JAK1 Pseudokinase V666G Mutant Dominantly Impairs JAK3 Phosphorylation and IL-2 Signaling
Abstract
1. Introduction
2. Results
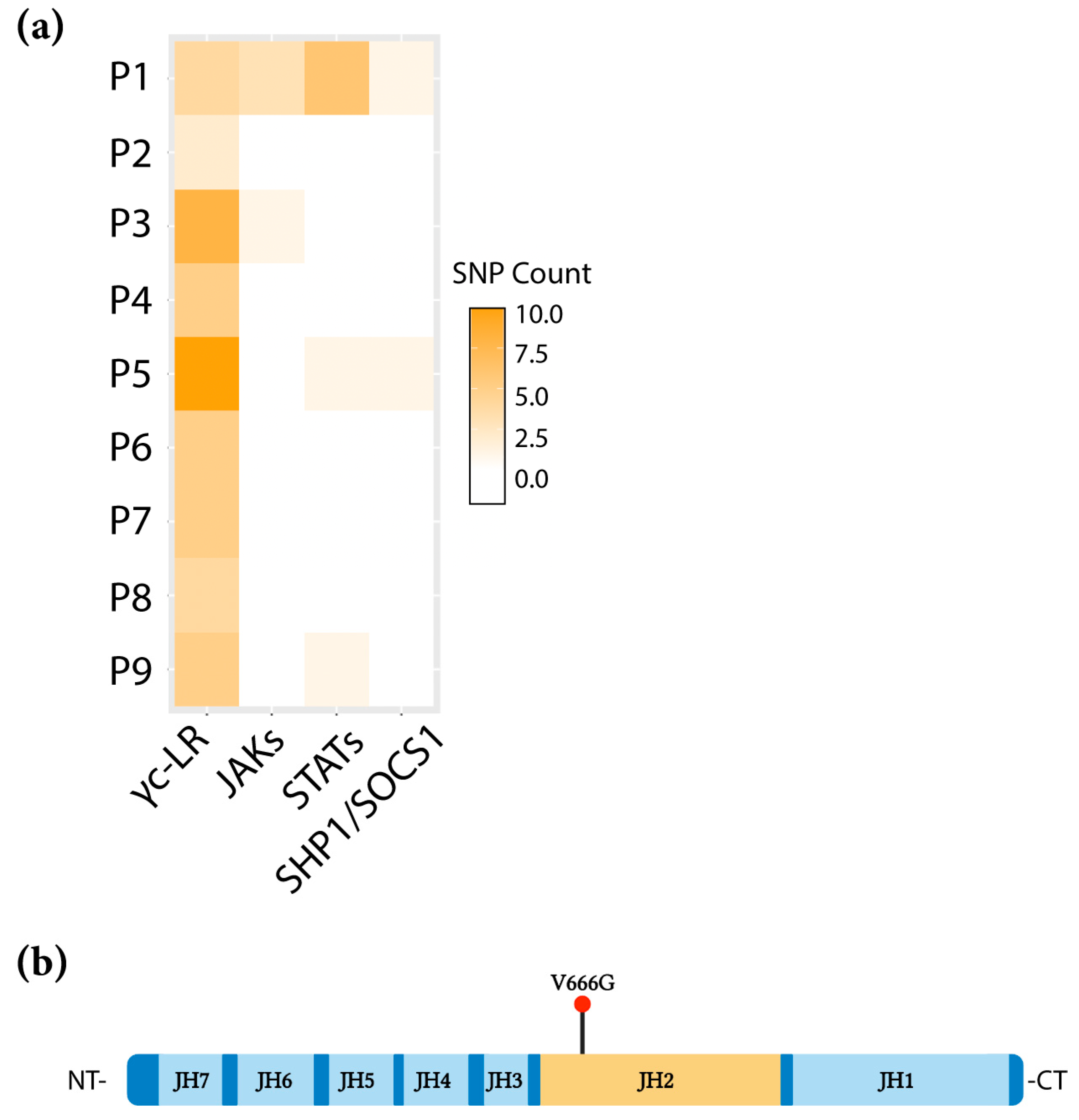
2.1. Identification of the JAK1 V666G Mutation
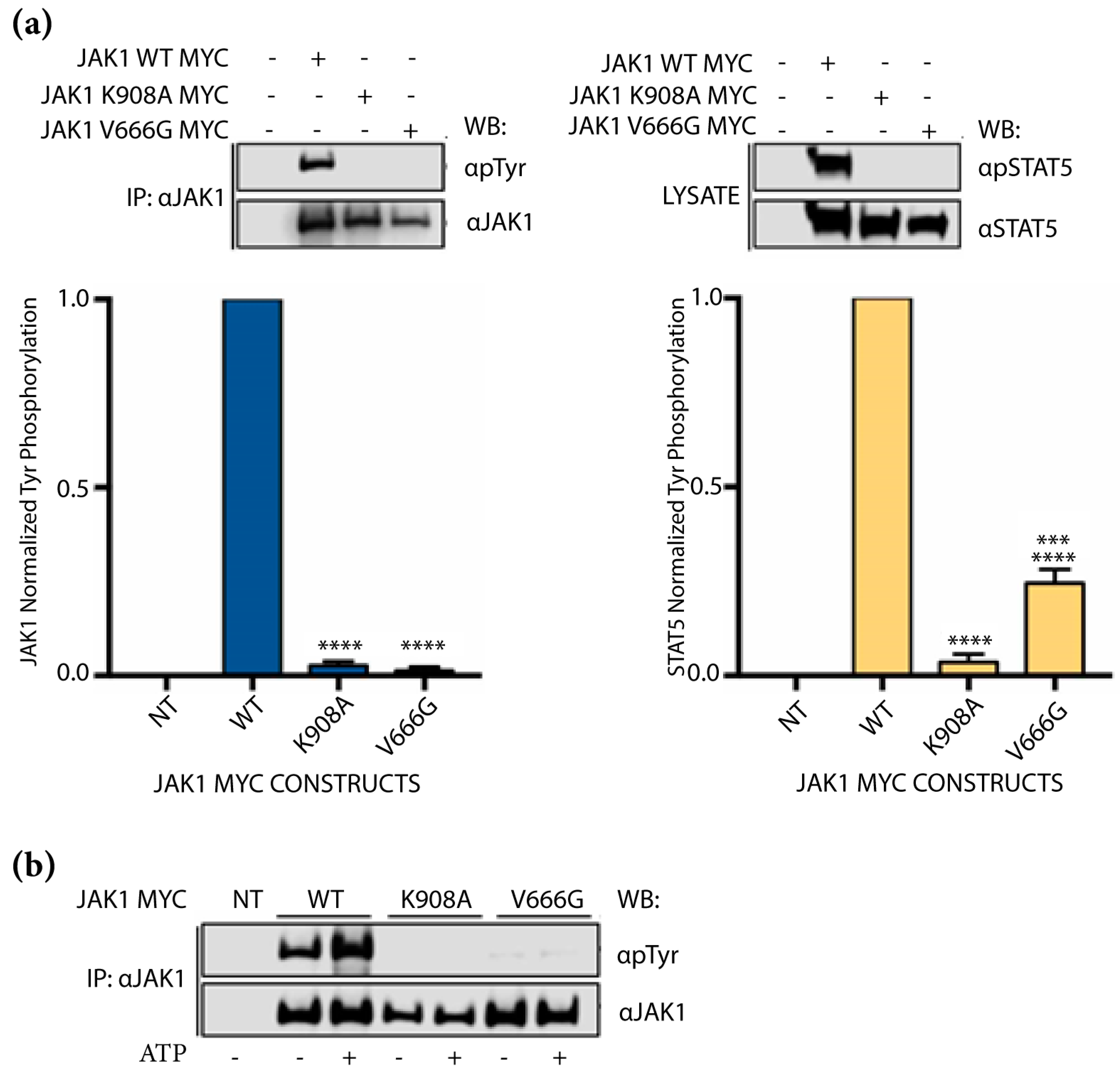
2.2. Abrogation of JAK1 V666G Auto-Activation and Substrate Phosphorylation
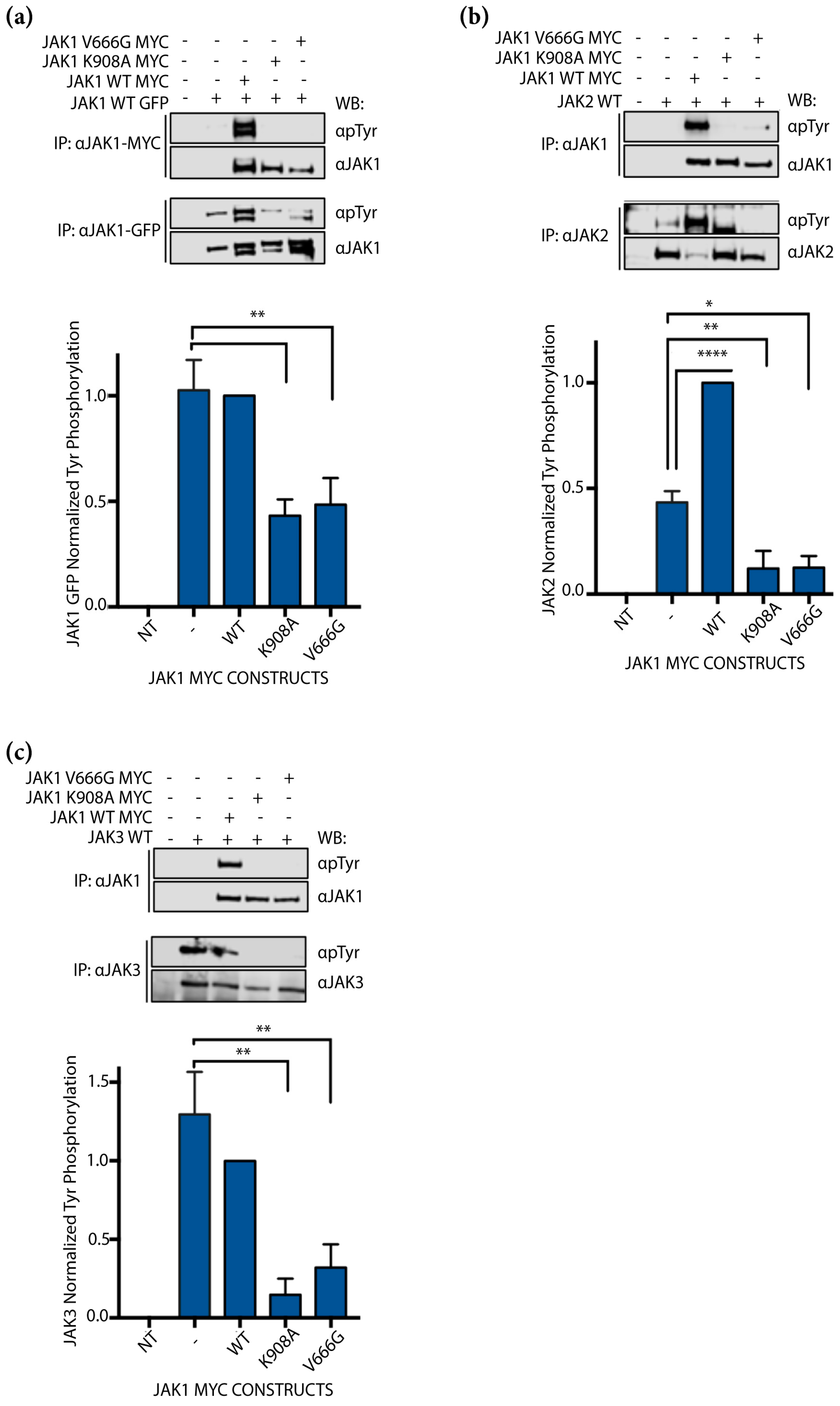
2.3. JAK1 V666G Inhibits Cross-Activation of JAK1, JAK2, and JAK3 Dimer Partners
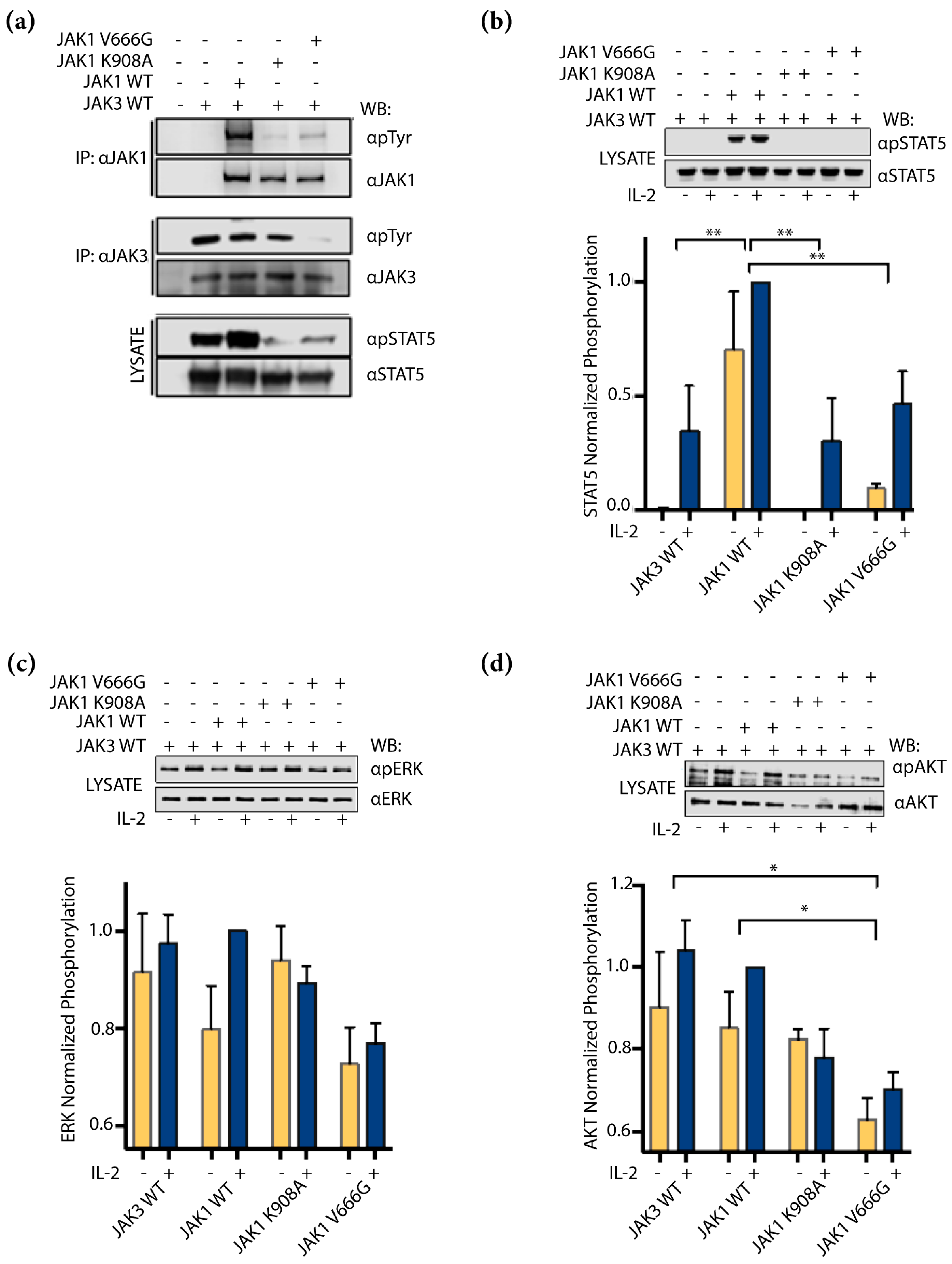
2.4. JAK1 V666G Inhibits Trans-Activation of JAK3 and IL-2 Signaling
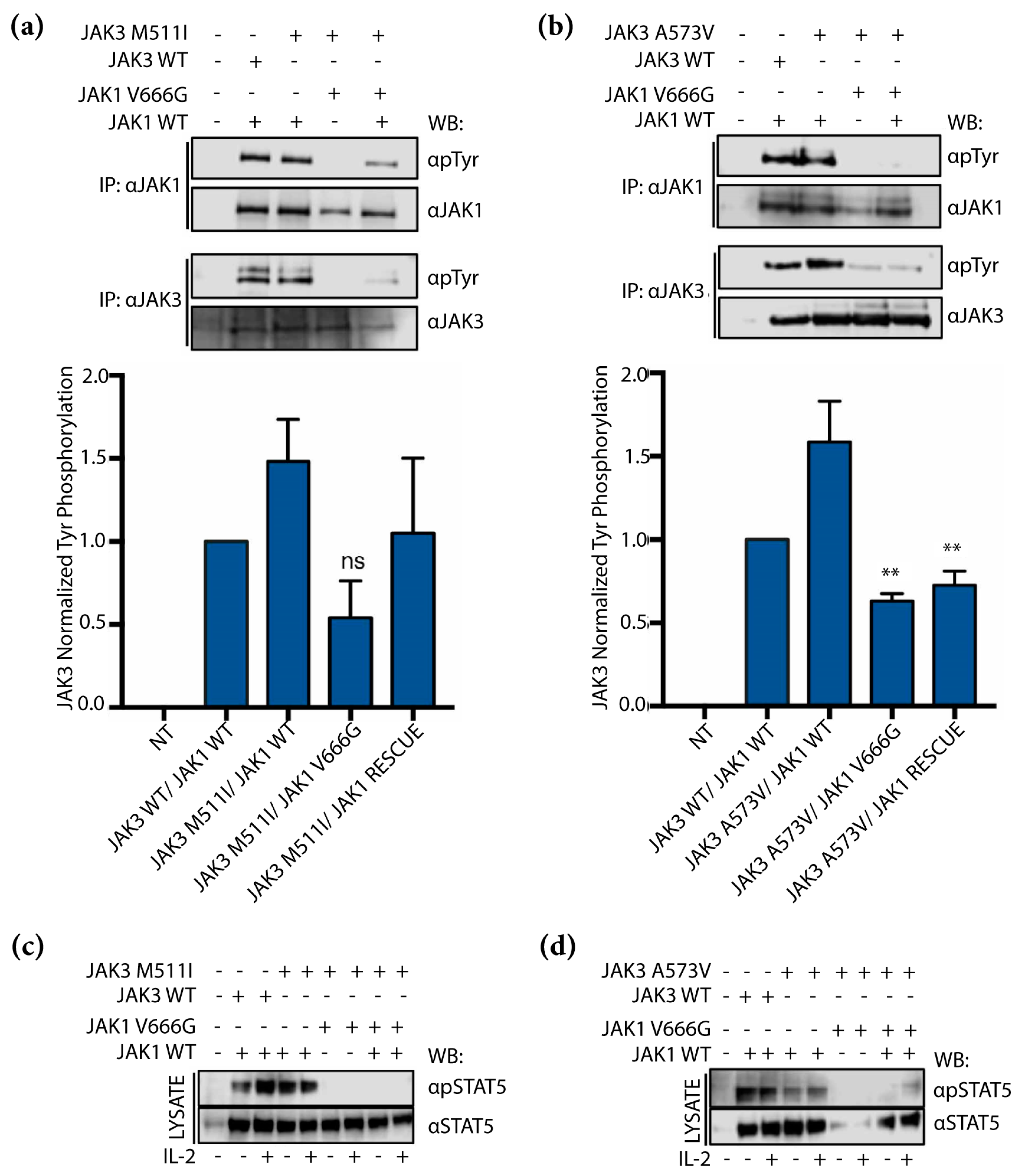
2.5. JAK1 V666G Inhibits Tyr Phosphorylation of the JAK3 M511I- and A573V-Activating Mutants
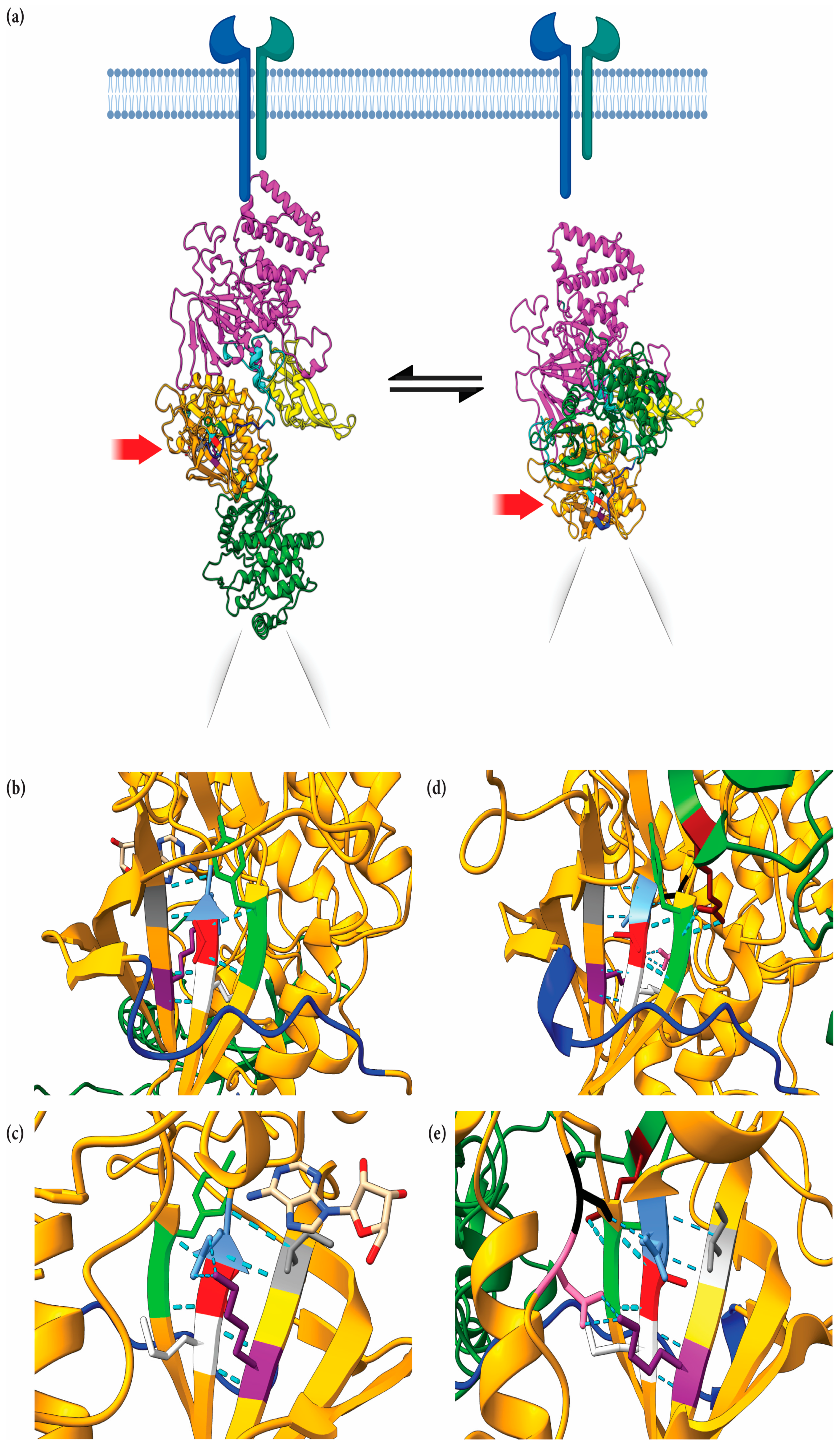
2.6. Structural Implications of JAK1 V666 Modeled in Open- and Closed-Kinase Confirmations
3. Discussion
4. Materials and Methods
4.1. Patient Samples, WES and Analysis
4.2. Plasmids and Site-Directed Mutagenesis
4.3. Cell Culture, Transfections for Auto-Activation, Cross-Activation and IL-2 Trans-Activation Experiments
4.4. Western Blot Analysis
4.5. JAK1 Modeling
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murray, P.J. The JAK-STAT signaling pathway: Input and output integration. J. Immunol. 2007, 178, 2623–2629. [Google Scholar] [CrossRef]
