Interferons and Resistance Mechanisms in Tumors and Pathogen-Driven Diseases—Focus on the Major Histocompatibility Complex (MHC) Antigen Processing Pathway
Abstract
1. Important Features of the Family of Interferons
2. Interferon Signal Transduction Pathways and Their Components
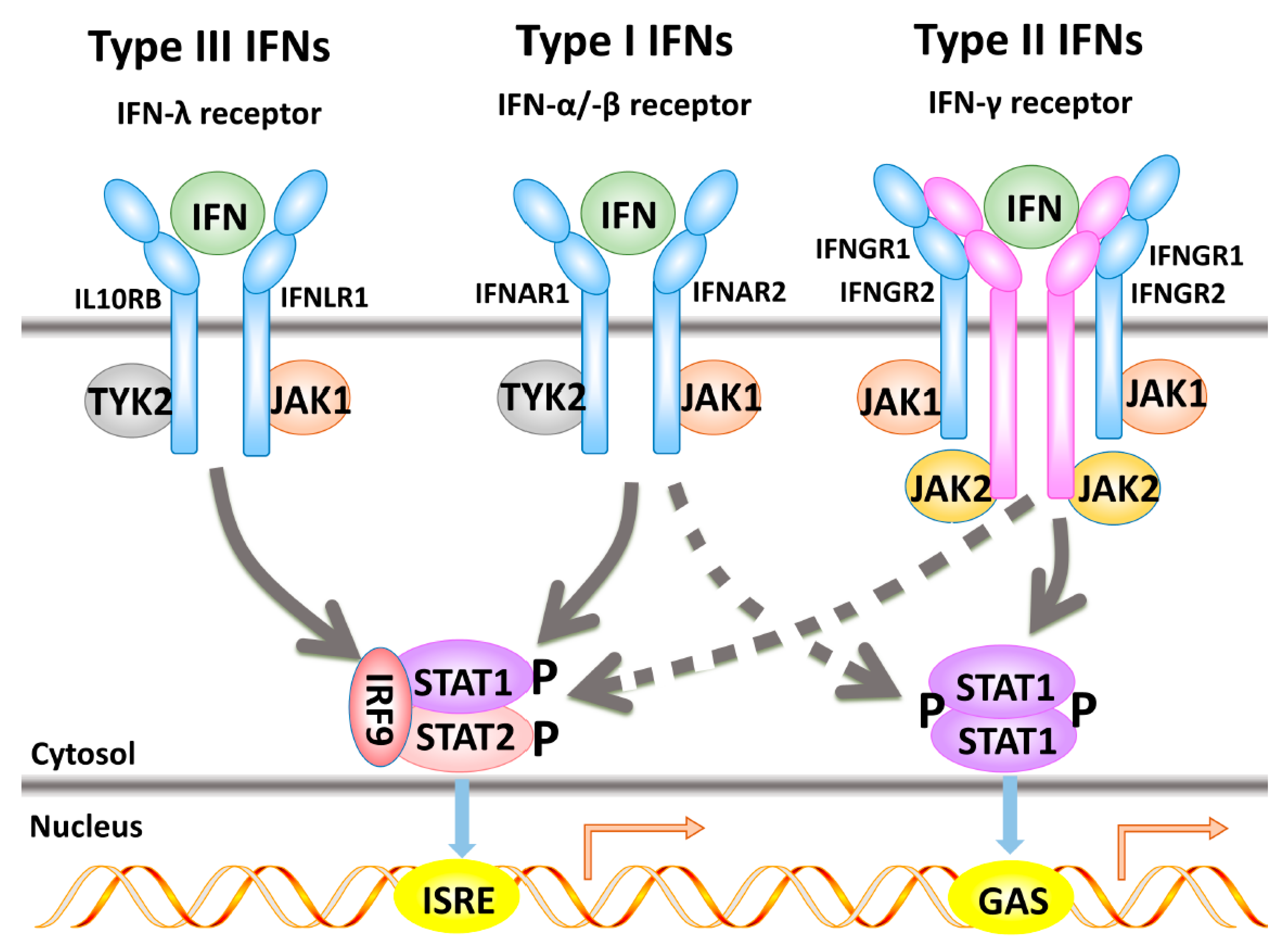
2.1. Type I Interferon-α-Induced Signal Transduction Pathways
2.2. Type II Interferon-Induced Signal Transduction Cascade
2.3. Type III-Induced Interferon Signal Transduction
2.4. Interferon Regulatory Factors
3. The Major Histocompatibility Class I and Class II Antigen Processing and Presentation Pathways
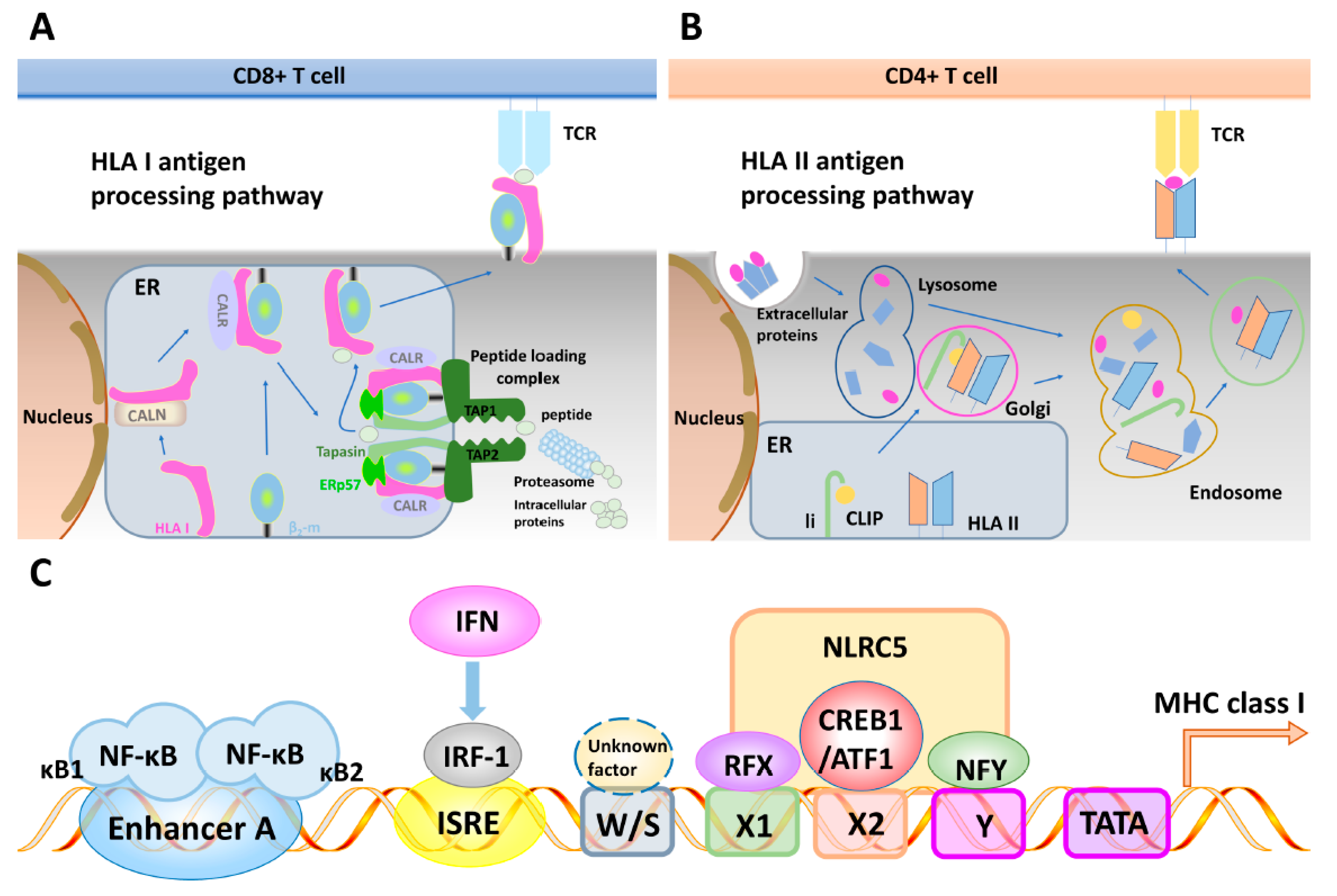
3.1. General Features of the MHC Antigen Processing Machinery
3.2. Regulatory Elements of the Major Histocompatibility Complex Class I and II Antigen Processing Machinery Promoters and Their Interferon Inducibility
3.3. Mechanisms of Cancer Cell Immune Evasion Mediated by Impaired Major Histocompatibility Class I and Antigen Processing Machinery Component Expression
4. Defects in the Interferon Pathways
4.1. Frequency of Defective IFN Inducibility of APM Components in Tumor Cells
4.2. Mechanisms of Defective Interferon Inducibility of the Expression of Major Histocompatibility Complex Class I Antigen Processing Machinery Components in Tumors
4.3. Involvement of the Interferon Pathways in Tumor Surveillance In Vivo
4.4. Effect of Interferon Signaling on the Expression of Major Histocompatibility Complex Class II Components in Tumors
4.5. Inborn Errors of Type I Interferon Immunity and Their Phenocopies among Humans
4.6. Inborn Errors of Type II Interferon Immunity
4.7. Human Autosomal Recessive Complete STAT1 and STAT2 Deficiencies
4.8. Major Histocompatibility Complex Deficiencies in the Context of Infection and Tumors
5. Clinical Relevance of Interferons
5.1. Clinical Relevance of Aberrant Interferon Signaling in Tumors and Upon Viral Infection
5.2. Other Clinical Applications of Interferons
5.3. Immunotherapy Resistance—Association with Defects in the IFN Pathway
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Isaacs, A.; Lindenmann, J. Virus interference. I. The interferon. By A. Isaacs and J. Lindenmann, 1957. J. Interferon Res. 1987, 7, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Amadori, M. The role of IFN-alpha as homeostatic agent in the inflammatory response: A balance between danger and response? J. Interferon Cytokine Res. 2007, 27, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Baccala, R.; Kono, D.H.; Theofilopoulos, A.N. Interferons as pathogenic effectors in autoimmunity. Immunol. Rev. 2005, 204, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Theofilopoulos, A.N.; Baccala, R.; Beutler, B.; Kono, D.H. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 2005, 23, 307–335. [Google Scholar] [CrossRef]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef] [PubMed]
- Barajas, J.; Caballero, K.L.; Rodriguez, O.; Radeva, P. Cardiac Phase Extraction in IVUS Sequences using 1-D Gabor Filters. In Proceedings of the 29th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Lyon, France, 22–26 August 2007; pp. 343–346. [Google Scholar] [CrossRef]
- Snell, L.M.; McGaha, T.L.; Brooks, D.G. Type I Interferon in Chronic Virus Infection and Cancer. Trends Immunol. 2017, 38, 542–557. [Google Scholar] [CrossRef]
- Decker, T.; Müller, M.; Stockinger, S. The Yin and Yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 2005, 5, 675–687. [Google Scholar] [CrossRef]
- Sheppard, P.; Kindsvogel, W.; Xu, W.; Henderson, K.; Schlutsmeyer, S.; Whitmore, T.E.; Kuestner, R.; Garrigues, U.; Birks, C.; Roraback, J.; et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 2003, 4, 63–68. [Google Scholar] [CrossRef]
- Kotenko, S.V.; Gallagher, G.; Baurin, V.V.; Lewis-Antes, A.; Shen, M.; Shah, N.K.; Langer, J.A.; Sheikh, F.; Dickensheets, H.; Donnelly, R.P. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 2003, 4, 69–77. [Google Scholar] [CrossRef]
- Wack, A.; Terczyńska-Dyla, E.; Hartmann, R. Guarding the frontiers: The biology of type III interferons. Nat. Immunol. 2015, 16, 802–809. [Google Scholar] [CrossRef]
- Uzé, G.; Monneron, D. IL-28 and IL-29: Newcomers to the interferon family. Biochimie 2007, 89, 729–734. [Google Scholar] [CrossRef]
- Kotenko, S.V. IFN-lambdas. Curr. Opin. Immunol. 2011, 23, 583–590. [Google Scholar] [CrossRef]
- Osterlund, P.I.; Pietilä, T.E.; Veckman, V.; Kotenko, S.V.; Julkunen, I. IFN regulatory factor family members differentially regulate the expression of type III IFN (IFN-lambda) genes. J. Immunol. 2007, 179, 3434–3442. [Google Scholar] [CrossRef]
- Trinchieri, G. Cytokines and cytokine receptors. Immunol. Rev. 2004, 202, 5–7. [Google Scholar] [CrossRef]
- Schroder, K.; Hertzog, P.J.; Ravasi, T.; Hume, D.A. Interferon-gamma: An overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004, 75, 163–189. [Google Scholar] [CrossRef]
- Walter, M.R. The Role of Structure in the Biology of Interferon Signaling. Front. Immunol. 2020, 11, 606489. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Kerr, I.M.; Williams, B.R.G.; Silverman, R.H.; Schreiber, R.D. HOW CELLS RESPOND TO INTERFERONS. Annu. Rev. Biochem. 1998, 67, 227–264. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef]
- Li, Z.; Metze, D.; Nashan, D.; Müller-Tidow, C.; Serve, H.L.; Poremba, C.; Luger, T.A.; Böhm, M. Expression of SOCS-1, suppressor of cytokine signalling-1, in human melanoma. J. Investig. Dermatol. 2004, 123, 737–745. [Google Scholar] [CrossRef]
- García-Sastre, A. Ten Strategies of Interferon Evasion by Viruses. Cell Host Microbe 2017, 22, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, T.H. IRF and STAT Transcription Factors—From Basic Biology to Roles in Infection, Protective Immunity, and Primary Immunodeficiencies. Front. Immunol. 2018, 9, 3047. [Google Scholar] [CrossRef] [PubMed]
- Ramana, C.V.; Gil, M.P.; Schreiber, R.D.; Stark, G.R. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002, 23, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Platanias, L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef]
- Yu, H.; Jove, R. The STATs of cancer—New molecular targets come of age. Nat. Rev. Cancer 2004, 4, 97–105. [Google Scholar] [CrossRef]
- Shin-Ya, M.; Hirai, H.; Satoh, E.; Kishida, T.; Asada, H.; Aoki, F.; Tsukamoto, M.; Imanishi, J.; Mazda, O. Intracellular interferon triggers Jak/Stat signaling cascade and induces p53-dependent antiviral protection. Biochem. Biophys. Res. Commun. 2005, 329, 1139–1146. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Zhou, Z.; Hamming, O.J.; Ank, N.; Paludan, S.R.; Nielsen, A.L.; Hartmann, R. Type III Interferon (IFN) Induces a Type I IFN-Like Response in a Restricted Subset of Cells through Signaling Pathways Involving both the Jak-STAT Pathway and the Mitogen-Activated Protein Kinases. J. Virol. 2007, 81, 7749–7758. [Google Scholar] [CrossRef]
- Kwon, M.J.; Yao, Y.; Walter, M.J.; Holtzman, M.J.; Chang, C.H. Role of PKCdelta in IFN-gamma-inducible CIITA gene expression. Mol. Immunol. 2007, 44, 2841–2849. [Google Scholar] [CrossRef]
- Lazear, H.M.; Nice, T.; Diamond, M.S. Interferon-lambda: Immune Functions at Barrier Surfaces and Beyond. Immunity 2015, 43, 15–28. [Google Scholar] [CrossRef]
