Identification of Optimal Reference Genes for qRT-PCR Normalization for Physical Activity Intervention and Omega-3 Fatty Acids Supplementation in Humans
Abstract
1. Introduction
2. Results
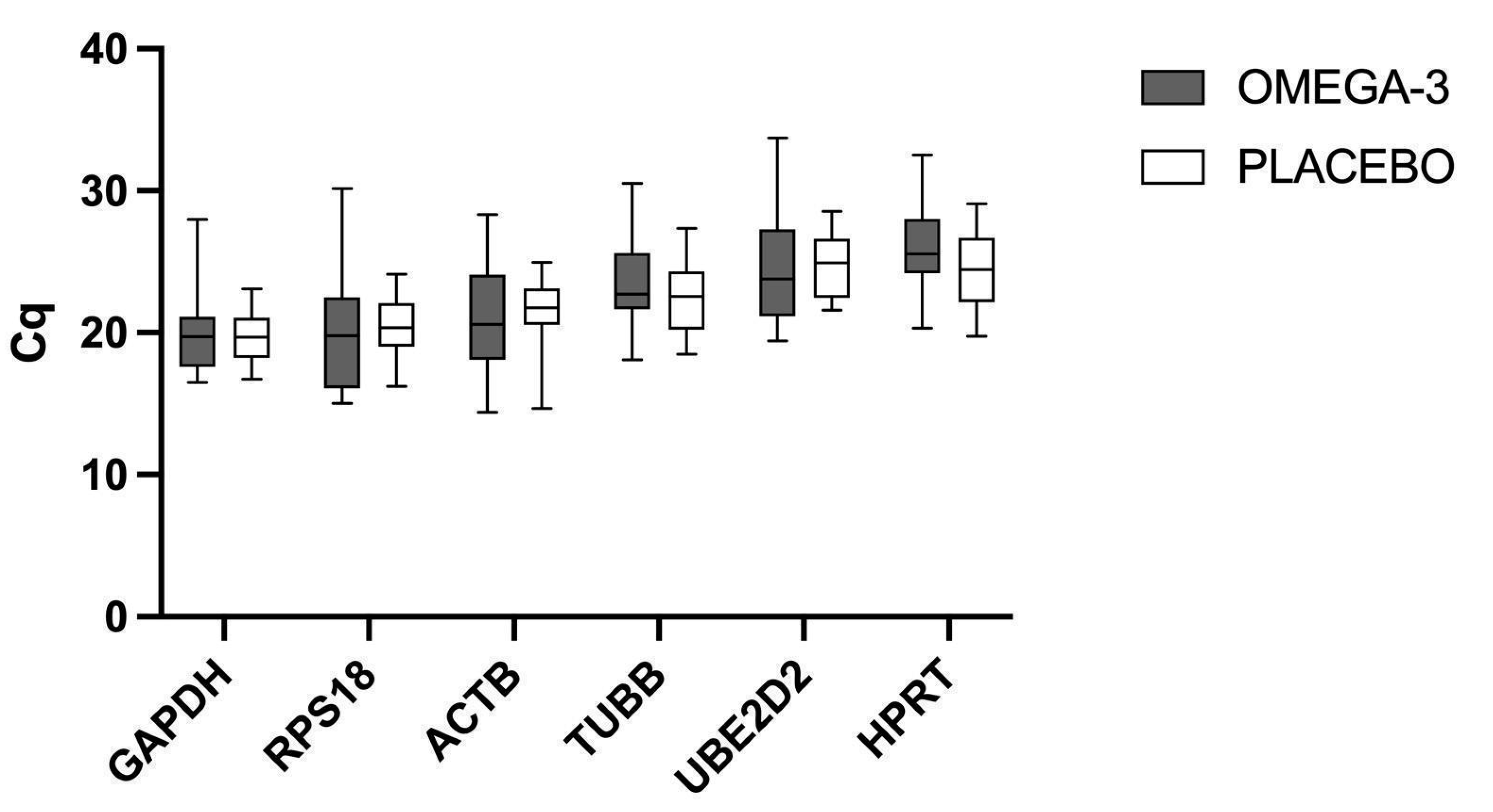
2.1. mRNA Levels of the Candidate Reference Genes
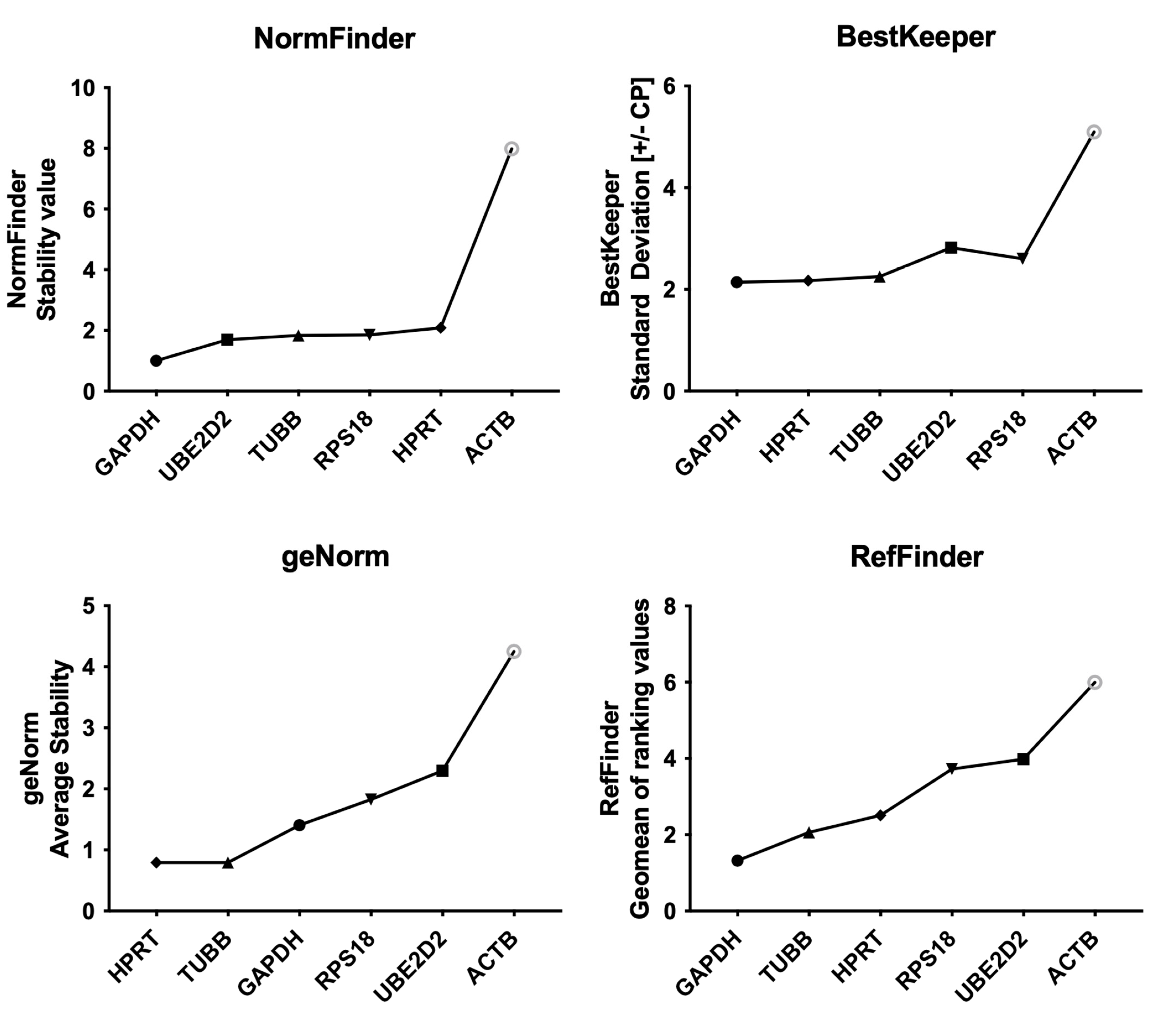
2.2. Evaluation of Candidate Reference Genes’ Expression after 12-Week Intervention
3. Discussion
4. Materials and Methods
4.1. Ethics
4.2. Study Setting and Subjects
4.3. Blood Collection, RNA Extraction and Reverse Transcription
4.4. Selection of Potential Reference Genes and Primer Design
4.5. Quantitative Real-Time Polymerase Chain Reaction
4.6. Evaluation of Stable Reference Genes for leukocytes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACTB | beta-actin |
| Average STDEV | Average of standard deviation |
| Cq | quantification cycle |
| Ct | cycle threshold |
| DHA | docosahexaenoic acid |
| DNA | Deoxyribonucleic acid |
| EPA | eicosapentaenoic acid |
| GAPDH | Glyceraldehyde-3-phosphate Dehydrogenase |
| HPRT1 | Hypoxanthine Phosphoribosyltransferase 1 |
| MCTs | Medium Chain Triglycerides |
| MIQE | Minimum Information for Publication of Quantitative Real-Time PCR Experiments |
| n-3 PUFAs | omega-3 polyunsaturated fatty acids |
| PBMC | Peripheral Blood Mononuclear Cell |
| qPCR | Quantitative Polymerase Chain Reaction |
| qRT-PCR | Quantitative Real-time Polymerase Chain Reaction |
| RBCL | Red Blood Cell Lysis Buffer |
| RDML | Real-time PCR Data Markup Language |
| RGs | reference genes |
| RPS18 | Ribosomal Protein S18 |
| SD | standard deviation |
| TUBB | Tubulin Beta Class I |
| UBE2D2 | Ubiquitin-conjugating enzyme E2 D2 |
| VO2peak | peak oxygen uptake |
References
- Singh, K.P.; Miaskowski, C.; Dhruva, A.A.; Flowers, E.; Kober, K.M. Mechanisms and Measurement of Changes in Gene Expression. Biol. Res. Nurs. 2018, 20, 369–382. [Google Scholar] [CrossRef]
- Baine, M.J.; Mallya, K.; Batra, S.K. Quantitative Real-Time PCR Expression Analysis of Peripheral Blood Mononuclear Cells in Pancreatic Cancer Patients. In Pancreatic Cancer: Methods and Protocols; Methods in Molecular Biology; Su, G.H., Ed.; Humana Press: Totowa, NJ, USA, 2013; pp. 157–173. ISBN 978-1-62703-287-2. [Google Scholar]