- Ghoreschi, K.; Laurence, A.; O’Shea, J.J. Janus kinases in immune cell signaling. Immunol. Rev. 2009, 228, 273–287. [Google Scholar] [CrossRef]
- Fujii, H. Mechanisms of Signal Transduction from Receptors of Type I and Type II Cytokines. J. Immunotoxicol. 2007, 4, 69–76. [Google Scholar] [CrossRef]
- Haan, C.; Kreis, S.; Margue, C.; Behrmann, I. Jaks and cytokine receptors—An intimate relationship. Biochem. Pharmacol. 2006, 72, 1538–1546. [Google Scholar] [CrossRef] [PubMed]
- Cornejo, M.G.; Boggon, T.J.; Mercher, T. JAK3: A two-faced player in hematological disorders. Int. J. Biochem. Cell Biol. 2009, 41, 2376–2379. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lupardus, P.; Laporte, S.L.; Garcia, K.C. Structural biology of shared cytokine receptors. Annu. Rev. Immunol. 2009, 27, 29–60. [Google Scholar] [CrossRef] [PubMed]
- Witthuhn, B.A.; Williams, M.D.; Kerawalla, H.; Uckun, F.M. Differential substrate recognition capabilities of Janus family protein tyrosine kinases within the interleukin 2 receptor (IL2R) system: Jak3 as a potential molecular target for treatment of leukemias with a hyperactive Jak-Stat signaling machinery. Leuk. Lymphoma 1999, 32, 289–297. [Google Scholar] [CrossRef]
