Bridging Gaps in HDR Improvement: The Role of MAD2L2, SCAI, and SCR7
Abstract
1. Introduction
2. Results
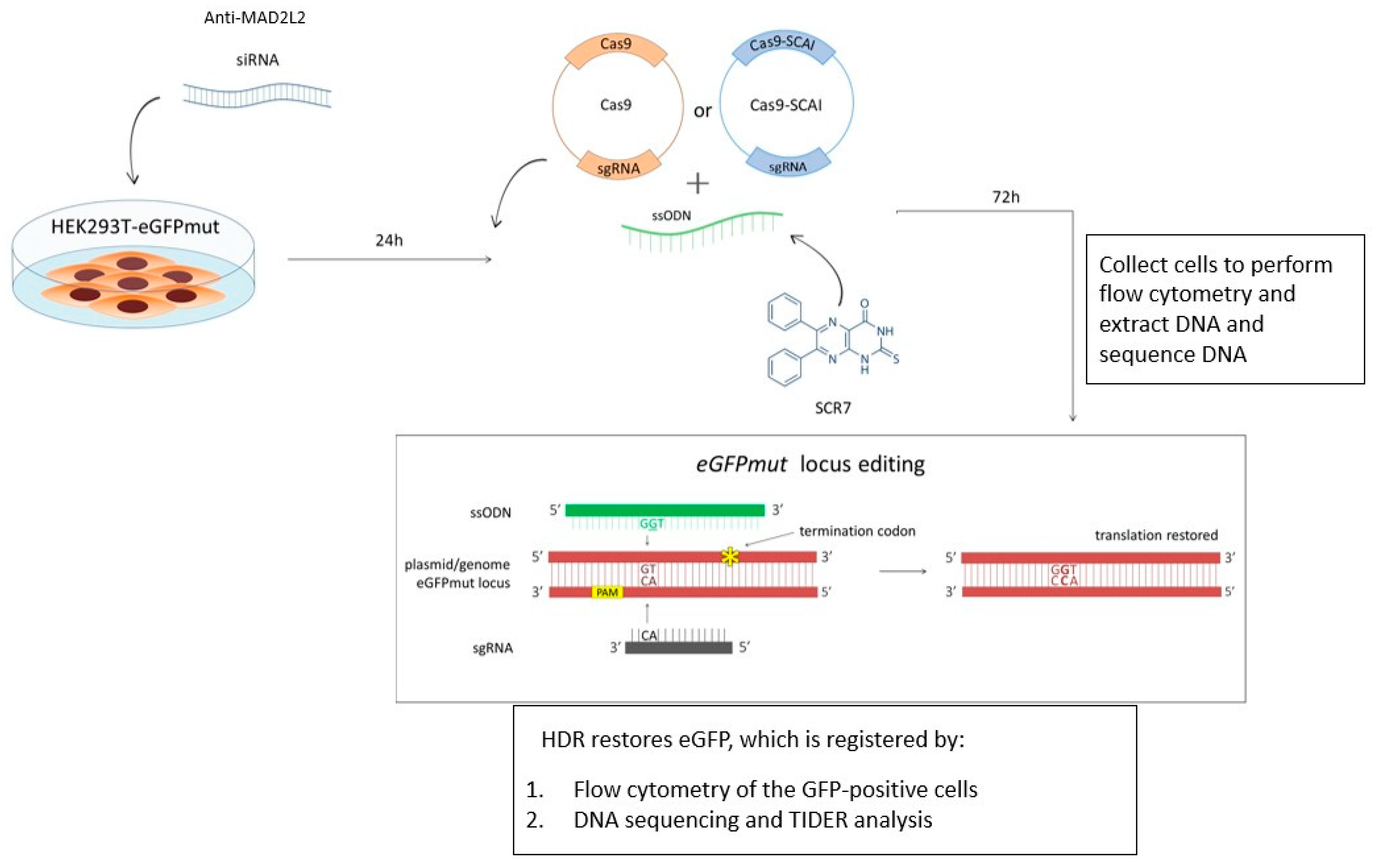
2.1. Design
2.2. Knockdown of MAD2L2 Enhances CRISPR-Mediated HDR
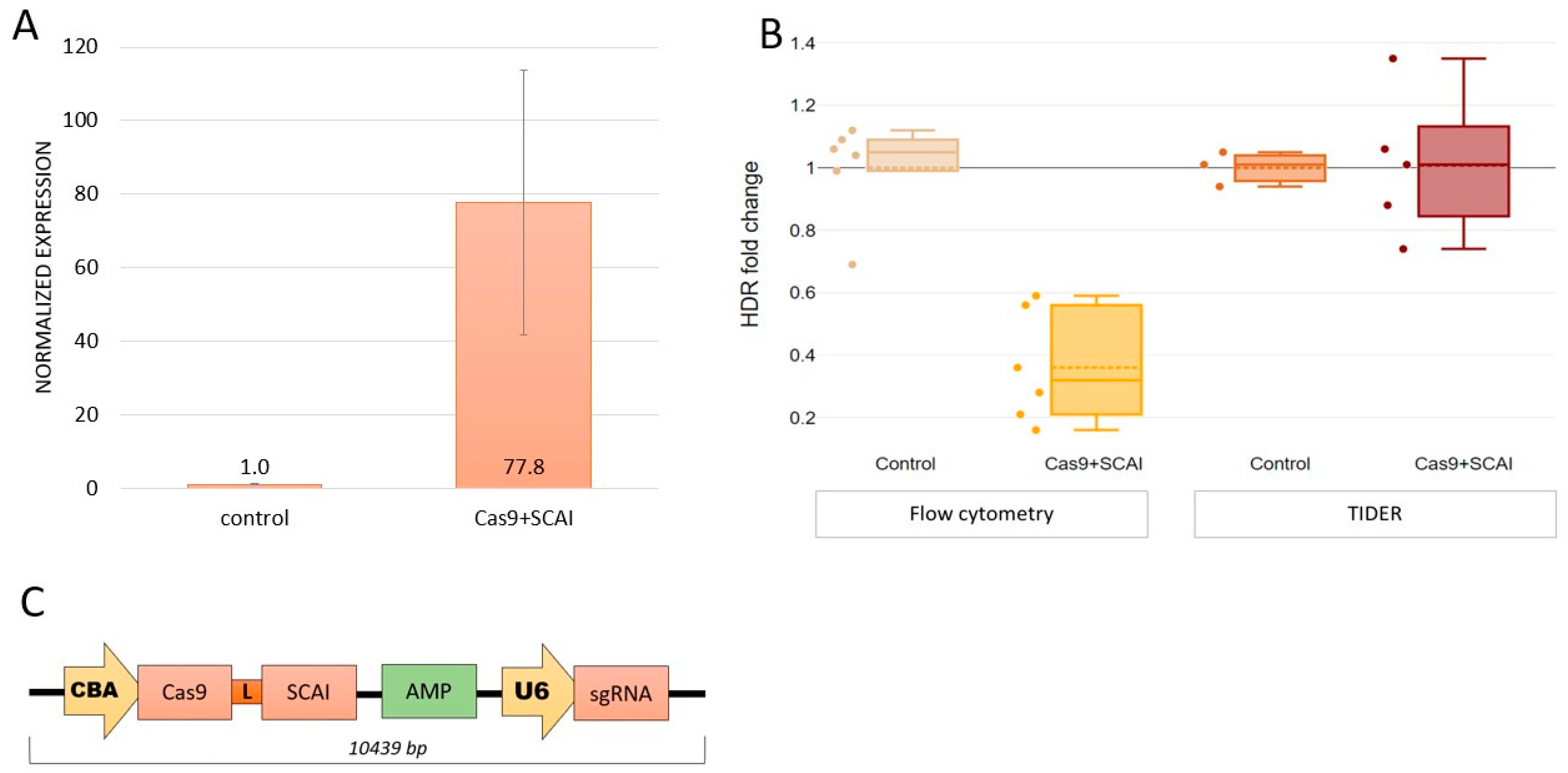
2.3. Cas9-SCAI Fusion Does Not Increase HDR Efficiency
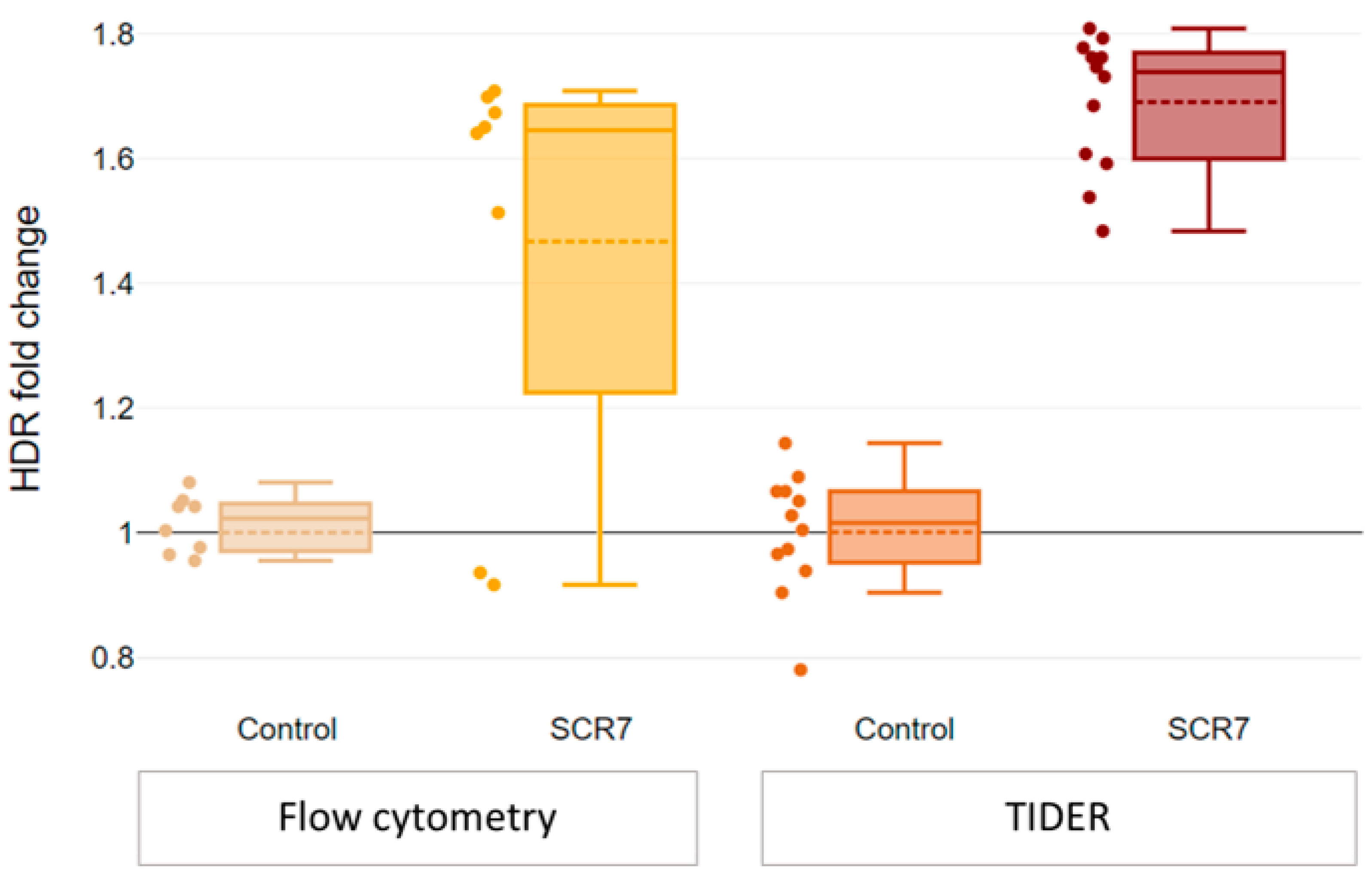
2.4. SCR7 Enhances CRISPR-Mediated HDR
3. Discussion
4. Materials and Methods
4.1. Model Development
4.2. CRISPR/Cas System and Oligonucleotides
4.3. Cell Culture and Transfection
4.4. Nucleic Acid Manipulations
4.5. Measurement of HDR Efficiency
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fradet-Turcotte, A.; Canny, M.D.; Escribano-Díaz, C.; Orthwein, A.; Leung, C.C.; Huang, H.; Landry, M.C.; Kitevski-LeBlanc, J.; Noordermeer, S.M.; Sicheri, F.; et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 2013, 499, 50–54. [Google Scholar] [CrossRef]
- Verma, P.; Greenberg, R.A. Noncanonical views of homology-directed DNA repair. Genes Dev. 2016, 30, 1138–1154. [Google Scholar] [CrossRef] [PubMed]
- Panier, S.; Boulton, S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014, 15, 7–18. [Google Scholar] [CrossRef]
- Hu, Z.; Shi, Z.; Guo, X.; Jiang, B.; Wang, G.; Luo, D.; Chen, Y.; Zhu, Y.S. Ligase IV inhibitor SCR7 enhances gene editing directed by CRISPR–Cas9 and ssODN in human cancer cells. Cell Biosci. 2018, 8, 12. [Google Scholar] [CrossRef]
- Liu, M.; Rehman, S.; Tang, X.; Gu, K.; Fan, Q.; Chen, D.; Ma, W. Methodologies for Improving HDR Efficiency. Front. Genet. 2019, 9, 691. [Google Scholar] [CrossRef] [PubMed]