- Oturai, D.B.; Søndergaard, H.B.; Börnsen, L.; Sellebjerg, F.; Romme Christensen, J. Identification of Suitable Reference Genes for Peripheral Blood Mononuclear Cell Subset Studies in Multiple Sclerosis. Scand. J. Immunol. 2016, 83, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.-H.; Chou, L.-S.; Chou, S.-J.; Wang, J.-H.; Stott, J.; Blanchard, M.; Jen, I.-F.; Yang, W.-C. Selection of Suitable Reference Genes for Normalization of Quantitative RT-PCR in Peripheral Blood Samples of Bottlenose Dolphins (Tursiops truncatus). Sci. Rep. 2015, 5, 15425. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.; et al. The Need for Transparency and Good Practices in the QPCR Literature. Nat. Methods 2013, 10, 1063–1067. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-Time RT-PCR Normalisation; Strategies and Considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Riedel, G.; Rüdrich, U.; Fekete-Drimusz, N.; Manns, M.P.; Vondran, F.W.R.; Bock, M. An Extended ΔCT-Method Facilitating Normalisation with Multiple Reference Genes Suited for Quantitative RT-PCR Analyses of Human Hepatocyte-Like Cells. PLoS ONE 2014, 9, e93031. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Connolly, P.H.; Caiozzo, V.J.; Zaldivar, F.; Nemet, D.; Larson, J.; Hung, S.; Heck, J.D.; Hatfield, G.W.; Cooper, D.M. Effects of Exercise on Gene Expression in Human Peripheral Blood Mononuclear Cells. J. Appl. Physiol. 2004, 97, 1461–1469. [Google Scholar] [CrossRef]
- Tremblay, B.L.; Guénard, F.; Rudkowska, I.; Lemieux, S.; Couture, P.; Vohl, M.-C. Epigenetic Changes in Blood Leukocytes Following an Omega-3 Fatty Acid Supplementation. Clin. Epigenetics 2017, 9, 43. [Google Scholar] [CrossRef]
- Calder, P.C. Very Long Chain Omega-3 (n-3) Fatty Acids and Human Health. Eur. J. Lipid Sci. Technol. 2014, 116, 1280–1300. [Google Scholar] [CrossRef]
- Lewis, N.A.; Daniels, D.; Calder, P.C.; Castell, L.M.; Pedlar, C.R. Are There Benefits from the Use of Fish Oil Supplements in Athletes? A Systematic Review. Adv. Nutr. Bethesda Md. 2020, 11, 1300–1314. [Google Scholar] [CrossRef]
- Tomczyk, M.; Jost, Z.; Chroboczek, M.; Urbański, R.; Calder, P.C.; Fisk, H.L.; Sprengel, M.; Antosiewicz, J. Effects of 12 Wk of Omega-3 Fatty Acid Supplementation in Long-Distance Runners. Med. Sci. Sports Exerc. 2023, 55, 216. [Google Scholar] [CrossRef] [PubMed]
- Ruscica, M.; Sirtori, C.R.; Carugo, S.; Calder, P.C.; Corsini, A. Omega-3 and Cardiovascular Prevention—Is This Still a Choice? Pharmacol. Res. 2022, 182, 106342. [Google Scholar] [CrossRef]