- Koch, S.; Finotto, S. Role of Interferon-lambda in Allergic Asthma. J. Innate Immun. 2015, 7, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Lew, D.; Decker, T.; Kessler, D.; Darnell, J. Synergistic interaction between interferon-alpha and interferon-gamma through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 1990, 9, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu, S.N.; Croze, E.; Murti, A.; Wang, C.; Basu, L.; Hollander, D.; Russell-Harde, D.; Betts, M.; Garcia-Martinez, V.; Mullersman, J.E. Expression and signaling specificity of the IFNAR chain of the type I interferon receptor complex. Proc. Natl. Acad. Sci. USA 1995, 92, 10487–10491. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Caraglia, M.; Marra, M.; Pelaia, G.; Maselli, R.; Caputi, M.; Marsico, S.A.; Abbruzzese, A. Alpha-interferon and its effects on signal transduction pathways. J. Cell. Physiol. 2004, 202, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Uddin, S.; Lekmine, F.; Sassano, A.; Rui, H.; Fish, E.N.; Platanias, L.C. Role of Stat5 in Type I interferon-signaling and transcriptional regulation. Biochem. Biophys. Res. Commun. 2003, 308, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, Y.; Nishibori, T.; Su, L.; Arduini, R.M.; Baker, D.P.; David, M. Cutting edge: Role of STAT1, STAT3, and STAT5 in IFN-alpha beta responses in T lymphocytes. J. Immunol. 2005, 174, 609–613. [Google Scholar] [CrossRef]
- Zanin, N.; de Lesegno, C.V.; Lamaze, C.; Blouin, C.M. Interferon Receptor Trafficking and Signaling: Journey to the Cross Roads. Front. Immunol. 2021, 11, 615603. [Google Scholar] [CrossRef]
- Ivashkiv, L.B. IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558. [Google Scholar] [CrossRef]
- Ramsauer, K.; Sadzak, I.; Porras, A.; Pilz, A.; Nebreda, A.R.; Decker, T.; Kovarik, P. p38 MAPK enhances STAT1-dependent transcription independently of Ser-727 phosphorylation. Proc. Natl. Acad. Sci. USA 2002, 99, 12859–12864. [Google Scholar] [CrossRef]
- Harada, H.; Fujita, T.; Miyamoto, M.; Kimura, Y.; Maruyama, M.; Furia, A.; Miyata, T.; Taniguchi, T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 1989, 58, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Meinke, A.; Barahmand-Pour, F.; Wöhrl, S.; Stoiber, D.; Decker, T. Activation of different Stat5 isoforms contributes to cell-type-restricted signaling in response to interferons. Mol. Cell. Biol. 1996, 16, 6937–6944. [Google Scholar] [CrossRef] [PubMed]
- Woldman, I.; Varinou, L.; Ramsauer, K.; Rapp, B.; Decker, T. The Stat1 binding motif of the interferon-gamma receptor is sufficient to mediate Stat5 activation and its repression by SOCS3. J. Biol. Chem. 2001, 276, 45722–45728. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; MacDuff, D.A.; Imanaka, N.; Gainey, M.D.; Shrestha, B.; Eitson, J.L.; Mar, K.B.; Richardson, R.B.; Ratushny, A.V.; Litvak, V.; et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014, 505, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Recent advances in antiviral interferon-stimulated gene biology [version 1; peer review: 2 approved]. F1000Research 2018, 7, 309. [Google Scholar] [CrossRef] [PubMed]
- Odendall, C.; Kagan, J.C. The unique regulation and functions of type III interferons in antiviral immunity. Curr. Opin. Virol. 2015, 12, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.W.; Forero, A. Beyond Good and Evil: Molecular Mechanisms of Type I and III IFN Functions. J. Immunol. 2022, 208, 247–256. [Google Scholar] [CrossRef]
- Ozato, K.; Tailor, P.; Kubota, T. The Interferon Regulatory Factor Family in Host Defense: Mechanism of Action. J. Biol. Chem. 2007, 282, 20065–20069. [Google Scholar] [CrossRef]
- Negishi, H.; Taniguchi, T.; Yanai, H. The Interferon (IFN) Class of Cytokines and the IFN Regulatory Factor (IRF) Transcription Factor Family. Cold Spring Harb. Perspect. Biol. 2017, 10, a028423. [Google Scholar] [CrossRef]
- Feng, H.; Zhang, Y.-B.; Gui, J.-F.; Lemon, S.M.; Yamane, D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 2021, 17, e1009220. [Google Scholar] [CrossRef]
- Dersh, D.; Hollý, J.; Yewdell, J.W. A few good peptides: MHC class I-based cancer immunosurveillance and immunoevasion. Nat. Rev. Immunol. 2020, 21, 116–128. [Google Scholar] [CrossRef] [PubMed]
- Boon, T.; Coulie, P.G.; van den Eynde, B. Tumor antigens recognized by T cells. Immunol. Today 1997, 18, 267–268. [Google Scholar] [CrossRef] [PubMed]
- Jongsma, M.; Neefjes, J.; Spaapen, R. Playing hide and seek: Tumor cells in control of MHC class I antigen presentation. Mol. Immunol. 2021, 136, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Rock, K.L.; York, I.; Goldberg, A.L. Post-proteasomal antigen processing for major histocompatibility complex class I presentation. Nat. Immunol. 2004, 5, 670–677. [Google Scholar] [CrossRef]
- Kloetzel, P.M. Generation of major histocompatibility complex class I antigens: Functional interplay between proteasomes and TPPII. Nat. Immunol. 2004, 5, 661–669. [Google Scholar] [CrossRef]
- Abele, R.; Tampé, R. Modulation of the antigen transport machinery TAP by friends and enemies. FEBS Lett. 2005, 580, 1156–1163. [Google Scholar] [CrossRef]
- Chang, S.C.; Momburg, F.; Bhutani, N.; Goldberg, A.L. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc. Natl. Acad. Sci. USA 2005, 102, 17107–17112. [Google Scholar] [CrossRef]
- Blees, A.; Januliene, D.; Hofmann, T.; Koller, N.; Schmidt, C.; Trowitzsch, S.; Moeller, A.; Tampé, R. Structure of the human MHC-I peptide-loading complex. Nature 2017, 551, 525–528. [Google Scholar] [CrossRef]
- Cresswell, P.; Bangia, N.; Dick, T.; Diedrich, G. The nature of the MHC class I peptide loading complex. Immunol. Rev. 1999, 172, 21–28. [Google Scholar] [CrossRef]
- Jiang, J.; Taylor, D.K.; Kim, E.J.; Boyd, L.F.; Ahmad, J.; Mage, M.G.; Truong, H.V.; Woodward, C.H.; Sgourakis, N.G.; Cresswell, P.; et al. Structural mechanism of tapasin-mediated MHC-I peptide loading in antigen presentation. Nat. Commun. 2022, 13, 1–13. [Google Scholar] [CrossRef]
- Cox, J.H.; Yewdell, J.W.; Eisenlohr, L.C.; Johnson, P.R.; Bennink, J.R. Antigen Presentation Requires Transport of MHC Class I Molecules from the Endoplasmic Reticulum. Science 1990, 247, 715–718. [Google Scholar] [CrossRef]
- Jensen, P.E. Recent advances in antigen processing and presentation. Nat. Immunol. 2007, 8, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Seliger, B.S.; Ferrone, H.L. A Class I Antigen Processing Machinery Defects in Cancer Cells-Frequency, Functional Significance, and Clinical Relevance with Special Emphasis on Their Role in T Cell-Based Immunotherapy of Malignant Disease. Methods Mol. Biol. 2020, 2055, 325–350. [Google Scholar]
- Seliger, B.; Kloor, M.; Ferrone, S. HLA class II antigen-processing pathway in tumors: Molecular defects and clinical relevance. Oncoimmunology 2017, 6, e1171447. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.Y.; Kwek, S.S.; Raju, S.S.; Li, T.; McCarthy, E.; Chow, E.; Aran, D.; Ilano, A.; Pai, C.-C.S.; Rancan, C.; et al. Intratumoral CD4+ T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell 2020, 181, 1612–1625.e13. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, M.L.; Cook, R.S.; Johnson, D.B.; Balko, J.M. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin. Cancer Res. 2019, 25, 2392–2402. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, S.; Zhang, X.; Jia, K.; Deng, J.; Zhou, C.; He, Y. Major histocompatibility complex class II molecule in non-small cell lung cancer diagnosis, prognosis and treatment. OncoTargets Ther. 2019, 12, 7281–7288. [Google Scholar] [CrossRef] [PubMed]
- Hearing, M. Prefrontal-accumbens opioid plasticity: Implications for relapse and dependence. Pharmacol. Res. 2018, 139, 158–165. [Google Scholar] [CrossRef]
- Buetow, K.H.; Meador, L.R.; Menon, H.; Lu, Y.-K.; Brill, J.; Cui, H.; Roe, D.J.; DiCaudo, D.J.; Hastings, K.T. High GILT Expression and an Active and Intact MHC Class II Antigen Presentation Pathway Are Associated with Improved Survival in Melanoma. J. Immunol. 2019, 203, 2577–2587. [Google Scholar] [CrossRef]
