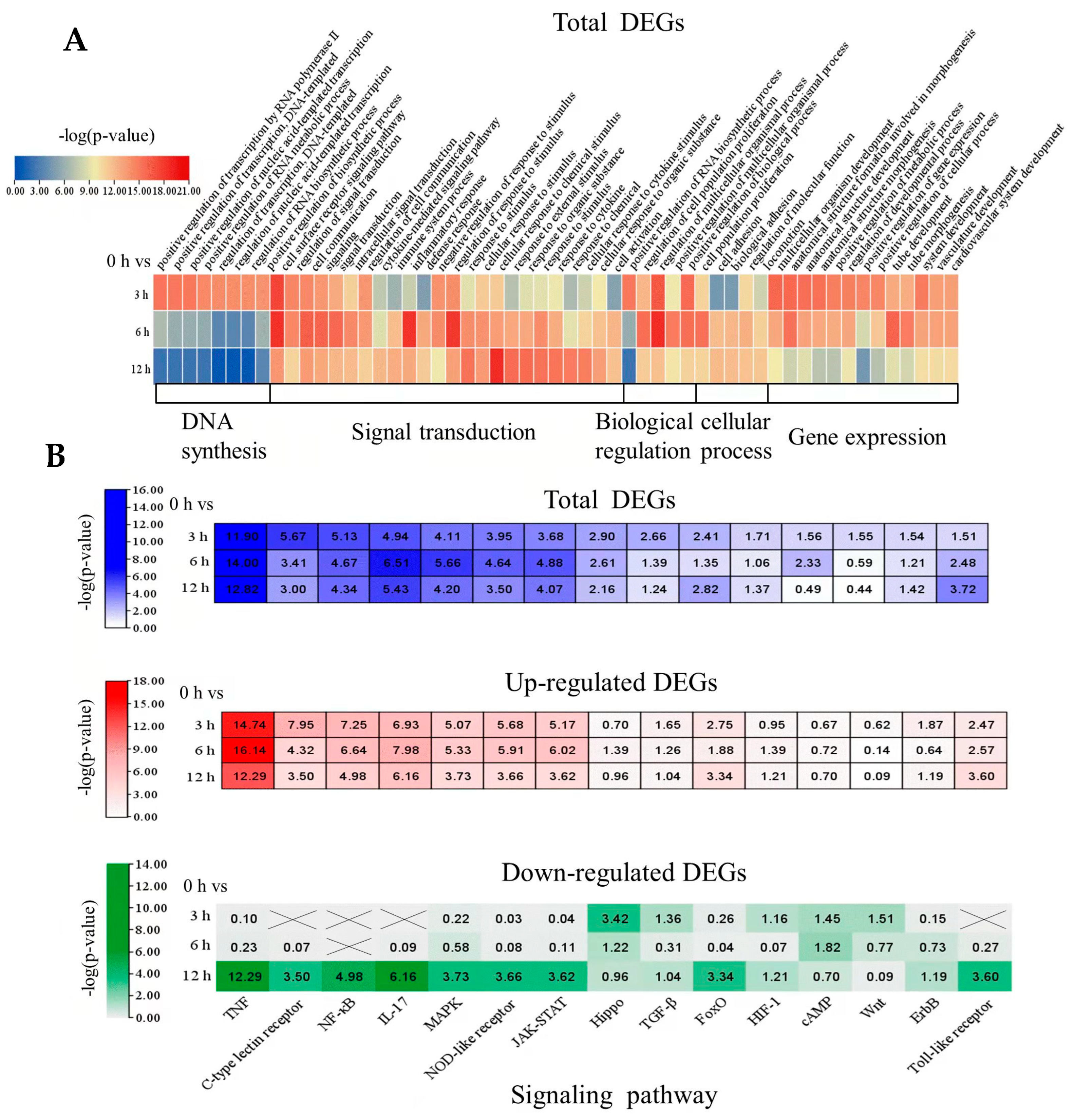
2.4. Identifying Biological Processes and Cell Signaling Responses to Oleodaphnone
2.5. PPI Network Analysis and Identification of Hub Genes
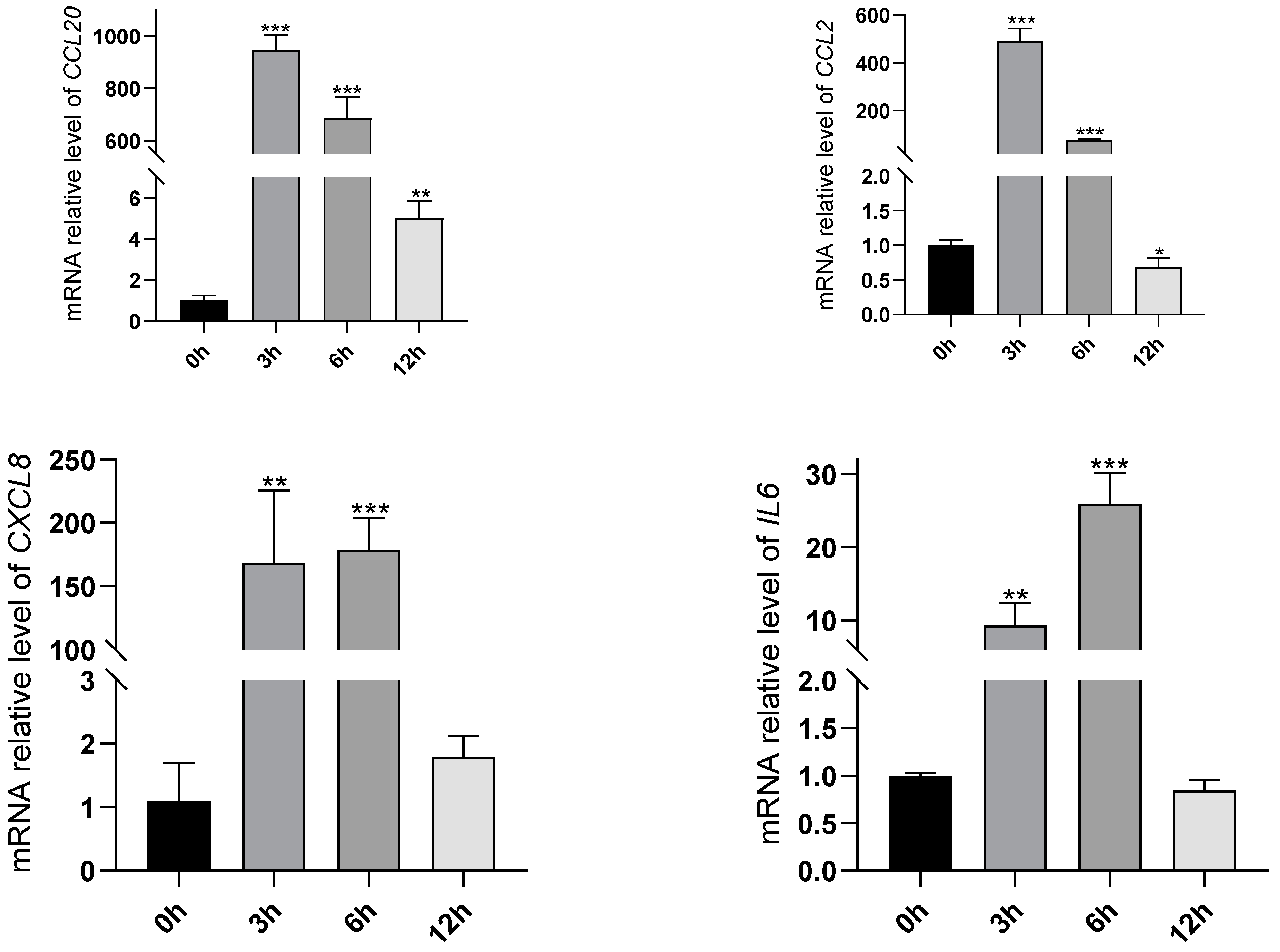
2.6. Validation on the Changes of TNF and IL-17 Signaling Pathways Induced by Oleodaphnone
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedure
4.2. Plant Material
4.3. Extraction and Isolation
4.4. Cell
4.5. HIV Latency-Reversing Assays
4.6. Transcriptome Analysis
4.7. Bioinformatics Analysis
4.8. RT-qPCR Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| HIV | Human immunodeficiency virus |
| AIDS | Acquired immunodeficiency syndrome |
| cART | Combination antiretroviral therapy |
| LRAs | Latency-reversing agents |
| NMR | Nuclear magnetic resonance |
| HRESIMS | High resolution electrospray ionization mass spectroscopy |
| HSQC | Heteronuclear signal quantum coherence |
| 1H-1H COSY | 1H-1H Correlation spectroscopy |
| HMBC | Heteronuclear multiple bond correlation |
| ROESY | Rotating-Frame Overhauser Enhancement and Exchange Spectroscopy |
| ECD | Electronic Circular Dichroism |
| PCA | Principal component analysis |
| DEGs | Different expression genes |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PPI | Protein–protein interaction |
| BP | Biological process |
| CC | Cellular component |
| MF | Molecular Function |
| PKC | protein kinase C |
| P-TEFb | Positive transcription elongation factor b |
| HDACi | Histone deacetylase inhibitor |
| HPLC | High Performance Liquid Chromatography |
| LH-20 | Sephadex LH-20 |
| s | Singlet (Spectroscopic) |
| d | Doublet (Spectroscopic) |
| J | Coupling constant (in NMR) |
| δ | Chemical shift in parts per million downfield from TMS |
| 2D NMR | Two dimensional NMR |
References