- Bouwens, M.; van de Rest, O.; Dellschaft, N.; Bromhaar, M.G.; de Groot, L.C.; Geleijnse, J.M.; Müller, M.; Afman, L.A. Fish-Oil Supplementation Induces Antiinflammatory Gene Expression Profiles in Human Blood Mononuclear Cells. Am. J. Clin. Nutr. 2009, 90, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper--Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. MiRDeepFinder: A MiRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Cheng, W.-C.; Chang, C.-W.; Chen, C.-R.; Tsai, M.-L.; Shu, W.-Y.; Li, C.-Y.; Hsu, I.C. Identification of Reference Genes across Physiological States for QRT-PCR through Microarray Meta-Analysis. PLoS ONE 2011, 6, e17347. [Google Scholar] [CrossRef]
- Giri, A.; Sundar, I.K. Evaluation of Stable Reference Genes for QPCR Normalization in Circadian Studies Related to Lung Inflammation and Injury in Mouse Model. Sci. Rep. 2022, 12, 1764. [Google Scholar] [CrossRef]
- Elagizi, A.; Kachur, S.; Carbone, S.; Lavie, C.J.; Blair, S.N. A Review of Obesity, Physical Activity, and Cardiovascular Disease. Curr. Obes. Rep. 2020, 9, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Djuricic, I.; Calder, P.C. Beneficial Outcomes of Omega-6 and Omega-3 Polyunsaturated Fatty Acids on Human Health: An Update for 2021. Nutrients 2021, 13, 2421. [Google Scholar] [CrossRef] [PubMed]
- Browning, L.M.; Walker, C.G.; Mander, A.P.; West, A.L.; Madden, J.; Gambell, J.M.; Young, S.; Wang, L.; Jebb, S.A.; Calder, P.C. Incorporation of Eicosapentaenoic and Docosahexaenoic Acids into Lipid Pools When given as Supplements Providing Doses Equivalent to Typical Intakes of Oily Fish. Am. J. Clin. Nutr. 2012, 96, 748–758. [Google Scholar] [CrossRef]
- Myhrstad, M.C.W.; Ulven, S.M.; Günther, C.-C.; Ottestad, I.; Holden, M.; Ryeng, E.; Borge, G.I.; Kohler, A.; Brønner, K.W.; Thoresen, M.; et al. Fish Oil Supplementation Induces Expression of Genes Related to Cell Cycle, Endoplasmic Reticulum Stress and Apoptosis in Peripheral Blood Mononuclear Cells: A Transcriptomic Approach. J. Intern. Med. 2014, 276, 498–511. [Google Scholar] [CrossRef]
- Schmidt, S.; Stahl, F.; Mutz, K.-O.; Scheper, T.; Hahn, A.; Schuchardt, J.P. Transcriptome-Based Identification of Antioxidative Gene Expression after Fish Oil Supplementation in Normo- and Dyslipidemic Men. Nutr. Metab. 2012, 9, 45. [Google Scholar] [CrossRef]
- Leal, M.F.; Astur, D.C.; Debieux, P.; Arliani, G.G.; Franciozi, C.E.S.; Loyola, L.C.; Andreoli, C.V.; Smith, M.C.; Pochini, A. de C.; Ejnisman, B.; et al. Identification of Suitable Reference Genes for Investigating Gene Expression in Anterior Cruciate Ligament Injury by Using Reverse Transcription-Quantitative PCR. PLoS ONE 2015, 10, e0133323. [Google Scholar] [CrossRef]
- Ren, G.; Juhl, M.; Peng, Q.; Fink, T.; Porsborg, S.R. Selection and Validation of Reference Genes for QPCR Analysis of Differentiation and Maturation of THP-1 Cells into M1 Macrophage-like Cells. Immunol. Cell Biol. 2022, 100, 822–829. [Google Scholar] [CrossRef]