- Smirnikhina, S.A.; Anuchina, A.A.; Lavrov, A.V. Ways of improving precise knock-in by genome-editing technologies. Hum. Genet. 2019, 138, 1–19. [Google Scholar] [CrossRef]
- Canny, M.D.; Moatti, N.; Wan, L.C.; Fradet-Turcotte, A.; Krasner, D.; Mateos-Gomez, P.A.; Zimmermann, M.; Orthwein, A.; Juang, Y.C.; Zhang, W.; et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat. Biotechnol. 2018, 36, 95–102. [Google Scholar] [CrossRef]
- Shao, S.; Ren, C.; Liu, Z.; Bai, Y.; Chen, Z.; Wei, Z.; Wang, X.; Zhang, Z.; Xu, K. Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int. J. Biochem. Cell Biol. 2017, 92, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Hashimoto, H.; Murakumo, Y.; Kobayashi, S.; Kogame, T.; Unzai, S.; Akashi, S.; Takeda, S.; Shimizu, T.; Sato, M. Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase ζ and REV1. J. Biol. Chem. 2010, 285, 12299–12307. [Google Scholar] [CrossRef]
- Simonetta, M.; de Krijger, I.; Serrat, J.; Moatti, N.; Fortunato, D.; Hoekman, L.; Bleijerveld, O.B.; Altelaar, A.M.; Jacobs, J.J. H4K20me2 distinguishes pre-replicative from post-replicative chromatin to appropriately direct DNA repair pathway choice by 53BP1-RIF1-MAD2L2. Cell Cycle 2018, 17, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Chapman, J.R.; Brandsma, I.; Yuan, J.; Mistrik, M.; Bouwman, P.; Bartkova, J.; Gogola, E.; Warmerdam, D.; Barazas, M.; et al. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature 2015, 521, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Brandt, D.T.; Baarlink, C.; Kitzing, T.M.; Kremmer, E.; Ivaska, J.; Nollau, P.; Grosse, R. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of β1-integrin. Nat. Cell Biol. 2009, 11, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Gasparics, Á.; Kökény, G.; Fintha, A.; Bencs, R.; Mózes, M.M.; Ágoston, E.I.; Buday, A.; Ivics, Z.; Hamar, P.; Győrffy, B.; et al. Alterations in SCAI Expression during Cell Plasticity, Fibrosis and Cancer. Pathol. Oncol. Res. POR 2018, 24, 641–651. [Google Scholar] [CrossRef]
- Isobe, S.Y.Y.; Nagao, K.; Nozaki, N.; Kimura, H.; Obuse, C. Inhibition of RIF1 by SCAI Allows BRCA1-Mediated Repair. Cell Rep. 2017, 20, 297–307. [Google Scholar] [CrossRef]
- Lieber, M.R. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End Joining Pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Nambiar, M.; Sharma, S.; Karki, S.S.; Goldsmith, G.; Hegde, M.; Kumar, S.; Pandey, M.; Singh, R.K.; Ray, P.; et al. An inhibitor of nonhomologous end-joining abrogates double-strand break repair and impedes cancer progression. Cell 2012, 151, 1474–1487. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Dougan, S.K.; Truttmann, M.C.; Bilate, A.M.; Ingram, J.R.; Ploegh, H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015, 33, 538–542. [Google Scholar] [CrossRef]
- Robert, F.; Barbeau, M.; Éthier, S.; Dostie, J.; Pelletier, J. Pharmacological inhibition of DNA-PK stimulates Cas9-mediated genome editing. Genome Med. 2015, 7, 93. [Google Scholar] [CrossRef]