- Kim, A.; Sadegh-Nasseri, S. Determinants of immunodominance for CD4 T cells. Curr. Opin. Immunol. 2015, 34, 9–15. [Google Scholar] [CrossRef]
- Landsverk, O.J.B.; Bakke, O.; Gregers, T.F. MHC II and the Endocytic Pathway: Regulation by Invariant Chain. Scand. J. Immunol. 2009, 70, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Cresswell, P.; Roche, P. Invariant chain–MHC class II complexes: Always odd and never invariant. Immunol. Cell Biol. 2014, 92, 471–472. [Google Scholar] [CrossRef] [PubMed]
- Rocha, N.; Neefjes, J. MHC class II molecules on the move for successful antigen presentation. EMBO J. 2007, 27, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kropshofer, H.; Vogt, A.B.; Moldenhauer, G.; Hammer, J.; Blum, J.S.; Hämmerling, G.J. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 1996, 15, 6144–6154. [Google Scholar] [CrossRef] [PubMed]
- Roche, P.A.; Furuta, K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat. Rev. Immunol. 2015, 15, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Wearsch, P.A.; Cresswell, P. Pathways of Antigen Processing. Annu. Rev. Immunol. 2013, 31, 443–473. [Google Scholar] [CrossRef]
- Dengjel, J.; Schoor, O.; Fischer, R.; Reich, M.; Kraus, M.; Müller, M.; Kreymborg, K.; Altenberend, F.; Brandenburg, J.; Kalbacher, H.; et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 7922–7927. [Google Scholar] [CrossRef]
- Schmid, D.; Pypaert, M.; Münz, C. Antigen-Loading Compartments for Major Histocompatibility Complex Class II Molecules Continuously Receive Input from Autophagosomes. Immunity 2007, 26, 79–92. [Google Scholar] [CrossRef]
- Howcroft, T.K.; Raval, A.; Weissman, J.D.; Gegonne, A.; Singer, D.S. Distinct Transcriptional Pathways Regulate Basal and Activated Major Histocompatibility Complex Class I Expression. Mol. Cell. Biol. 2003, 23, 3377–3391. [Google Scholar] [CrossRef]
- Wright, K.L.; White, L.C.; Kelly, A.; Beck, S.; Trowsdale, J.; Ting, J.P. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J. Exp. Med. 1995, 181, 1459–1471. [Google Scholar] [CrossRef]
- Bukur, J.; Herrmann, F.; Handke, D.; Recktenwald, C.; Seliger, B. Identification of E2F1 as an Important Transcription Factor for the Regulation of Tapasin Expression. J. Biol. Chem. 2010, 285, 30419–30426. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, D.; Pützer, B.M.; Lipinski, K.S.; Schmücker, U.; Esche, H. A multiprotein complex consisting of the cellular coactivator p300, AP-1/ATF, as well as NF-kappaB is responsible for the activation of the mouse major histocompatibility class I (H-2K(b)) enhancer A. Gene Expr. 1999, 8, 1–18. [Google Scholar] [PubMed]
- van den Elsen, P.J. Expression Regulation of Major Histocompatibility Complex Class I and Class II Encoding Genes. Front. Immunol. 2011, 2, 48. [Google Scholar] [CrossRef] [PubMed]
- Jongsma, M.L.; Guarda, G.; Spaapen, R.M. The regulatory network behind MHC class I expression. Mol. Immunol. 2019, 113, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee-Kishore, M.; Kishore, R.; Hicklin, D.J.; Marincola, F.M.; Ferrone, S. Different Requirements for Signal Transducer and Activator of Transcription 1α and Interferon Regulatory Factor 1 in the Regulation of Low Molecular Mass Polypeptide 2 and Transporter Associated with Antigen Processing 1 Gene Expression. J. Biol. Chem. 1998, 273, 16177–16183. [Google Scholar] [CrossRef] [PubMed]
- Brucet, M.; Marqués, L.; Sebastián, C.; Lloberas, J.; Celada, A. Regulation of murine Tap1 and Lmp2 genes in macrophages by interferon gamma is mediated by STAT1 and IRF-1. Genes Immun. 2004, 5, 26–35. [Google Scholar] [CrossRef]
- Dimasuay, K.G.; Sanchez, A.; Schaefer, N.; Polanco, J.; Ferrington, D.; Chu, H.W. Immunoproteasomes as a novel antiviral mechanism in rhinovirus-infected airways. Clin. Sci. 2018, 132, 1711–1723. [Google Scholar] [CrossRef]
- Benhammadi, M.; Mathé, J.; Dumont-Lagacé, M.; Kobayashi, K.S.; Gaboury, L.; Brochu, S.; Perreault, C. IFN-lambda Enhances Constitutive Expression of MHC Class I Molecules on Thymic Epithelial Cells. J. Immunol. 2020, 205, 1268–1280. [Google Scholar] [CrossRef]
- Ting, J.P.-Y.; Trowsdale, J. Genetic Control of MHC Class II Expression. Cell 2002, 109, S21–S33. [Google Scholar] [CrossRef]
- van den Elsen, P.J.; Holling, T.M.; Kuipers, H.F.; van der Stoep, N. Transcriptional regulation of antigen presentation. Curr. Opin. Immunol. 2004, 16, 67–75. [Google Scholar] [CrossRef]
- Hake, S.B.; Masternak, K.; Kammerbauer, C.; Janzen, C.; Reith, W.; Steimle, V. CIITA Leucine-Rich Repeats Control Nuclear Localization, In Vivo Recruitment to the Major Histocompatibility Complex (MHC) Class II Enhanceosome, and MHC Class II Gene Transactivation. Mol. Cell. Biol. 2000, 20, 7716–7725. [Google Scholar] [CrossRef] [PubMed]
- Mühlethaler-Mottet, A.; Otten, L.A.; Steimle, V.; Mach, B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997, 16, 2851–2860. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, K.; Sisk, T.J.; Inohara, N.; Yee, C.S.; Kennell, J.; Cho, M.-C.; Yannie, P.J.; Núñez, G.; Chang, C.-H. Dendritic Cell-specific MHC Class II Transactivator Contains a Caspase Recruitment Domain That Confers Potent Transactivation Activity. J. Biol. Chem. 2001, 276, 19089–19093. [Google Scholar] [CrossRef] [PubMed]
- Muhlethaler-Mottet, A.; Di Berardino, W.; Otten, L.A.; Mach, B. Activation of the MHC class II transactivator CIITA by interferon-gamma requires cooperative interaction between Stat1 and USF-1. Immunity 1998, 8, 157–166. [Google Scholar] [CrossRef]
- Dong, Y.; Rohn, W.M.; Benveniste, E. N IFN-gamma regulation of the type IV class II transactivator promoter in astrocytes. J. Immunol. 1999, 162, 4731–4739. [Google Scholar] [CrossRef]
- Londhe, M.; Davie, J.K. Gamma interferon modulates myogenesis through the major histocompatibility complex class II transactivator, CIITA. Mol. Cell. Biol. 2011, 31, 2854–2866. [Google Scholar] [CrossRef]
- Chen, H.; Gilbert, C.A.; Hudson, J.A.; Bolick, S.C.; Wright, K.L.; Piskurich, J.F. Positive regulatory domain I-binding factor 1 mediates repression of the MHC class II transactivator (CIITA) type IV promoter. Mol. Immunol. 2007, 44, 1461–1470. [Google Scholar] [CrossRef]
- Lapierre, L.A.; Fiers, W.; Pober, J.S. Three distinct classes of regulatory cytokines control endothelial cell MHC antigen expression. Interactions with immune gamma interferon differentiate the effects of tumor necrosis factor and lymphotoxin from those of leukocyte alpha and fibroblast beta interferons. J. Exp. Med. 1988, 167, 794–804. [Google Scholar] [CrossRef]
- Drukker, M.; Katz, G.; Urbach, A.; Schuldiner, M.; Markel, G.; Itskovitz-Eldor, J.; Reubinoff, B.; Mandelboim, O.; Benvenisty, N. Characterization of the expression of MHC proteins in human embryonic stem cells. Proc. Natl. Acad. Sci. USA 2002, 99, 9864–9869. [Google Scholar] [CrossRef]
- Keller, A.D.; Maniatis, T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991, 5, 868–879. [Google Scholar] [CrossRef]
- Elias, S.; Robertson, E.J.; Bikoff, E.K.; Mould, A.W. Blimp-1/PRDM1 is a critical regulator of Type III Interferon responses in mammary epithelial cells. Sci. Rep. 2018, 8, 237. [Google Scholar] [CrossRef] [PubMed]
- Garrido, F.; Algarra, I. MHC antigens and tumor escape from immune surveillance. Adv. Cancer Res. 2001, 83, 117–158. [Google Scholar] [CrossRef] [PubMed]
- Aptsiauri, N.; Garrido, F. The Challenges of HLA Class I Loss in Cancer Immunotherapy: Facts and Hopes. Clin. Cancer Res. 2022, 28, 5021–5029. [Google Scholar] [CrossRef]