- Rasmussen, T.A.; Tolstrup, M.; Sgaard, O.S. Reversal of latency as part of a cure for HIV-1. Trends Microbiol. 2015, 24, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Jiao, Y.Y.; Zhang, L.W.; Li, S.F. Research progress on natural product human immunodeficiency virus-1 (HIV-1) latency reactivation agents. Zhongcaoyao 2021, 52, 297–308. [Google Scholar]
- Kulkosky, J.; Culnan, D.M.; Roman, J.; Dornadula, G.; Schnell, M.; Boyd, M.R.; Pomerantz, R.J. Prostratin: Activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 2001, 98, 3006–3015. [Google Scholar] [CrossRef]
- Wender, P.A.; Kee, J.M.; Warrington, J.M. Practical synthesis of prostratin, DPP, and their analogs, adjuvant leads against latent HIV. Science 2008, 320, 649–652. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Ho, P.; Yu, J.; Zhu, L.; Lee, K.H.; Chen, C.H. Picomolar dichotomous activity of gnidimacrin against HIV-1. PLoS ONE 2011, 6, e26677. [Google Scholar] [CrossRef]
- Lai, W.; Huang, L.; Zhu, L.; Ferrari, G.; Chan, C.; Li, W.; Lee, K.H.; Chen, C.H. Gnidimacrin, a potent anti-HIV diterpene, can eliminate latent HIV-1 ex vivo by activation of protein kinase C β. J. Med. Chem. 2015, 58, 8638–8646. [Google Scholar] [CrossRef]
- Li, S.F.; Jiao, Y.Y.; Zhang, Z.Q.; Chao, J.B.; Jia, J.; Shi, X.L.; Zhang, L.W. Diterpenes from buds of Wikstroemia chamaedaphne showing anti-hepatitis B virus activities. Phytochemistry 2018, 151, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, S.F.; Zhang, L.W.; Chao, J.B. Chemical constituents from flowers of Wikstroemia chamaedaphne and their anti-hepatitis B virus activity. Zhongcaoyao 2017, 48, 1292–1297. [Google Scholar]
- Li, S.F.; Wang, X.Y.; Li, G.L.; Jiao, Y.Y.; Wang, W.H.; Wu, X.K.; Zhang, L.W. Potential HIV latency-reversing agents with STAT1-activating activity from the leaves of Wikstroemia chamaedaphne. Phytochemistry 2022, 203, 113395. [Google Scholar] [CrossRef]