- Kirken, R.A.; Rui, H.; Malabarba, M.G.; Howard, O.M.; Kawamura, M.; O'Shea, J.J.; Farrar, W.L. Activation of JAK3, but not JAK1, is critical for IL-2-induced proliferation and STAT5 recruitment by a COOH-terminal region of the IL-2 receptor beta-chain. Cytokine 1995, 7, 689–700. [Google Scholar] [CrossRef]
- Haan, C.; Rolvering, C.; Raulf, F.; Kapp, M.; Druckes, P.; Thoma, G.; Behrmann, I.; Zerwes, H.G. Jak1 has a dominant role over Jak3 in signal transduction through gammac-containing cytokine receptors. Chem. Biol. 2011, 18, 314–323. [Google Scholar] [CrossRef]
- Bains, T.; Heinrich, M.C.; Loriaux, M.M.; Beadling, C.; Nelson, D.; Warrick, A.; Neff, T.L.; Tyner, J.W.; Dunlap, J.; Corless, C.L.; et al. Newly described activating JAK3 mutations in T-cell acute lymphoblastic leukemia. Leukemia 2012, 26, 2144–2146. [Google Scholar] [CrossRef]
- Greenplate, A.; Wang, K.; Tripathi, R.M.; Palma, N.; Ali, S.M.; Stephens, P.J.; Miller, V.A.; Shyr, Y.; Guo, Y.; Reddy, N.M.; et al. Genomic Profiling of T-Cell Neoplasms Reveals Frequent JAK1 and JAK3 Mutations with Clonal Evasion from Targeted Therapies. JCO Precis. Oncol. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.G.; Yang, Y.L.; Payne-Turner, D.; Lin, W.; Files, J.K.; Dickerson, K.; Gu, Z.; Taunton, J.; Janke, L.J.; Chen, T.; et al. Oncogenic role and therapeutic targeting of ABL-class and JAK-STAT activating kinase alterations in Ph-like ALL. Blood Adv. 2017, 1, 1657–1671. [Google Scholar] [PubMed]
- Mullighan, C.G.; Zhang, J.; Harvey, R.C.; Collins-Underwood, J.R.; Schulman, B.A.; Phillips, L.A.; Tasian, S.K.; Loh, M.L.; Su, X.; Liu, W.; et al. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2009, 106, 9414–9418. [Google Scholar] [CrossRef]
- Vicente, C.; Schwab, C.; Broux, M.; Geerdens, E.; Degryse, S.; Demeyer, S.; Lahortiga, I.; Elliott, A.; Chilton, L.; La Starza, R.; et al. Targeted sequencing identifies associations between IL7R-JAK mutations and epigenetic modulators in T-cell acute lymphoblastic leukemia. Haematologica 2015, 100, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Tarlock, K.; Chang, B.; Cooper, T.; Gross, T.; Gupta, S.; Neudorf, S.; Adlard, K.; Ho, P.A.; McGoldrick, S.; Watt, T.; et al. Sorafenib treatment following hematopoietic stem cell transplant in pediatric FLT3/ITD acute myeloid leukemia. Pediatr. Blood Cancer 2015, 62, 1048–1054. [Google Scholar] [CrossRef]