- Tarze, A.; Deniaud, A.; Le Bras, M.; Maillier, E.; Molle, D.; Larochette, N.; Zamzami, N.; Jan, G.; Kroemer, G.; Brenner, C. GAPDH, a Novel Regulator of the pro-Apoptotic Mitochondrial Membrane Permeabilization. Oncogene 2007, 26, 2606–2620. [Google Scholar] [CrossRef]
- Zala, D.; Hinckelmann, M.-V.; Yu, H.; Lyra da Cunha, M.M.; Liot, G.; Cordelières, F.P.; Marco, S.; Saudou, F. Vesicular Glycolysis Provides On-Board Energy for Fast Axonal Transport. Cell 2013, 152, 479–491. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Rajendran, M.; Rostovtseva, T.; Hool, L. How Cytoskeletal Proteins Regulate Mitochondrial Energetics in Cell Physiology and Diseases. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20210324. [Google Scholar] [CrossRef]
- Hirose, E.; Yokoya, A.; Kawamura, K.; Suzuki, K. Analysis of Differentially Expressed Genes on Human X Chromosome Harboring Large Deletion Induced by X-Rays. J. Radiat. Res. 2023, 64, 300–303. [Google Scholar] [CrossRef]
- Ilin, A.A.; Malygin, A.A.; Karpova, G.G. Ribosomal Protein S18e as a Putative Molecular Staple for the 18S RRNA 3′-Major Domain Core. Biochim. Biophys. Acta 2011, 1814, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Sferra, A.; Petrini, S.; Bellacchio, E.; Nicita, F.; Scibelli, F.; Dentici, M.L.; Alfieri, P.; Cestra, G.; Bertini, E.S.; Zanni, G. TUBB Variants Underlying Different Phenotypes Result in Altered Vesicle Trafficking and Microtubule Dynamics. Int. J. Mol. Sci. 2020, 21, 1385. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.O.; Lee, C.K.; Ryu, K.S.; Chi, S.W. The RING Domain of Mitochondrial E3 Ubiquitin Ligase 1 and Its Complex with Ube2D2: Crystallization and X-Ray Diffraction. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2020, F76, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Spiegelaere, W.D.; Dern-Wieloch, J.; Weigel, R.; Schumacher, V.; Schorle, H.; Nettersheim, D.; Bergmann, M.; Brehm, R.; Kliesch, S.; Vandekerckhove, L.; et al. Reference Gene Validation for RT-QPCR, a Note on Different Available Software Packages. PLoS ONE 2015, 10, e0122515. [Google Scholar] [CrossRef]
- Ledderose, C.; Heyn, J.; Limbeck, E.; Kreth, S. Selection of Reliable Reference Genes for Quantitative Real-Time PCR in Human T Cells and Neutrophils. BMC Res. Notes 2011, 4, 427. [Google Scholar] [CrossRef]
- Xue, W.; Wang, L.; Li, X.; Tang, M.; Li, J.; Ding, B.; Kawabata, S.; Li, Y.; Zhang, Y. Evaluation of Reference Genes for Quantitative PCR in Eustoma Grandiflorum under Different Experimental Conditions. Horticulturae 2022, 8, 164. [Google Scholar] [CrossRef]