- Paschen, A.; Méndez, R.M.; Jimenez, P.; Sucker, A.; Ruiz-Cabello, F.; Song, M.; Garrido, F.; Schadendorf, D. Complete loss of HLA class I antigen expression on melanoma cells: A result of successive mutational events. Int. J. Cancer 2002, 103, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Ramal, L.M.; Maleno, I.; Cabrera, T.; Collado, A.; Ferron, A.; Lopez-Nevot, M.A.; Garrido, F. Molecular strategies to define HLA haplotype loss in microdissected tumor cells. Hum. Immunol. 2000, 61, 1001–1012. [Google Scholar] [CrossRef]
- Jiménez, P.; Cabrera, T.; Méndez, R.; Esparza, C.; Cózar, J.; Tallada, M.; López-Nevot, M.A.; Ruiz-Cabello, F.; Garrido, F. A nucleotide insertion in exon 4 is responsible for the absence of expression of an HLA-A*0301 allele in a prostate carcinoma cell line. Immunogenetics 2001, 53, 606–610. [Google Scholar] [CrossRef]
- Perez, B.; Benitez, R.; Fernandez, M.A.; Oliva, M.R.; Soto, J.L.; Serrano, S.; Lopez Nevot, M.A.; Garrido, F. A new beta 2 microglobulin mutation found in a melanoma tumor cell line. Tissue Antigens 1999, 53, 569–572. [Google Scholar] [CrossRef]
- Maleno, I.; Cabrera, C.M.; Cabrera, T.; Paco, L.; López-Nevot, M.A.; Collado, A.; Garrido, F. Distribution of HLA class I altered phenotypes in colorectal carcinomas: High frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Immunogenetics 2004, 56, 244–253. [Google Scholar] [CrossRef]
- Garrido, F.; Cabrera, T.; Aptsiauri, N. “Hard” and “soft” lesions underlying the HLA class I alterations in cancer cells: Implications for immunotherapy. Int. J. Cancer 2010, 127, 249–256. [Google Scholar] [CrossRef]
- Chang, C.-C.; Campoli, M.; Restifo, N.P.; Wang, X.; Ferrone, S. Immune Selection of Hot-Spot β2-Microglobulin Gene Mutations, HLA-A2 Allospecificity Loss, and Antigen-Processing Machinery Component Down-Regulation in Melanoma Cells Derived from Recurrent Metastases following Immunotherapy. J. Immunol. 2005, 174, 1462–1471. [Google Scholar] [CrossRef]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Seliger, B.; Ritz, U.; Soldano, F. Molecular mechanisms of HLA class I antigen abnormalities following viral infection and transformation. Int. J. Cancer 2005, 118, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.A.; Ruiz-Cabello, F.; Oliva, M.R.; Cabrera, T.; Jimenez, P.; López Nevot, M.A.; Garrido, F. Beta2-microglobulin gene mutation is not a common mechanism of HLA class I total loss in human tumors. Int. J. Clin. Lab. Res. 2000, 30, 87–92. [Google Scholar] [PubMed]
- Galassi, C.; Vitale, I.; Galluzzi, L. Using epigenetic modifiers to target cancer stem cell immunoevasion. Cancer Cell 2021, 39, 1573–1575. [Google Scholar] [CrossRef]
- Jhunjhunwala, S.; Hammer, C.; Delamarre, L. Antigen presentation in cancer: Insights into tumour immunogenicity and immune evasion. Nat. Rev. Cancer 2021, 21, 298–312. [Google Scholar] [CrossRef] [PubMed]
- Tajima, K.; Ito, Y.; Demachi, A.; Nishida, K.; Akatsuka, Y.; Tsujimura, K.; Hida, T.; Morishima, Y.; Kuwano, H.; Mitsudomi, T.; et al. Interferon-gamma differentially regulates susceptibility of lung cancer cells to telomerase-specific cytotoxic T lymphocytes. Int. J. Cancer 2004, 110, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Seliger, B.; Hammers, S.; Höhne, A.; Zeidler, R.; Knuth, A.; Gerharz, C.D.; Huber, C. IFN-gamma-mediated coordinated transcriptional regulation of the human TAP-1 and LMP-2 genes in human renal cell carcinoma. Clin. Cancer Res. 1997, 3, 573–578. [Google Scholar]
- Duncan, C.J.; Randall, R.E.; Hambleton, S. Genetic Lesions of Type I Interferon Signalling in Human Antiviral Immunity. Trends Genet. 2020, 37, 46–58. [Google Scholar] [CrossRef]
- Alspach, E.; Lussier, D.M.; Schreiber, R.D. Interferon gamma and Its Important Roles in Promoting and Inhibiting Spontaneous and Therapeutic Cancer Immunity. Cold Spring Harb Perspect Biol. 2019, 11, a028480. [Google Scholar] [CrossRef]
- Kaplan, D.H.; Shankaran, V.; Dighe, A.S.; Stockert, E.; Aguet, M.; Old, L.J.; Schreiber, R.D. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. USA 1998, 95, 7556–7561. [Google Scholar] [CrossRef]
- Rodriguez, T.; Mendez, R.; Del Campo, A.; Aptsiauri, N.; Martin, J.; Orozco, G.; Pawelec, G.; Schadendorf, D.; Ruiz-Cabello, F.; Garrido, F. Patterns of constitutive and IFN-gamma inducible expression of HLA class II molecules in human melanoma cell lines. Immunogenetics 2007, 59, 123–133. [Google Scholar] [CrossRef]
- Grasso, C.S.; Tsoi, J.; Onyshchenko, M.; Abril-Rodriguez, G.; Ross-Macdonald, P.; Wind-Rotolo, M.; Champhekar, A.; Medina, E.; Torrejon, D.Y.; Shin, D.S.; et al. Conserved Interferon-gamma Signaling Drives Clinical Response to Immune Checkpoint Blockade Therapy in Melanoma. Cancer Cell 2020, 38, 500–515.e3. [Google Scholar] [CrossRef]
- Respa, A.; Bukur, J.; Ferrone, S.; Pawelec, G.; Zhao, Y.; Wang, E.; Marincola, F.M.; Seliger, B. Association of IFN-gamma signal transduction defects with impaired HLA class I antigen processing in melanoma cell lines. Clin. Cancer Res. 2011, 17, 2668–2678. [Google Scholar] [CrossRef]
- Meyer, S.; Handke, D.; Mueller, A.; Biehl, K.; Kreuz, M.; Bukur, J.; Koehl, U.; Lazaridou, M.-F.; Berneburg, M.; Steven, A.; et al. Distinct Molecular Mechanisms of Altered HLA Class II Expression in Malignant Melanoma. Cancers 2021, 13, 3907. [Google Scholar] [CrossRef]
- von Locquenghien, M.; Rozalén, C.; Celià-Terrassa, T. Interferons in cancer immunoediting: Sculpting metastasis and immunotherapy response. J. Clin. Investig. 2021, 131, e143296. [Google Scholar] [CrossRef] [PubMed]
- Abril, E.; Mendez, R.E.; García, A.; Serrano, A.; Cabrera, T.; Garrido, F.; Ruiz-Cabello, F. Characterization of a gastric tumor cell line defective in MHC class I inducibility by both alpha- and gamma-interferon. Tissue Antigens 1996, 47, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Holko, M.; Williams, B.R.; Banerjee, D.; Chadalavada, R.S.; Bourdon, V.; Korkola, J.E.; Motzer, R.J.; Chaganti, R.; Leaman, D.W.; Chawla-Sarkar, M.; et al. Functional Annotation of IFN-α-Stimulated Gene Expression Profiles from Sensitive and Resistant Renal Cell Carcinoma Cell Lines. J. Interf. Cytokine Res. 2006, 26, 534–547. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, T.; Méndez, R.; Del Campo, A.; Jiménez, P.; Aptsiauri, N.; Garrido, F.; Ruiz-Cabello, F. Distinct mechanisms of loss of IFN.N-gamma mediated HLA class I inducibility in two melanoma cell lines. BMC Cancer 2007, 7, 34. [Google Scholar] [CrossRef]
- Hayashi, T.; Kobayashi, Y.; Kohsaka, S.; Sano, K. The mutation in the ATP-binding region of JAK1, identified in human uterine leiomyosarcomas, results in defective interferon-gamma inducibility of TAP1 and LMP2. Oncogene 2006, 25, 4016–4026. [Google Scholar] [CrossRef]
- Kang, Y.H.; Biswas, A.; Field, M.; Snapper, S.B. STAT1 signaling shields T cells from NK cell-mediated cytotoxicity. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Freeman, A.J.; Vervoort, S.J.; Ramsbottom, K.M.; Kelly, M.J.; Michie, J.; Pijpers, L.; Johnstone, R.W.; Kearney, C.J.; Oliaro, J. Natural Killer Cells Suppress T Cell-Associated Tumor Immune Evasion. Cell Rep. 2019, 28, 2784–2794.e5. [Google Scholar] [CrossRef]
- Zhao, W.; Cha, E.N.; Lee, C.; Park, C.Y.; Schindler, C. Stat2-Dependent Regulation of MHC Class II Expression. J. Immunol. 2007, 179, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Moro, H.; Otero, D.C.; Tanabe, Y.; David, M. T Cell-Intrinsic and -Extrinsic Contributions of the IFNAR/STAT1-Axis to Thymocyte Survival. PLoS ONE 2011, 6, e24972. [Google Scholar] [CrossRef] [PubMed]
- Sexl, V.; Kovacic, B.; Piekorz, R.; Moriggl, R.; Stoiber, D.; Hoffmeyer, A.; Liebminger, R.; Kudlacek, O.; Weisz, E.; Rothammer, K.; et al. Jak1 deficiency leads to enhanced Abelson-induced B-cell tumor formation. Blood 2003, 101, 4937–4943. [Google Scholar] [CrossRef] [PubMed]