- Li, S.F.; Liang, X.; Wu, X.K.; Gao, X.; Zhang, L.W. Discovering the mechanisms of wikstroelide E as a potential HIV-latency-reversing agent by transcriptome profiling. J. Nat. Prod. 2021, 84, 1022–1033. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Q.B.; Hou, Z.L.; Shi, S.C.; Ren, H.; Yao, G.D.; Lin, B.; Huang, X.X.; Song, S.J. Discovery of guaiane-type sesquiterpenoids from the roots of Daphne genkwa with neuroprotective effects. Bioorg. Chem. 2020, 95, 103545. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Taninaka, H.; Takaishi, Y.; Honda, G.; Imakura, Y.; Sezik, E.; Yesilada, E. Terpenoids and aromatic compounds from Daphne oleoides ssp. oleoides. Phytochemistry 1999, 52, 1525–1529. [Google Scholar] [CrossRef]
- Kimata, J.T.; Rice, A.P.; Wang, J. Challenges and strategies for the eradication of the HIV reservoir. Curr. Opin. Immunol. 2016, 42, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Van Lint, C.; Bouchat, S.; Marcello, A. HIV-1 transcription and latency: An update. Retrovirology 2013, 10, 67. [Google Scholar] [CrossRef]
- Bandera, A.; Gori, A.; Clerici, M.; Sironi, M. Phylogenies in ART: HIV reservoirs, HIV latency and drug resistance. Curr. Opin. Pharmacol. 2019, 48, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Dahabieh, M.S.; Battivelli, E.; Verdin, E. Understanding HIV latency: The road to an HIV cure. Annu. Rev. Med. 2015, 66, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Kalvatchev, Z.; Walder, R.; Garzaro, D. Different effects of phorbol ester derivates on human immunodeficiency virus 1 replication in lymphocytic and monocytic human cells. Acta Virol. 1997, 5, 289–292. [Google Scholar]
- De la Torre-Tarazona, H.E.; Jiménez, R.; Bueno, P. 4-Deoxyphorbol inhibits HIV-1 infection in synergism with antiretroviral drugs and reactivates viral reservoirs through PKC/MEK activation synergizing with vorinostat. Biochem. Pharmacol. 2020, 177, 113937. [Google Scholar] [CrossRef] [PubMed]
- Shang, H.T.; Ding, J.W.; Yu, S.Y.; Wu, T.; Zhang, Q.L.; Liang, F.J. Progress and challenges in the use of latent HIV-1 reactivating agents. Acta Pharmacol. Sin. 2015, 36, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Este, J.A.; Cihlar, T. Current status and challenges of antiretroviral research and therapy. Antivir. Res. 2010, 85, 25–33. [Google Scholar] [CrossRef]