- Nguyen, K.; Devidas, M.; Cheng, S.-C.; La, M.; Raetz, E.A.; Carroll, W.L.; Winick, N.J.; Hunger, S.P.; Gaynon, P.S.; Loh, M.L. Factors influencing survival after relapse from acute lymphoblastic leukemia: A Children’s Oncology Group study. Leukemia 2008, 22, 2142–2150. [Google Scholar] [CrossRef]
- Roskoski, R., Jr. Janus kinase (JAK) inhibitors in the treatment of neoplastic and inflammatory disorders. Pharmacol. Res. 2022, 183, 106362. [Google Scholar] [CrossRef]
- Ding, Y.Y.; Stern, J.W.; Jubelirer, T.F.; Wertheim, G.B.; Lin, F.; Chang, F.; Gu, Z.; Mullighan, C.G.; Li, Y.; Harvey, R.C.; et al. Clinical efficacy of ruxolitinib and chemotherapy in a child with Philadelphia chromosome-like acute lymphoblastic leukemia with GOLGA5-JAK2 fusion and induction failure. Haematologica 2018, 103, e427–e431. [Google Scholar] [CrossRef]
- Loh, M.L.; Tasian, S.K.; Rabin, K.R.; Brown, P.; Magoon, D.; Reid, J.M.; Chen, X.; Ahern, C.H.; Weigel, B.J.; Blaney, S.M. A phase 1 dosing study of ruxolitinib in children with relapsed or refractory solid tumors, leukemias, or myeloproliferative neoplasms: A Children’s Oncology Group phase 1 consortium study (ADVL1011). Pediatr. Blood Cancer 2015, 62, 1717–1724. [Google Scholar] [CrossRef]
- Vainchenker, W.; Leroy, E.; Gilles, L.; Marty, C.; Plo, I.; Constantinescu, S.N. JAK inhibitors for the treatment of myeloproliferative neoplasms and other disorders. F1000Research 2018, 7, 82. [Google Scholar] [CrossRef]