- Brinckmann, A.; Axer, S.; Jakschies, D.; Dallmann, I.; Grosse, J.; Patzelt, T.; Bernier, T.; Emmendoerffer, A.; Atzpodien, J. Interferon-α resistance in renal carcinoma cells is associated with defective induction of signal transducer and activator of transcription 1 which can be restored by a supernatant of phorbol 12-myristate 13-acetate stimulated peripheral blood mononuclear cells. Br. J. Cancer 2002, 86, 449–455. [Google Scholar] [CrossRef] [PubMed]
- E Dovhey, S.; Ghosh, N.S.; Wright, K.L. Loss of interferon-gamma inducibility of TAP1 and LMP2 in a renal cell carcinoma cell line. Cancer Res. 2000, 60, 5789–5796. [Google Scholar]
- Ilangumaran, S.; Finan, D.; La Rose, J.; Raine, J.; Silverstein, A.; De Sepulveda, P.; Rottapel, R. A positive regulatory role for suppressor of cytokine signaling 1 in IFN-gamma-induced MHC class II expression in fibroblasts. J. Immunol. 2002, 169, 5010–5020. [Google Scholar] [CrossRef]
- Waiboci, L.W.; Ahmed, C.M.; Mujtaba, M.G.; Flowers, L.O.; Martin, J.P.; Haider, M.I.; Johnson, H.M. Both the Suppressor of Cytokine Signaling 1 (SOCS-1) Kinase Inhibitory Region and SOCS-1 Mimetic Bind to JAK2 Autophosphorylation Site: Implications for the Development of a SOCS-1 Antagonist. J. Immunol. 2007, 178, 5058–5068. [Google Scholar] [CrossRef]
- Yu, S.-J.; Long, Z.-W. Effect of SOCS1 silencing on proliferation and apoptosis of melanoma cells: An in vivo and in vitro study. Tumor Biol. 2017, 39, 1010428317694315. [Google Scholar] [CrossRef]
- Komyod, W.; Böhm, M.; Metze, D.; Heinrich, P.C.; Behrmann, I. Constitutive Suppressor of Cytokine Signaling 3 Expression Confers a Growth Advantage to a Human Melanoma Cell Line. Mol. Cancer Res. 2007, 5, 271–281. [Google Scholar] [CrossRef]
- Fojtova, M.; Boudny, V.; Kovarik, A.; Lauerova, L.; Adamkova, L.; Souckova, K.; Jarkovsky, J.; Kovarik, J. Development of IFN-gamma resistance is associated with attenuation of SOCS genes induction and constitutive expression of SOCS 3 in melanoma cells. Br. J. Cancer 2007, 97, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Braathen, R.; Hohman, V.S.; Brandtzaeg, P.; Johansen, F.-E. Secretory Antibody Formation: Conserved Binding Interactions between J Chain and Polymeric Ig Receptor from Humans and Amphibians. J. Immunol. 2007, 178, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Abril, E.; Real, L.M.; Serrano, A.; Jimenez, P.; García, A.; Canton, J.; Trigo, I.; Garrido, F.; Ruiz-Cabello, F. Unresponsiveness to interferon associated with STAT1 protein deficiency in a gastric adenocarcinoma cell line. Cancer Immunol. Immunother. 1998, 47, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Xi, S.; Dyer, K.F.; Kimak, M.; Zhang, Q.; Gooding, W.E.; Chaillet, J.R.; Chai, R.L.; Ferrell, R.E.; Zamboni, B.; Hunt, J.; et al. Decreased STAT1 Expression by Promoter Methylation in Squamous Cell Carcinogenesis. Gynecol. Oncol. 2006, 98, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Duncan, T.J.; Rolland, P.; Deen, S.; Scott, I.V.; Liu, D.T.; Spendlove, I.; Durrant, L.G. Loss of IFN gamma receptor is an independent prognostic factor in ovarian cancer. Clin. Cancer Res. 2007, 13, 4139–4145. [Google Scholar] [CrossRef]
- Sato, A.; Ohtsuki, M.; Hata, M.; Kobayashi, E.; Murakami, T. Antitumor activity of IFN-lambda in murine tumor models. J. Immunol. 2006, 176, 7686–7694. [Google Scholar] [CrossRef]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The Immunobiology of Cancer Immunosurveillance and Immunoediting. Immunity 2004, 21, 137–148. [Google Scholar] [CrossRef]
- Bui, J.D.; Carayannopoulos, L.N.; Lanier, L.L.; Yokoyama, W.M.; Schreiber, R.D. IFN-Dependent Down-Regulation of the NKG2D Ligand H60 on Tumors. J. Immunol. 2006, 176, 905–913. [Google Scholar] [CrossRef]
- Marqués, L.; Brucet, M.; Lloberas, J.; Celada, A. STAT1 Regulates Lipopolysaccharide- and TNF-α-Dependent Expression of Transporter Associated with Antigen Processing 1 and Low Molecular Mass Polypeptide 2 Genes in Macrophages by Distinct Mechanisms. J. Immunol. 2004, 173, 1103–1110. [Google Scholar] [CrossRef]
- Yang, D.; Stewart, T.J.; Smith, K.K.; Georgi, D.; Abrams, S.I.; Liu, K. Downregulation of IFN-gammaR in association with loss of Fas function is linked to tumor progression. Int. J. Cancer 2008, 122, 350–362. [Google Scholar] [CrossRef]
- Bianchini, F.; Mannini, A.; Mugnai, G.; Ruggieri, S.; Calorini, L. Expression of a metastatic phenotype in IFNs-primed/TNFalpha-activated B16 murine melanoma cells: Role of JAK1/PKCdelta signal transduction factors. Clin. Exp. Metastasis 2006, 23, 203–208. [Google Scholar] [CrossRef]
- Gocher, A.M.; Workman, C.J.; Vignali, D.A.A. Interferon-gamma: Teammate or opponent in the tumour microenvironment? Nat. Rev. Immunol. 2022, 22, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.S.; Kim, D.W.; Hwang, J.H.; Jung, H.S.; Suh, J.M.; Park, Y.J.; Chung, H.K.; Song, J.H.; Park, K.C.; Park, S.H.; et al. Regulation of Signal Transducer and Activator of Transcription 1 (STAT1) and STAT1-Dependent Genes by RET/PTC (Rearranged in Transformation/Papillary Thyroid Carcinoma) Oncogenic Tyrosine Kinases. Mol. Endocrinol. 2004, 18, 2672–2684. [Google Scholar] [CrossRef] [PubMed]
- Rahat, M.A.; Chernichovski, I.; Lahat, N. Increased binding of IFN regulating factor 1 mediates the synergistic induction of CIITA by IFN-gamma and tumor necrosis factor-alpha in human thyroid carcinoma cells. Int. Immunol. 2001, 13, 1423–1432. [Google Scholar] [CrossRef] [PubMed]
- Gialitakis, M.; Kretsovali, A.; Spilianakis, C.; Kravariti, L.; Mages, J.; Hoffmann, R.; Hatzopoulos, A.K.; Papamatheakis, J. Coordinated changes of histone modifications and HDAC mobilization regulate the induction of MHC class II genes by Trichostatin A. Nucleic Acids Res. 2006, 34, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Chou, S.-D.; Khan, A.N.H.; Magner, W.J.; Tomasi, T.B. Histone acetylation regulates the cell type specific CIITA promoters, MHC class II expression and antigen presentation in tumor cells. Int. Immunol. 2005, 17, 1483–1494. [Google Scholar] [CrossRef]
- Satoh, A.; Toyota, M.; Ikeda, H.; Morimoto, Y.; Akino, K.; Mita, H.; Suzuki, H.; Sasaki, Y.; Kanaseki, T.; Takamura, Y.; et al. Epigenetic inactivation of class II transactivator (CIITA) is associated with the absence of interferon-gamma-induced HLA-DR expression in colorectal and gastric cancer cells. Oncogene 2004, 23, 8876–8886. [Google Scholar] [CrossRef]
- Meyts, I.; Casanova, J. Viral infections in humans and mice with genetic deficiencies of the type I IFN response pathway. Eur. J. Immunol. 2021, 51, 1039–1061. [Google Scholar] [CrossRef]
- Ciancanelli, M.J.; Huang, S.X.L.; Luthra, P.; Garner, H.; Itan, Y.; Volpi, S.; Lafaille, F.G.; Trouillet, C.; Schmolke, M.; Albrecht, R.A.; et al. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 2015, 348, 448–453. [Google Scholar] [CrossRef]
- Hernandez, N.; Melki, I.; Jing, H.; Habib, T.; Huang, S.S.; Danielson, J.; Kula, T.; Drutman, S.; Belkaya, S.; Rattina, V.; et al. Life-threatening influenza pneum.monitis in a child with inherited IRF9 deficiency. J. Exp. Med. 2018, 215, 2567–2585. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bastard, P.; Karbuz, A.; Gervais, A.; Tayoun, A.A.; Aiuti, A.; Belot, A.; Bolze, A.; Gaudet, A.; Bondarenko, A.; et al. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature 2022, 603, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Asano, T.; Boisson, B.; Onodi, F.; Matuozzo, D.; Moncada-Velez, M.; Maglorius Renkilaraj, M.R.L.; Zhang, P.; Meertens, L.; Bolze, A.; Materna, M.; et al. X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 2021, 6, eabl4348. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, N.; Bucciol, G.; Moens, L.; Le Pen, J.; Shahrooei, M.; Goudouris, E.; Shirkani, A.; Changi-Ashtiani, M.; Rokni-Zadeh, H.; Sayar, E.H.; et al. Inherited IFNAR1 deficiency in otherwise healthy patients with adverse reaction to measles and yellow fever live vaccines. J. Exp. Med. 2019, 216, 2057–2070. [Google Scholar] [CrossRef]