- Roy, J.G.; McElhaney, J.E.; Verschoor, C.P. Reliable Reference Genes for the Quantification of MRNA in Human T-Cells and PBMCs Stimulated with Live Influenza Virus. BMC Immunol. 2020, 21, 4. [Google Scholar] [CrossRef]
- Jeon, R.-H.; Lee, W.-J.; Son, Y.-B.; Bharti, D.; Shivakumar, S.B.; Lee, S.-L.; Rho, G.-J. PPIA, HPRT1, and YWHAZ Genes Are Suitable for Normalization of MRNA Expression in Long-Term Expanded Human Mesenchymal Stem Cells. BioMed Res. Int. 2019, 2019, e3093545. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Luo, D.; Liao, D.J. Pseudogenes as Weaknesses of ACTB (Actb) and GAPDH (Gapdh) Used as Reference Genes in Reverse Transcription and Polymerase Chain Reactions. PLoS ONE 2012, 7, e41659. [Google Scholar] [CrossRef]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate QPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Waldenström, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef]
- Grzybkowska, A.; Anczykowska, K.; Ratkowski, W.; Aschenbrenner, P.; Antosiewicz, J.; Bonisławska, I.; Żychowska, M. Changes in Serum Iron and Leukocyte MRNA Levels of Genes Involved in Iron Metabolism in Amateur Marathon Runners—Effect of the Running Pace. Genes 2019, 10, 460. [Google Scholar] [CrossRef] [PubMed]
- Żychowska, M.; Grzybkowska, A.; Wiech, M.; Urbański, R.; Pilch, W.; Piotrowska, A.; Czerwińska-Ledwig, O.; Antosiewicz, J. Exercise Training and Vitamin C Supplementation Affects Ferritin MRNA in Leukocytes without Affecting Prooxidative/Antioxidative Balance in Elderly Women. Int. J. Mol. Sci. 2020, 21, 6469. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-Step Method of RNA Isolation by Acid Guanidinium Thiocyanate-Phenol-Chloroform Extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Desjardins, P.; Conklin, D. NanoDrop Microvolume Quantitation of Nucleic Acids. J. Vis. Exp. JoVE 2010, 45, e2565. [Google Scholar] [CrossRef]
- Ceriani, C.; Streeter, G.S.; Lemu, K.J.; James, K.S.; Ghofrani, S.; Allard, B.; Shook-Sa, B.E.; Margolis, D.M.; Archin, N.M. Defining Stable Reference Genes in HIV Latency Reversal Experiments. J. Virol. 2021, 95, e02305-20. [Google Scholar] [CrossRef]
- Geigges, M.; Gubser, P.M.; Unterstab, G.; Lecoultre, Y.; Paro, R.; Hess, C. Reference Genes for Expression Studies in Human CD8+ Naïve and Effector Memory T Cells under Resting and Activating Conditions. Sci. Rep. 2020, 10, 9411. [Google Scholar] [CrossRef]