- Kothiwale, S.; Borza, C.; Pozzi, A.; Meiler, J. Quantitative Structure-Activity Relationship Modeling of Kinase Selectivity Profiles. Molecules 2017, 22, 1576. [Google Scholar] [CrossRef]
- Cohen, S.B. JAK inhibitors and VTE risk: How concerned should we be? Nat. Rev. Rheumatol. 2021, 17, 133–134. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. Janus kinase (JAK) inhibitors in the treatment of inflammatory and neoplastic diseases. Pharmacol. Res. 2016, 111, 784–803. [Google Scholar] [CrossRef]
- Chimalakonda, A.; Burke, J.; Cheng, L.; Catlett, I.; Tagen, M.; Zhao, Q.; Patel, A.; Shen, J.; Girgis, I.G.; Banerjee, S.; et al. Selectivity Profile of the Tyrosine Kinase 2 Inhibitor Deucravacitinib Compared with Janus Kinase 1/2/3 Inhibitors. Dermatol. Ther. 2021, 11, 1763–1776. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration Approves Sotyktu™ (deucravacitinib), Oral Treatment for Adults with Moderate-to-Severe Plaque Psoriasis. Available online: https://news.bms.com/news/details/2022/U.S.-Food-and-Drug-Administration-Approves-Sotyktu-deucravacitinib-Oral-Treatment-for-Adults-with-Moderate-to-Severe-Plaque-Psoriasis/default.aspx (accessed on 1 February 2023).