- Bastard, P.; Manry, J.; Chen, J.; Rosain, J.; Seeleuthner, Y.; AbuZaitun, O.; Lorenzo, L.; Khan, T.; Hasek, M.; Hernandez, N.; et al. Herpes simplex encephalitis in a patient with a distinctive form of inherited IFNAR1 deficiency. J. Clin. Investig. 2021, 131, e139980. [Google Scholar] [CrossRef]
- Zhang, S.-Y. Herpes simplex virus encephalitis of childhood: Inborn errors of central nervous system cell-intrinsic immunity. Hum. Genet. 2020, 139, 911–918. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.-H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Auto-antibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef]
- Zhang, Q.; Pizzorno, A.; Miorin, L.; Bastard, P.; Gervais, A.; Le Voyer, T.; Bizien, L.; Manry, J.; Rosain, J.; Philippot, Q.; et al. Autoantibodies against type I IFNs in patients with critical influenza pneumonia. J. Exp. Med. 2022, 219, e20220514. [Google Scholar] [CrossRef]
- Bustamante, J. Mendelian susceptibility to mycobacterial disease: Recent discoveries. Hum. Genet. 2020, 139, 993–1000. [Google Scholar] [CrossRef]
- Le Voyer, T.; Neehus, A.-L.; Yang, R.; Ogishi, M.; Rosain, J.; Alroqi, F.; Alshalan, M.; Blumental, S.; Al Ali, F.; Khan, T.; et al. Inherited deficiency of stress granule ZNFX1 in patients with monocytosis and mycobacterial disease. Proc. Natl. Acad. Sci. USA 2021, 118, e2102804118. [Google Scholar] [CrossRef]
- Noma, K.; Mizoguchi, Y.; Tsumura, M.; Okada, S. Mendelian susceptibility to mycobacterial diseases: State of the art. Clin. Microbiol. Infect. 2022, 28, 1429–1434. [Google Scholar] [CrossRef] [PubMed]
- Ogishi, M.; Yang, R.; Rodriguez, R.; Golec, D.P.; Martin, E.; Philippot, Q.; Bohlen, J.; Pelham, S.J.; Arias, A.A.; Khan, T.; et al. Inherited human ITK deficiency impairs IFN-gamma immunity and underlies tuberculosis. J. Exp. Med. 2023, 220, e20220484. [Google Scholar] [CrossRef] [PubMed]
- Ogishi, M.; Arias, A.A.; Yang, R.; Han, J.E.; Zhang, P.; Rinchai, D.; Halpern, J.; Mulwa, J.; Keating, N.; Chrabieh, M.; et al. Impaired IL-23-dependent induction of IFN-gamma underlies mycobacterial disease in patients with inherited TYK2 deficiency. J. Exp. Med. 2022, 219, e20220094. [Google Scholar] [CrossRef] [PubMed]
- Ogishi, M.; Yang, R.; Aytekin, C.; Langlais, D.; Bourgey, M.; Khan, T.; Al Ali, F.; Rahman, M.; Delmonte, O.M.; Chrabieh, M.; et al. Inherited PD-1 deficiency underlies tuberculosis and autoimmunity in a child. Nat. Med. 2021, 27, 1646–1654. [Google Scholar] [CrossRef] [PubMed]
- Kerner, G.; Rosain, J.; Guérin, A.; Al-Khabaz, A.; Oleaga-Quintas, C.; Rapaport, F.; Massaad, M.J.; Ding, J.Y.; Khan, T.; Al Ali, F.; et al. Inherited human IFN-gamma deficiency underlies mycobacterial disease. J. Clin. Investig. 2020, 130, 3158–3171. [Google Scholar] [CrossRef] [PubMed]
- Boisson-Dupuis, S. The monogenic basis of human tuberculosis. Hum. Genet. 2020, 139, 1001–1009. [Google Scholar] [CrossRef]
- Boisson-Dupuis, S.; Ramirez-Alejo, N.; Li, Z.; Patin, E.; Rao, G.; Kerner, G.; Lim, C.K.; Krementsov, D.N.; Hernandez, N.; Ma, C.S.; et al. Tuberculosis and impaired IL-23-dependent IFN-gamma immunity in humans homozygous for a common TYK2 missense variant. Sci. Immunol. 2018, 3, eaau8714. [Google Scholar] [CrossRef]
- Kerner, G.; Ramirez-Alejo, N.; Seeleuthner, Y.; Yang, R.; Ogishi, M.; Cobat, A.; Patin, E.; Quintana-Murci, L.; Boisson-Dupuis, S.; Casanova, J.-L.; et al. Homozygosity for TYK2 P1104A underlies tuberculosis in about 1% of patients in a cohort of European ancestry. Proc. Natl. Acad. Sci. USA 2019, 116, 10430–10434. [Google Scholar] [CrossRef]
- Huang, S.; Hendriks, W.; Althage, A.; Hemmi, S.; Bluethmann, H.; Kamijo, R.; Vilček, J.; Zinkernagel, R.M.; Aguet, M. Immune response in mice that lack the interferon-gamma receptor. Science 1993, 259, 1742–1745. [Google Scholar] [CrossRef]
- Dalton, D.K.; Pitts-Meek, S.; Keshav, S.; Figari, I.S.; Bradley, A.; Stewart, T.A. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science 1993, 259, 1739–1742. [Google Scholar] [CrossRef]
- Rosain, J.; Neehus, A.L.; Manry, J.; Yang, R.; Le Pen, J.; Daher, W.; Liu, Z.; Chan, Y.H.; Tahuil, N.; Türel, Ö.; et al. Human IRF1 governs macrophagic IFN-gamma immunity to mycobacteria. Cell 2023, 186, 621–645.e33. [Google Scholar] [CrossRef] [PubMed]
- Puel, A.; Bastard, P.; Bustamante, J.; Casanova, J.-L. Human autoantibodies underlying infectious diseases. J. Exp. Med. 2022, 219, e20211387. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Barker, D.J.; Georgiou, X.; Cooper, M.A.; Flicek, P.; Marsh, S.G.E. IPD-IMGT/HLA Database. Nucleic Acids Res. 2020, 48, D948–D955. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Rahman, M.; Ahmed, I.; Al Ali, F.; Jithesh, P.V.; Marr, N. Human leukocyte antigen class II gene diversity tunes antibody repertoires to common pathogens. Front. Immunol. 2022, 13, 856497. [Google Scholar] [CrossRef] [PubMed]
- Greenbaum, J.; Sidney, J.; Chung, J.; Brander, C.; Peters, B.; Sette, A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics 2011, 63, 325–335. [Google Scholar] [CrossRef]
- Bodewes, R.; Geelhoed-Mieras, M.; Nieuwkoop, N.; Hanson, J.; David, C.; Fouchier, R.; Osterhaus, A.; Rimmelzwaan, G. Redundancy of the influenza A virus-specific cytotoxic T lymphocyte response in HLA-B*2705 transgenic mice limits the impact of a mutation in the immunodominant NP383–391 epitope on influenza pathogenesis. Virus Res. 2011, 155, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Jalali-Najafabadi, F.; Stadler, M.; Dand, N.; Jadon, D.; Soomro, M.; Ho, P.; Marzo-Ortega, H.; Helliwell, P.; Korendowych, E.; Simpson, M.A.; et al. Application of information theoretic feature selection and machine learning methods for the development of genetic risk prediction models. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bousfiha, A.; Moundir, A.; Tangye, S.G.; Picard, C.; Jeddane, L.; Al-Herz, W.; Rundles, C.C.; Franco, J.L.; Holland, S.M.; Klein, C.; et al. The 2022 Update of IUIS Phenotypical Classification for Human Inborn Errors of Immunity. J. Clin. Immunol. 2022, 42, 1508–1520. [Google Scholar] [CrossRef]
- Shklovskaya, E.; Rizos, H. MHC Class I Deficiency in Solid Tumors and Therapeutic Strategies to Overcome It. Int. J. Mol. Sci. 2021, 22, 6741. [Google Scholar] [CrossRef]
- Ishikawa, T.; Higuchi, K.; Kubota, T.; Seki, K.-I.; Honma, T.; Yoshida, T.; Kamimura, T. Combination PEG-IFN a-2b/Ribavirin Therapy Following Treatment of Hepatitis C Virus-Associated Hepatocellular Carcinoma is Capable of Improving Hepatic Functional Reserve and Survival. Hepato-Gastroenterology 2011, 59, 529–532. [Google Scholar] [CrossRef]
- Lasfar, A.; Zloza, A.; Cohen-Solal, K.A. IFN-lambda therapy: Current status and future perspectives. Drug Discov. Today 2016, 21, 167–171. [Google Scholar] [CrossRef] [PubMed]
- George, P.M.; Badiger, R.; Alazawi, W.; Foster, G.R.; Mitchell, J.A. Pharmacology and therapeutic potential of interferons. Pharmacol. Ther. 2012, 135, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Caraglia, M.; Marra, M.; Tagliaferri, P.; Lamberts, S.W.J.; Zappavigna, S.; Misso, G.; Cavagnini, F.; Facchini, G.; Abbruzzese, A.; Hofland, L.J.; et al. Emerging strategies to strengthen the anti-tumour activity of type I interferons: Overcoming survival pathways. Curr. Cancer Drug Targets 2009, 9, 690–704. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Zhu, B.; Chen, D. Type I interferon-mediated tumor immunity and its role in immunotherapy. Cell. Mol. Life Sci. 2022, 79, 1–24. [Google Scholar] [CrossRef]