- Eisele, E.; Siliciano, R.F. Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 2012, 37, 377–388. [Google Scholar] [CrossRef]
- Swaminathan, G.; Pascual, D.; Rival, G.; Perales-Linares, R.; Martin-Garcia, J.; Navas-Martin, S. Hepatitis C virus core protein enhances HIV-1 replication in human macrophages through TLR2, JNK, and MEK1/2-dependent upregulation of TNF-α and IL-6. FEBS Lett. 2014, 588, 3501–3510. [Google Scholar] [CrossRef] [PubMed]
- Lazdins, J.K.; Grell, M.; Walker, M.R.; Woods-Cook, K.; Scheurich, P.; Pfizenmaier, K. Membrane tumor necrosis factor (TNF) induced cooperative signaling of TNFR60 and TNFR80 favors induction of cell death rather than virus production in HIV-infected T cells. J. Exp. Med. 1997, 185, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Hodges, A.; Sharrocks, K.; Edelmann, M.; Baban, D.; Moris, A.; Schwartz, O.; Drakesmith, H.; Davies, K.; Kessler, B.; McMichael, A.; et al. Activation of the lectin DC-SIGN induces an immature dendritic cell phenotype triggering Rho-GTPase activity required for HIV-1 replication. Nat. Immunol. 2007, 8, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Mayr, L.; Su, B.; Moog, C. Langerhans Cells: The ‘Yin and Yang’ of HIV Restriction and Transmission. Trends Microbiol. 2017, 25, 170–172. [Google Scholar] [CrossRef]
- Castedo, M.; Perfettini, J.L.; Andreau, K.; Roumier, T.; Piacentini, M.; Kroemer, G. Mitochondrial apoptosis induced by the HIV-1 envelope. Ann. N. Y. Acad. Sci. 2003, 1010, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Jiang, X.; Cao, L.; Gu, Q.; Teng, X.; He, L. Diverse sesquiterpenoids from the roots of Croton crassifolius and their inhibitory effects on ferroptosis. Chem. Biodivers. 2022, 19, e202101028. [Google Scholar] [CrossRef]