- Roskoski, R., Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2021 update. Pharmacol. Res. 2021, 105463. [Google Scholar] [CrossRef] [PubMed]
- Lupardus, P.J.; Ultsch, M.; Wallweber, H.; Bir Kohli, P.; Johnson, A.R.; Eigenbrot, C. Structure of the pseudokinase-kinase domains from protein kinase TYK2 reveals a mechanism for Janus kinase (JAK) autoinhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 8025–8030. [Google Scholar] [CrossRef]
- Tasian, S.K.; Loh, M.L.; Hunger, S.P. Philadelphia chromosome-like acute lymphoblastic leukemia. Blood 2017, 130, 2064–2072. [Google Scholar] [CrossRef] [PubMed]
- Degryse, S.; Bornschein, S.; de Bock, C.E.; Leroy, E.; Vanden Bempt, M.; Demeyer, S.; Jacobs, K.; Geerdens, E.; Gielen, O.; Soulier, J.; et al. Mutant JAK3 signaling is increased by loss of wild-type JAK3 or by acquisition of secondary JAK3 mutations in T-ALL. Blood 2018, 131, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ding, L.; Holmfeldt, L.; Wu, G.; Heatley, S.L.; Payne-Turner, D.; Easton, J.; Chen, X.; Wang, J.; Rusch, M.; et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 2012, 481, 157–163. [Google Scholar] [CrossRef]
- Grant, A.H.; Ayala-Marin, Y.M.; Mohl, J.E.; Robles-Escajeda, E.; Rodriguez, G.; Dutil, J.; Kirken, R.A. The Genomic Landscape of a Restricted ALL Cohort from Patients Residing on the U.S./Mexico Border. Int. J. Environ. Res. Public Health 2021, 18, 7345. [Google Scholar] [CrossRef]
- Bandapalli, O.R.; Schuessele, S.; Kunz, J.B.; Rausch, T.; Stutz, A.M.; Tal, N.; Geron, I.; Gershman, N.; Izraeli, S.; Eilers, J.; et al. The activating STAT5B N642H mutation is a common abnormality in pediatric T-cell acute lymphoblastic leukemia and confers a higher risk of relapse. Haematologica 2014, 99, e188–e192. [Google Scholar] [CrossRef] [PubMed]
- Hammaren, H.M.; Virtanen, A.T.; Raivola, J.; Silvennoinen, O. The regulation of JAKs in cytokine signaling and its breakdown in disease. Cytokine 2019, 118, 48–63. [Google Scholar] [CrossRef]
- Martinez, G.S.; Ross, J.A.; Kirken, R.A. Transforming Mutations of Jak3 (A573V and M511I) Show Differential Sensitivity to Selective Jak3 Inhibitors. Clin. Cancer Drugs 2016, 3, 131–137. [Google Scholar] [CrossRef]
- Glassman, C.R.; Tsutsumi, N.; Saxton, R.A.; Lupardus, P.J.; Jude, K.M.; Garcia, K.C. Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation. Science 2022, 376, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Liau, N.P.D.; Laktyushin, A.; Lucet, I.S.; Murphy, J.M.; Yao, S.; Whitlock, E.; Callaghan, K.; Nicola, N.A.; Kershaw, N.J.; Babon, J.J. The molecular basis of JAK/STAT inhibition by SOCS1. Nat. Commun. 2018, 9, 1558. [Google Scholar] [CrossRef]
- Hamaguchi, H.; Amano, Y.; Moritomo, A.; Shirakami, S.; Nakajima, Y.; Nakai, K.; Nomura, N.; Ito, M.; Higashi, Y.; Inoue, T. Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor. Bioorg. Med. Chem. 2018, 26, 4971–4983. [Google Scholar] [CrossRef] [PubMed]
- Wrobleski, S.T.; Moslin, R.; Lin, S.; Zhang, Y.; Spergel, S.; Kempson, J.; Tokarski, J.S.; Strnad, J.; Zupa-Fernandez, A.; Cheng, L.; et al. Highly Selective Inhibition of Tyrosine Kinase 2 (TYK2) for the Treatment of Autoimmune Diseases: Discovery of the Allosteric Inhibitor BMS-986165. J. Med. Chem. 2019, 62, 8973–8995. [Google Scholar] [CrossRef]
- Lupardus, P.J.; Skiniotis, G.; Rice, A.J.; Thomas, C.; Fischer, S.; Walz, T.; Garcia, K.C. Structural snapshots of full-length Jak1, a transmembrane gp130/IL-6/IL-6Ralpha cytokine receptor complex, and the receptor-Jak1 holocomplex. Structure 2011, 19, 45–55. [Google Scholar] [CrossRef]
- Hammaren, H.M.; Ungureanu, D.; Grisouard, J.; Skoda, R.C.; Hubbard, S.R.; Silvennoinen, O. ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation. Proc. Natl. Acad. Sci. USA 2015, 112, 4642–4647. [Google Scholar] [CrossRef]
- Ungureanu, D.; Wu, J.; Pekkala, T.; Niranjan, Y.; Young, C.; Jensen, O.N.; Xu, C.F.; Neubert, T.A.; Skoda, R.C.; Hubbard, S.R.; et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat. Struct. Mol. Biol. 2011, 18, 971–976. [Google Scholar] [CrossRef]
- Canté-Barrett, K.; Uitdehaag, J.C.; Meijerink, J.P. Structural modeling of JAK1 mutations in T-cell acute lymphoblastic leukemia reveals a second contact site between pseudokinase and kinase domains. Haematologica 2016, 101, e189–e191. [Google Scholar] [CrossRef]
- Raivola, J.; Haikarainen, T.; Silvennoinen, O. Characterization of JAK1 Pseudokinase Domain in Cytokine Signaling. Cancers 2019, 12, 78. [Google Scholar] [CrossRef]
- Degryse, S.; de Bock, C.E.; Cox, L.; Demeyer, S.; Gielen, O.; Mentens, N.; Jacobs, K.; Geerdens, E.; Gianfelici, V.; Hulselmans, G.; et al. JAK3 mutants transform hematopoietic cells through JAK1 activation, causing T-cell acute lymphoblastic leukemia in a mouse model. Blood 2014, 124, 3092–3100. [Google Scholar] [CrossRef] [PubMed]
- Albacker, L.A.; Wu, J.; Smith, P.; Warmuth, M.; Stephens, P.J.; Zhu, P.; Yu, L.; Chmielecki, J. Loss of function JAK1 mutations occur at high frequency in cancers with microsatellite instability and are suggestive of immune evasion. PLoS ONE 2017, 12, e0176181. [Google Scholar] [CrossRef] [PubMed]