- Usarek, E.; Barańczyk-Kuźma, A.; Kaźmierczak, B.; Gajewska, B.; Kuźma-Kozakiewicz, M. Validation of QPCR Reference Genes in Lymphocytes from Patients with Amyotrophic Lateral Sclerosis. PLoS ONE 2017, 12, e0174317. [Google Scholar] [CrossRef]
- Kaszubowska, L.; Wierzbicki, P.M.; Karsznia, S.; Damska, M.; Ślebioda, T.J.; Foerster, J.; Kmieć, Z. Optimal Reference Genes for QPCR in Resting and Activated Human NK Cells—Flow Cytometric Data Correspond to QPCR Gene Expression Analysis. J. Immunol. Methods 2015, 422, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, A.; Jamwal, M.; Viswanathan, G.K.; Sharma, P.; Sachdeva, M.S.; Bansal, D.; Malhotra, P.; Das, R. Optimal Reference Gene Selection for Expression Studies in Human Reticulocytes. J. Mol. Diagn. 2018, 20, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Kozmus, C.E.P.; Potočnik, U. Reference Genes for Real-Time QPCR in Leukocytes from Asthmatic Patients before and after Anti-Asthma Treatment. Gene 2015, 570, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zuo, S.; Tang, J.; Zuo, C.; Jia, D.; Liu, Q.; Liu, G.; Zhu, Q.; Wang, Y.; Zhang, J.; et al. Inhibition of CRTH2-Mediated Th2 Activation Attenuates Pulmonary Hypertension in Mice. J. Exp. Med. 2018, 215, 2175–2195. [Google Scholar] [CrossRef] [PubMed]
- Spandidos, A.; Wang, X.; Wang, H.; Seed, B. PrimerBank: A Resource of Human and Mouse PCR Primer Pairs for Gene Expression Detection and Quantification. Nucleic Acids Res. 2010, 38, D792–D799. [Google Scholar] [CrossRef] [PubMed]
- Żychowska, M.; Grzybkowska, A.; Zasada, M.; Piotrowska, A.; Dworakowska, D.; Czerwińska-Ledwig, O.; Pilch, W.; Antosiewicz, J. Effect of Six Weeks 1000 Mg/Day Vitamin C Supplementation and Healthy Training in Elderly Women on Genes Expression Associated with the Immune Response—A Randomized Controlled Trial. J. Int. Soc. Sports Nutr. 2021, 18, 19. [Google Scholar] [CrossRef]
- Lefever, S.; Hellemans, J.; Pattyn, F.; Przybylski, D.R.; Taylor, C.; Geurts, R.; Untergasser, A.; Vandesompele, J.; on behalf of the RDML consortium. RDML: Structured Language and Reporting Guidelines for Real-Time Quantitative PCR Data. Nucleic Acids Res. 2009, 37, 2065–2069. [Google Scholar] [CrossRef] [PubMed]

| Gene Symbol | Gene Accession Number | Name | Function |
|---|---|---|---|
| ACTB | NM_001101 | Beta actin | Cytoskeletal protein |
| GAPDH | NM_002046 | Glyceraldehyde-3-phosphate dehydrogenase | Oxidoreductase in glycolysis and gluconeogenesis |
| RPS18 | NM_022551.3 | Ribosomal Protein S18 | Encodes a ribosomal protein that is a component of the 40S subunit |
| TUBB | NM_001293212.2 | Tubulin Beta Class I | Forms a dimer with alpha-tubulin and acts as a structural component of microtubules |