- Park, A. and A. Iwasaki, Type I and Type III Interferons–Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef]
- Jia, R.; Kralovics, R. Progress in elucidation of molecular pathophysiology of myeloproliferative neoplasms and its application to therapeutic decisions. Int. J. Hematol. 2019, 111, 182–191. [Google Scholar] [CrossRef]
- Parmar, S.; Platanias, L.C. Interferons: Mechanisms of action and clinical applications. Curr. Opin. Oncol. 2003, 15, 431–439. [Google Scholar] [CrossRef]
- Castro, F.; Cardoso, A.P.; Gonçalves, R.M.; Serre, K.; Oliveira, M.J. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 2018, 9, 847. [Google Scholar] [CrossRef]
- Zaidi, M.R.; Merlino, G. The two faces of interferon-gamma in cancer. Clin. Cancer Res. 2011, 17, 6118–6124. [Google Scholar] [CrossRef]
- Savage, C.O.; Brooks, C.J.; Harcourt, G.C.; Picard, J.K.; King, W.; Sansom, D.M.; Willcox, N. Human vascular endothelial cells process and present autoantigen to human T cell lines. Int. Immunol. 1995, 7, 471–479. [Google Scholar] [CrossRef]
- Bukowski, R.M. Natural history and therapy of metastatic renal cell carcinoma: The role of interleukin-2. Cancer 1997, 80, 1198–1220. [Google Scholar] [CrossRef]
- Umeda, T.; Niijima, T. Phase II study of alpha interferon on renal cell carcinoma. Summary of three collaborative trials. Cancer 1986, 58, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Quesada, J.R.; Rios, A.; Swanson, D.; Trown, P.; Gutterman, J.U. Antitumor activity of recombinant-derived interferon alpha in metastatic renal cell carcinoma. J. Clin. Oncol. 1985, 3, 1522–1528. [Google Scholar] [CrossRef] [PubMed]
- Logothetis, C. TREATMENT OF CHEMOTHERAPY-REFRACTORY METASTATIC UROTHELIAL TUMORS. Urol. Clin. N. Am. 1992, 19, 775–777. [Google Scholar] [CrossRef]
- Mucci, A.; Antonarelli, G.; Caserta, C.; Vittoria, F.M.; Desantis, G.; Pagani, R.; Greco, B.; Casucci, M.; Escobar, G.; Passerini, L.; et al. Myeloid cell-based delivery of IFN-gamma reprograms the leukemia microenvironment and induces anti-tumoral immune responses. EMBO Mol. Med. 2021, 13, e13598. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Kees, T.; Almeida, A.S.; Liu, B.; He, X.-Y.; Ng, D.; Han, X.; Spector, D.L.; McNeish, I.A.; Gimotty, P.; et al. Activating a collaborative innate-adaptive immune response to control metastasis. Cancer Cell 2021, 39, 1361–1374.e9. [Google Scholar] [CrossRef] [PubMed]
- Windbichler, G.H.; Hausmaninger, H.; Stummvoll, W.; Graf, A.H.; Kainz, C.; Lahodny, J.; Denison, U.; Müller-Holzner, E.; Marth, C. Interferon-gamma in the first-line therapy of ovarian cancer: A randomized phase III trial. Br. J. Cancer 2000, 82, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulos, A.; Constantinides, C.; Fokaeas, E.; Stravodimos, C.; Giannopoulou, M.; Kyroudi, A.; Gounaris, A. The immunomodulating effect of interferon-gamma intravesical instillations in preventing bladder cancer recurrence. Clin. Cancer Res. 2003, 9, 5550–5558. [Google Scholar]
- Lienard, D.; Eggermont, A.M.; Kroon, B.B.; Koops, H.S.; Lejeune, F.J. Isolated limb perfusion in primary and recurrent melanoma: Indications and results. Semin. Surg. Oncol. 1998, 14, 202–209. [Google Scholar] [CrossRef]
- Gleave, M.E.; Elhilali, M.; Fradet, Y.; Davis, I.; Venner, P.; Saad, F.; Klotz, L.H.; Moore, M.J.; Paton, V.; Bajamonde, A.; et al. Interferon Gamma-1b Compared with Placebo in Metastatic Renal-Cell Carcinoma. N. Engl. J. Med. 1998, 338, 1265–1271. [Google Scholar] [CrossRef]
- Wiesenfeld, M.; O’Connell, M.J.; Wieand, H.S.; Gonchoroff, N.J.; Donohue, J.H.; Fitzgibbons, R.J., Jr.; Krook, J.E.; Mailliard, J.A.; Gerstner, J.B.; Pazdur, R. Controlled clinical trial of interferon-gamma as postoperative surgical adjuvant therapy for colon cancer. J. Clin. Oncol. 1995, 13, 2324–2329. [Google Scholar] [CrossRef]
- Jett, J.R.; Maksymiuk, A.W.; Su, J.Q.; Mailliard, J.A.; Krook, J.E.; Tschetter, L.K.; Kardinal, C.G.; I Twito, D.; Levitt, R.; Gerstner, J.B. Phase III trial of recombinant interferon gamma in complete responders with small-cell lung cancer. J. Clin. Oncol. 1994, 12, 2321–2326. [Google Scholar] [CrossRef]
- Wong, L.H.; Krauer, K.G.; Hatzinisiriou, I.; Estcourt, M.J.; Hersey, P.; Tam, N.D.; Edmondson, S.; Devenish, R.J.; Ralph, S.J. Interferon-resistant Human Melanoma Cells Are Deficient in ISGF3 Components, STAT1, STAT2, and p48-ISGF3γ. J. Biol. Chem. 1997, 272, 28779–28785. [Google Scholar] [CrossRef] [PubMed]
- van de Vosse, E.; van Dissel, J.T. IFN-gammaR1 defects: Mutation update and description of the IFNGR1 variation database. Hum. Mutat. 2017, 38, 1286–1296. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Shi, L.Z.; Zhao, H.; Chen, J.; Xiong, L.; He, Q.; Chen, T.; Roszik, J.; Bernatchez, C.; Woodman, S.E.; et al. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell 2016, 167, 397–404.e9. [Google Scholar] [CrossRef]
- Cabrera, T.; Lara, E.; Romero, J.M.; Maleno, I.; Real, L.M.; Ruiz-Cabello, F.; Valero, P.; Camacho, F.M.; Garrido, F. HLA class I expression in metastatic melanoma correlates with tumor development during autologous vaccination. Cancer Immunol. Immunother. 2006, 56, 709–717. [Google Scholar] [CrossRef] [PubMed]
- Carretero, F.J.; del Campo, A.B.; Flores-Martín, J.F.; Mendez, R.; García-Lopez, C.; Cozar, J.M.; Adams, V.; Ward, S.; Cabrera, T.; Ruiz-Cabello, F.; et al. Frequent HLA class I alterations in human prostate cancer: Molecular mechanisms and clinical relevance. Cancer Immunol. Immunother. 2015, 65, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, J.; Fieschi, C.; Doffinger, R.; Feinberg, M.; Leclerc, T.; Boisson-Dupuis, S.; Picard, C.; Bustamante, J.; Chapgier, A.; Filipe-Santos, O.; et al. Bacillus Calmette Guerin triggers the IL-12/IFN-gamma axis by an IRAK-4- and NEMO-dependent, non-cognate interaction between monocytes, NK, and T lymphocytes. Eur. J. Immunol. 2004, 34, 3276–3284. [Google Scholar] [CrossRef]
- Saban, M.R.; Hellmich, H.L.; Simpson, C.; A Davis, C.; Lang, M.L.; A Ihnat, M.; A O’Donnell, M.; Wu, X.-R.; Saban, R. Repeated BCG treatment of mouse bladder selectively stimulates small GTPases and HLA antigens and inhibits single-spanning uroplakins. BMC Cancer 2007, 7, 204. [Google Scholar] [CrossRef]
- Kitamura, H.; Torigoe, T.; Honma, I.; Sato, E.; Asanuma, H.; Hirohashi, Y.; Sato, N.; Tsukamoto, T. Effect of Human Leukocyte Antigen Class I Expression of Tumor Cells on Outcome of Intravesical Instillation of Bacillus Calmette-Guerin Immunotherapy for Bladder Cancer. Clin. Cancer Res. 2006, 12, 4641–4644. [Google Scholar] [CrossRef]
- Kalbasi, A.; Tariveranmoshabad, M.; Hakimi, K.; Kremer, S.; Campbell, K.M.; Funes, J.M.; Vega-Crespo, A.; Parisi, G.; Champekar, A.; Nguyen, C.; et al. Uncoupling interferon signaling and antigen presentation to overcome immunotherapy resistance due to JAK1 loss in melanoma. Sci. Transl. Med. 2020, 12, eabb0152. [Google Scholar] [CrossRef] [PubMed]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.R.; Albright, A.; Cheng, J.D.; Kang, S.P.; Shankaran, V.; et al. IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Edington, H.D.; Rao, U.N.; Jukic, D.M.; Land, S.R.; Ferrone, S.; Kirkwood, J.M. Modulation of signal transducers and activators of transcription 1 and 3 signaling in melanoma by high-dose IFNalpha2b. Clin. Cancer Res. 2007, 13, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Carson, W.E. Interferon-alpha-induced activation of signal transducer and activator of transcription proteins in malignant melanoma. Clin. Cancer Res. 1998, 4, 2219–2228. [Google Scholar] [PubMed]