- Pan, J.; Su, J.C.; Liu, Y.H.; Deng, B.; Hu, Z.F.; Wu, J.L.; Xia, R.F.; Chen, C.; He, Q.; Chen, J.C.; et al. Stelleranoids A-M, guaiane-type sesquiterpenoids based on [5,7] bicyclic system from Stellera chamaejasme and their cytotoxic activity. Bioorg. Chem. 2021, 115, 105251. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhu, Q.; Luo, K.; Zhou, Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 2001, 414, 317–322. [Google Scholar] [CrossRef]
- Li, Z.; Guo, J.; Wu, Y.; Zhou, Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013, 41, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO::TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [PubMed]

| Sesquiterpene | 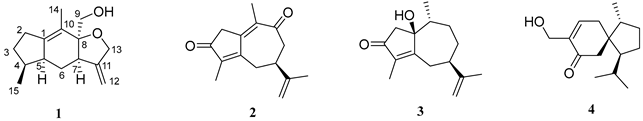 |
| Flavonoid | 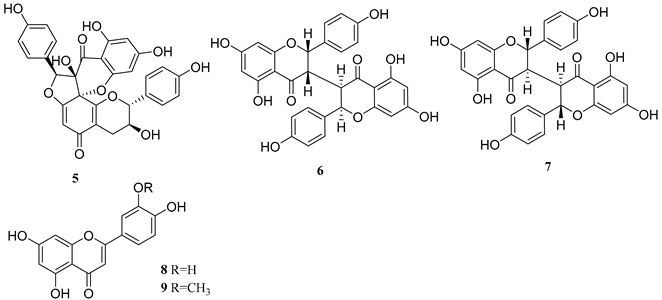 |
| Lignan | 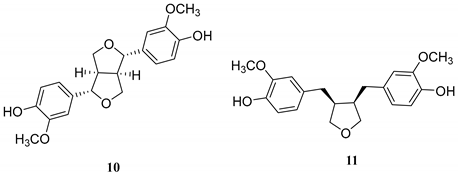 |
| NO. | 1 (CD3OD) | 2 (CD3OD) | 3 (CDCl3) | 4 (CD3OD) | ||||
|---|---|---|---|---|---|---|---|---|
| 1H | 13C | 13C | 1H | 13C | 1H | 13C | ||
| 1 | 144.5 | 144.8 | 83.4 | 1.32, overlap | 56.9 | |||
| 2 | 2.12, overlap | 26.6 | 3.18, q (21.0) | 40.6 | 2.61, d (18.0) | 52.3 | 1.67, overlap | 25.8 |
| 2.43, d (18.0) | 1.32, overlap | |||||||
| 3 | 1.65, overlap | 31.9 | 203.1 | 207.8 | 1.67, overlap | 29.8 | ||
| 1.41, dd (12.6, 7.8) | 1.32, overlap | |||||||
| 4 | 2.12, overlap | 36.0 | 145.1 | 139.1 | 1.67, overlap | 46.4 | ||
| 5 | 2.21, overlap | 44.2 | 163.9 | 174.6 | 48.2 | |||
| 6 | 1.61, overlap | 30.0 | 3.05, d (16.8) | 35.3 | 2.74, d (18.6) | 36.9 | 2.76, d (16.8) | 49.5 |
| 1.01, d (12.6) | 2.91, overlap | 2.62, d (18.0) | 2.24, d (16.8) | |||||
| 7 | 2.70, dd (13.2, 6.0) | 45.8 | 2.76, m | 39.3 | 2.27, overlap | 44.0 | 201.5 | |
| 8 | 86.2 | 2.91, overlap | 49.6 | 1.81, m | 31.3 | 138.0 | ||
| 1.54, m | ||||||||
| 9 | 3.43, d (10.8) | 63.5 | 201.5 | 1.65, m | 31.9 | 6.84, m | 145.9 | |
| 3.29, d (10.8) | 1.54, m | |||||||
| 10 | 123.7 | 131.6 | 2.27, overlap | 41.2 | 2.36, d (19.8) | 25.4 | ||