| UBE2D2 | NM_003339.3 | Ubiquitin-conjugating enzyme E2 D2 | Degradates misfolded, damaged, or short-lived proteins in eukaryotes |
| HPRT1 | NM_000194.3 | Hypoxanthine Phosphoribosyltransferase 1 | Plays a central role in the generation of purine nucleotides through the purine salvage pathway |
| Symbol | Primer Sequence | Amplicon Size | Source |
|---|---|---|---|
| ACTB | F: GAGAAAATCTGGCACCACACC | 177 | Chen et al., 2018 [55] |
| R: GGATAGCACAGCCTGGATAGCAA | |||
| GAPDH | F: TCTCCTCTGACTTCAACAGCGAC | 126 | Andersen et al., 2004 [17] |
| R: CCCTGTTGCTGTAGCCAAATTC | |||
| RPS18 | F: GCGGCGGAAAATAGCCTTTG | 139 | Spandidos et al., 2010 [56] |
| R: GATCACACGTTCCACCTCATC | |||
| TUBB | F: CTAGAACCTGGGACCATGGA | 191 | Żychowska et al., 2021 [57] |
| R: TGCAGGCAGTCACAGCTCT | |||
| UBE2D2 | F: GTACTCTTGTCCATCTGTTCTCTG | 120 | Roy et al., 2020 [40] |
| R: CCATTCCCGAGCTATTCTGTT | |||
| HPRT1 | F: CGAGATGTGATGAAGGAGATGG | 97 | Jeon et al., 2019 [41] |
| R: TGATGTAATCCAGCAGGTCAGC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grzybkowska, A.; Anczykowska, K.; Antosiewicz, J.; Olszewski, S.; Dzitkowska-Zabielska, M.; Tomczyk, M. Identification of Optimal Reference Genes for qRT-PCR Normalization for Physical Activity Intervention and Omega-3 Fatty Acids Supplementation in Humans. Int. J. Mol. Sci. 2023, 24, 6734. https://doi.org/10.3390/ijms24076734
Grzybkowska A, Anczykowska K, Antosiewicz J, Olszewski S, Dzitkowska-Zabielska M, Tomczyk M. Identification of Optimal Reference Genes for qRT-PCR Normalization for Physical Activity Intervention and Omega-3 Fatty Acids Supplementation in Humans. International Journal of Molecular Sciences. 2023; 24(7):6734. https://doi.org/10.3390/ijms24076734
Chicago/Turabian StyleGrzybkowska, Agata, Katarzyna Anczykowska, Jędrzej Antosiewicz, Szczepan Olszewski, Magdalena Dzitkowska-Zabielska, and Maja Tomczyk. 2023. "Identification of Optimal Reference Genes for qRT-PCR Normalization for Physical Activity Intervention and Omega-3 Fatty Acids Supplementation in Humans" International Journal of Molecular Sciences 24, no. 7: 6734. https://doi.org/10.3390/ijms24076734
APA StyleGrzybkowska, A., Anczykowska, K., Antosiewicz, J., Olszewski, S., Dzitkowska-Zabielska, M., & Tomczyk, M. (2023). Identification of Optimal Reference Genes for qRT-PCR Normalization for Physical Activity Intervention and Omega-3 Fatty Acids Supplementation in Humans. International Journal of Molecular Sciences, 24(7), 6734. https://doi.org/10.3390/ijms24076734