- Tarhini, A.A.; Gogas, H.; Kirkwood, J.M. IFN-α in the Treatment of Melanoma. J. Immunol. 2012, 189, 3789–3793. [Google Scholar] [CrossRef]
- Bracci, L.; Proietti, E.; Belardelli, F. IFN- and Novel Strategies of Combination Therapy for Cancer. Ann. N. Y. Acad. Sci. 2007, 1112, 256–268. [Google Scholar] [CrossRef]
- Sacchi, S.; Federico, M.; Vitolo, U.; Boccomini, C.; Vallisa, D.; Baldini, L.; Petrini, M.; Rupoli, S.; Di Raimondo, F.; Merli, F.; et al. Clinical activity and safety of combination immunotherapy with IFN-alpha 2a and Rituximab in patients with relapsed low grade non-Hodgkin’s lymphoma. Haematologica 2001, 86, 951–958. [Google Scholar]
- Rahimi Kalateh Shah Mohammad, G.; Ghahremanloo, A.; Soltani, A.; Fathi, E.; Hashemy, S.I. Cytokines as potential combination agents with PD-1/PD-L1 blockade for cancer treatment. J. Cell Physiol. 2020, 235, 5449–5460. [Google Scholar] [CrossRef]
- Lee, S.E.; Jang, G.-Y.; Lee, J.W.; Park, S.H.; Han, H.D.; Park, Y.-M.; Kang, T.H. Improvement of STING-mediated cancer immunotherapy using immune checkpoint inhibitors as a game-changer. Cancer Immunol. Immunother. 2022, 71, 3029–3042. [Google Scholar] [CrossRef]
- Rafique, I.; Kirkwood, J.M.; Tarhini, A.A. Immune Checkpoint Blockade and Interferon-α in Melanoma. Semin. Oncol. 2015, 42, 436–447. [Google Scholar] [CrossRef]
- Muss, H.B.; Caponera, M.; Zekan, P.J.; Jr, D.V.J.; Stuart, J.J.; Richards, F.; Cooper, M.R.; Levin, E.A.; Reich, S.D.; Capizzi, R.L. Recombinant gamma interferon in advanced breast cancer: A phase II trial. Investig. New Drugs 1986, 4, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Broun, M.; Bode, H.R. Characterization of the head organizer in hydra. Development 2002, 129, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Moss, A.J.; Benhorin, J. Prognosis and Management after a First Myocardial Infarction. N. Engl. J. Med. 1990, 322, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.L.; Escalante, N.K.; Jude, K.M.; Sotolongo Bellon, J.; Su, L.; Horton, T.M.; Tsutsumi, N.; Berardinelli, S.J.; Haltiwanger, R.S.; Piehler, J.; et al. Structure of the IFNgamma receptor complex guides design of biased agonists. Nature 2019, 567, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Stiff, A.; Carson, W., III. Investigations of interferon-lambda for the treatment of cancer. J. Innate Immun. 2015, 7, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Pingwara, R.; Kosmala, D.; Woźniak, N.; Orzechowski, A.; Mucha, J. IFN-lambda Modulates the Migratory Capacity of Canine Mammary Tumor Cells via Regulation of the Expression of Matrix Metalloproteinases and Their Inhibitors. Cells 2021, 10, 999. [Google Scholar] [CrossRef]
- Lasfar, A.; Gogas, H.; Zloza, A.; Kaufman, H.L.; Kirkwood, J.M. IFN-lambda cancer immunotherapy: New kid on the block. Immunotherapy 2016, 8, 877–888. [Google Scholar] [CrossRef]
- Hasegawa, K.; Tagawa, M.; Takagi, K.; Tsukamoto, H.; Tomioka, Y.; Suzuki, T.; Nishioka, Y.; Ohrui, T.; Numasaki, M. Anti-tumor immunity elicited by direct intratumoral administration of a recombinant adenovirus expressing either IL-28A/IFN-lambda2 or IL-29/IFN-lam.m.m.mbda1. Cancer Gene Ther. 2016, 23, 266–277. [Google Scholar] [CrossRef]
- Monk, P.D.; Marsden, R.J.; Tear, V.J.; Brookes, J.; Batten, T.N.; Mankowski, M.; Gabbay, F.J.; Davies, D.E.; Holgate, S.T.; Ho, L.P.; et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 2021, 9, 196–206. [Google Scholar] [CrossRef]
- Vinh, D.C.; Abel, L.; Bastard, P.; Cheng, M.P.; Condino-Neto, A.; Gregersen, P.K.; Haerynck, F.; Cicalese, M.P.; Hagin, D.; Soler-Palacín, P.; et al. Harnessing Type I IFN Immunity Against SARS-CoV-2 with Early Administration of IFN-beta. J. Clin. Immunol. 2021, 41, 1425–1442. [Google Scholar] [CrossRef]
- Berns, S.A.; Isakova, J.A.; Pekhtereva, P. The Therapeutic potential of interferon-gamma in tuberculosis. ADMET DMPK 2022, 10, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Dibeneditto, J.P.; Worobec, S.M. Exposure to hot environments can cause dermatological problems. Occup. Health Saf. (Waco, Tex.) 1985, 54, 35–38. [Google Scholar]
- Shin, D.S.; Zaretsky, J.M.; Escuin-Ordinas, H.; Garcia-Diaz, A.; Hu-Lieskovan, S.; Kalbasi, A.; Grasso, C.S.; Hugo, W.; Sandoval, S.; Torrejon, D.Y.; et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov. 2017, 7, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Huang, F.; Zhang, X.; Ojo, O.A.; Li, Y.; Trummell, H.Q.; Anderson, J.C.; Fiveash, J.; Bredel, M.; Yang, E.S.; et al. Selective suppression of melanoma lacking IFN-gamma pathway by JAK inhibition depends on T cells and host TNF signaling. Nat. Commun. 2022, 13, 5013. [Google Scholar] [CrossRef] [PubMed]
- Castiello, L.; Sestili, P.; Schiavoni, G.; Dattilo, R.; Monque, D.M.; Ciaffoni, F.; Iezzi, M.; Lamolinara, A.; Sistigu, A.; Moschella, F.; et al. Disruption of IFN-I Signaling Promotes HER2/Neu Tumor Progression and Breast Cancer Stem Cells. Cancer Immunol. Res. 2018, 6, 658–670. [Google Scholar] [CrossRef] [PubMed]
| Type I | Type II | Type III | |
|---|---|---|---|
| IFN-α | IFN-γ | IFN-λ | |
| chromosomal localization | 9p21 | 12q14 | 19q/3 |
| numbers | 13 α, β, ε, κ, ω in humans | 1 | 4 |
| receptor | IFN-αRI IFN-αRII ubiquitously expressed | IFN-γRI IFN-γRII ubiquitously expressed | IFN-λR1 IFN-10Rβ preferentially expressed on epithelial cells |
| Signal Transduction pathways | JAK1, TYK2, STAT1–5, PI3K, AKT, MAPK, NF-ĸB, p53 | JAK1, 2, STAT1, 3, 5, PI3K, AKT, MAPK, NF-ĸB | JAK1, STAT1–5, TyK2 |
| main function | anti-viral anti-proliferative | anti-bacterial anti-proliferative | anti-tumoral anti-proliferative |
| Categories of Lesions | Reversibility by IFN | Mechanisms |
|---|---|---|
| (a) irreversible or “hard” genetic lesions: | no |
|
| (b) irreversible or “soft” lesions: | no |
|
| (c) reversible or “soft” lesions: | yes |
|
| Molecule | Mechanisms | |
|---|---|---|
| receptor | IFN-R expression | loss |
| signal transduction | JAK/STAT/TYK2 | mutation, deletion |
| JAK/STAT | lack of activity | |
| STAT1, STAT3, JAK1, and JAK2, SOCS1/3 altered expression | loss or aberrant phosphorylation | |
| transcription factors | IRFs | reduced expression |
| IRF1 | impaired binding | |
| IRF1 | methylation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massa, C.; Wang, Y.; Marr, N.; Seliger, B. Interferons and Resistance Mechanisms in Tumors and Pathogen-Driven Diseases—Focus on the Major Histocompatibility Complex (MHC) Antigen Processing Pathway. Int. J. Mol. Sci. 2023, 24, 6736. https://doi.org/10.3390/ijms24076736
Massa C, Wang Y, Marr N, Seliger B. Interferons and Resistance Mechanisms in Tumors and Pathogen-Driven Diseases—Focus on the Major Histocompatibility Complex (MHC) Antigen Processing Pathway. International Journal of Molecular Sciences. 2023; 24(7):6736. https://doi.org/10.3390/ijms24076736
Chicago/Turabian StyleMassa, Chiara, Yuan Wang, Nico Marr, and Barbara Seliger. 2023. "Interferons and Resistance Mechanisms in Tumors and Pathogen-Driven Diseases—Focus on the Major Histocompatibility Complex (MHC) Antigen Processing Pathway" International Journal of Molecular Sciences 24, no. 7: 6736. https://doi.org/10.3390/ijms24076736
APA StyleMassa, C., Wang, Y., Marr, N., & Seliger, B. (2023). Interferons and Resistance Mechanisms in Tumors and Pathogen-Driven Diseases—Focus on the Major Histocompatibility Complex (MHC) Antigen Processing Pathway. International Journal of Molecular Sciences, 24(7), 6736. https://doi.org/10.3390/ijms24076736