| 2.13, d (19.8) | ||||||||
| 11 | 153.7 | 147.9 | 152.5 | 1.67, overlap | 29.4 | |||
| 12 | 4.96, s | 104.0 | 4.83, d (9.0) | 110.3 | 4.78, s | 109.4 | 0.78, t (9.6) | 24.2 |
| 4.84, s | 4.71, s | |||||||
| 13 | 4.31, d (13.2) | 68.9 | 1.82, s | 19.4 | 1.79, s | 20.5 | 0.95, t (9.6) | 21.6 |
| 4.21, d (13.2) | ||||||||
| 14 | 1.58, s | 12.3 | 1.98, s | 16.3 | 1.68, s | 14.6 | 0.87, m | 17.3 |
| 15 | 0.62, d (7.2) | 13.5 | 1.87, s | 7.8 | 0.77, d (7.2) | 7.8 | 4.24, s | 62.2 |
| Primer | Forward Primer (5′ to 3′) | Reverse Primer (5′to 3′) |
|---|---|---|
| CCL2 | AGAATCACCAGCAGCAAGTGTCC | TCCTGAACCCACTTCTGCTTGG |
| CCL20 | TGACTGCTGTCTTGGATACACAGA | TGATAGCATTGATGTCACAGCCT |
| CXCL8 | GTCCTTGTTCCACTGTGCCT | GCTTCCACATGTCCTCACAA |
| IL6 | AGACAGCCACTCACCTCTTCAG | TTCTGCCAGTGCCTCTTTGCTG |
| β-actin | AAGGATTCCTATGTGGGCGAC | CGTACAGGGATAGCACAGCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Wang, X.; Yang, Y.; Wu, X.; Zhang, L. Discovering the Mechanisms of Oleodaphnone as a Potential HIV Latency-Reversing Agent by Transcriptome Profiling. Int. J. Mol. Sci. 2023, 24, 7357. https://doi.org/10.3390/ijms24087357
Li S, Wang X, Yang Y, Wu X, Zhang L. Discovering the Mechanisms of Oleodaphnone as a Potential HIV Latency-Reversing Agent by Transcriptome Profiling. International Journal of Molecular Sciences. 2023; 24(8):7357. https://doi.org/10.3390/ijms24087357
Chicago/Turabian StyleLi, Shifei, Xiuyi Wang, Yuqin Yang, Xingkang Wu, and Liwei Zhang. 2023. "Discovering the Mechanisms of Oleodaphnone as a Potential HIV Latency-Reversing Agent by Transcriptome Profiling" International Journal of Molecular Sciences 24, no. 8: 7357. https://doi.org/10.3390/ijms24087357
APA StyleLi, S., Wang, X., Yang, Y., Wu, X., & Zhang, L. (2023). Discovering the Mechanisms of Oleodaphnone as a Potential HIV Latency-Reversing Agent by Transcriptome Profiling. International Journal of Molecular Sciences, 24(8), 7357. https://doi.org/10.3390/ijms24087357