- Cornish, A.L.; Chong, M.M.; Davey, G.M.; Darwiche, R.; Nicola, N.A.; Hilton, D.J.; Kay, T.W.; Starr, R.; Alexander, W.S. Suppressor of cytokine signaling-1 regulates signaling in response to interleukin-2 and other gamma c-dependent cytokines in peripheral T cells. J. Biol. Chem. 2003, 278, 22755–22761. [Google Scholar] [CrossRef]
- Calabrese, V.; Mallette, F.A.; Deschenes-Simard, X.; Ramanathan, S.; Gagnon, J.; Moores, A.; Ilangumaran, S.; Ferbeyre, G. SOCS1 links cytokine signaling to p53 and senescence. Mol. Cell 2009, 36, 754–767. [Google Scholar] [CrossRef]
- Toms, A.V.; Deshpande, A.; McNally, R.; Jeong, Y.; Rogers, J.M.; Kim, C.U.; Gruner, S.M.; Ficarro, S.B.; Marto, J.A.; Sattler, M.; et al. Structure of a pseudokinase-domain switch that controls oncogenic activation of Jak kinases. Nat. Struct. Mol. Biol. 2013, 20, 1221–1223. [Google Scholar] [CrossRef] [PubMed]
- Min, X.; Ungureanu, D.; Maxwell, S.; Hammaren, H.; Thibault, S.; Hillert, E.K.; Ayres, M.; Greenfield, B.; Eksterowicz, J.; Gabel, C.; et al. Structural and Functional Characterization of the JH2 Pseudokinase Domain of JAK Family Tyrosine Kinase 2 (TYK2). J. Biol. Chem. 2015, 290, 27261–27270. [Google Scholar] [CrossRef]
- Fabbro, D.; Cowan-Jacob, S.W.; Moebitz, H. Ten things you should know about protein kinases: IUPHAR Review 14. Br. J. Pharmacol. 2015, 172, 2675–2700. [Google Scholar] [CrossRef]
- Babon, J.J.; Lucet, I.S.; Murphy, J.M.; Nicola, N.A.; Varghese, L.N. The molecular regulation of Janus kinase (JAK) activation. Biochem. J. 2014, 462, 1–13. [Google Scholar] [CrossRef]
- Varghese, L.N.; Ungureanu, D.; Liau, N.P.; Young, S.N.; Laktyushin, A.; Hammaren, H.; Lucet, I.S.; Nicola, N.A.; Silvennoinen, O.; Babon, J.J.; et al. Mechanistic insights into activation and SOCS3-mediated inhibition of myeloproliferative neoplasm-associated JAK2 mutants from biochemical and structural analyses. Biochem. J. 2014, 458, 395–405. [Google Scholar] [CrossRef]
- Brooks, A.J.; Dai, W.; O'Mara, M.L.; Abankwa, D.; Chhabra, Y.; Pelekanos, R.A.; Gardon, O.; Tunny, K.A.; Blucher, K.M.; Morton, C.J.; et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 2014, 344, 1249783. [Google Scholar] [CrossRef] [PubMed]
- Hammarén, H.M.; Virtanen, A.T.; Silvennoinen, O. Nucleotide-binding mechanisms in pseudokinases. Biosci. Rep. 2015, 36, e00282. [Google Scholar] [CrossRef]
- Hari, S.B.; Merritt, E.A.; Maly, D.J. Sequence determinants of a specific inactive protein kinase conformation. Chem. Biol. 2013, 20, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Leung, M.-Y.; Knapka, J.A.; Wagler, A.E.; Rodriguez, G.; Kirken, R.A. OncoMiner: A Pipeline for Bioinformatics Analysis of Exonic Sequence Variants in Cancer. In Big Data Analytics in Genomics; Wong, K.-C., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 373–396. [Google Scholar]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Mitra, A.; Ross, J.A.; Rodriguez, G.; Nagy, Z.S.; Wilson, H.L.; Kirken, R.A. Signal transducer and activator of transcription 5b (Stat5b) serine 193 is a novel cytokine-induced phospho-regulatory site that is constitutively activated in primary hematopoietic malignancies. J. Biol. Chem. 2012, 287, 16596–16608. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.A.; Rodriguez, G.; Kirken, R.A. Analysis of Janus tyrosine kinase phosphorylation and activation. Methods Mol. Biol. 2013, 967, 3–20. [Google Scholar] [CrossRef]
- Cheng, H.; Ross, J.A.; Frost, J.A.; Kirken, R.A. Phosphorylation of human Jak3 at tyrosines 904 and 939 positively regulates its activity. Mol. Cell. Biol. 2008, 28, 2271–2282. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grant, A.H.; Rodriguez, A.C.; Rodriguez Moncivais, O.J.; Sun, S.; Li, L.; Mohl, J.E.; Leung, M.-Y.; Kirken, R.A.; Rodriguez, G. JAK1 Pseudokinase V666G Mutant Dominantly Impairs JAK3 Phosphorylation and IL-2 Signaling. Int. J. Mol. Sci. 2023, 24, 6805. https://doi.org/10.3390/ijms24076805
Grant AH, Rodriguez AC, Rodriguez Moncivais OJ, Sun S, Li L, Mohl JE, Leung M-Y, Kirken RA, Rodriguez G. JAK1 Pseudokinase V666G Mutant Dominantly Impairs JAK3 Phosphorylation and IL-2 Signaling. International Journal of Molecular Sciences. 2023; 24(7):6805. https://doi.org/10.3390/ijms24076805
Chicago/Turabian StyleGrant, Alice H., Alejandro C. Rodriguez, Omar J. Rodriguez Moncivais, Shengjie Sun, Lin Li, Jonathon E. Mohl, Ming-Ying Leung, Robert A. Kirken, and Georgialina Rodriguez. 2023. "JAK1 Pseudokinase V666G Mutant Dominantly Impairs JAK3 Phosphorylation and IL-2 Signaling" International Journal of Molecular Sciences 24, no. 7: 6805. https://doi.org/10.3390/ijms24076805
APA StyleGrant, A. H., Rodriguez, A. C., Rodriguez Moncivais, O. J., Sun, S., Li, L., Mohl, J. E., Leung, M.-Y., Kirken, R. A., & Rodriguez, G. (2023). JAK1 Pseudokinase V666G Mutant Dominantly Impairs JAK3 Phosphorylation and IL-2 Signaling. International Journal of Molecular Sciences, 24(7), 6805. https://doi.org/10.3390/ijms24076805