- Chu, V.T.; Weber, T.; Wefers, B.; Wurst, W.; Sander, S.; Rajewsky, K.; Kühn, R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015, 33, 543–548. [Google Scholar] [CrossRef]
- Li, G.; Zhang, X.; Zhong, C.; Mo, J.; Quan, R.; Yang, J.; Liu, D.; Li, Z.; Yang, H.; Wu, Z. Small molecules enhance CRISPR/Cas9-mediated homology-directed genome editing in primary cells. Sci. Rep. 2017, 7, 8943. [Google Scholar] [CrossRef]
- Song, J.; Yang, D.; Xu, J.; Zhu, T.; Chen, Y.E.; Zhang, J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016, 7, 10548. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, M.; Kraft, T.; Brenner, B.; Petersen, B.; Niemann, H.; Montag, J. Efficient Knock-in of a Point Mutation in Porcine Fibroblasts Using the CRISPR/Cas9-GMNN Fusion Gene. Genes 2018, 9, 296. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, E.K.; Chen, T.; Amendola, M.; van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014, 42, e168. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, E.K.; Kousholt, A.N.; Harmsen, T.; Leemans, C.; Chen, T.; Jonkers, J.; Van Steensel, B. Easy Quantification of Template-Directed CRISPR/Cas9 Editing. bioRxiv 2017, 218156. Available online: https://www.biorxiv.org/content/10.1101/218156v1 (accessed on 30 September 2022). [CrossRef]
- Stirling, D.R.; Swain-Bowden, M.J.; Lucas, A.M.; Carpenter, A.E.; Cimini, B.A.; Goodman, A. CellProfiler 4: Improvements in speed, utility and usability. BMC Bioinform. 2021, 22, 433. [Google Scholar] [CrossRef]
- Gan, G.N.; Wittschieben, J.P.; Wittschieben, B.Ø.; Wood, R.D. DNA polymerase zeta (pol ζ) in higher eukaryotes. Cell Res. 2008, 18, 174–183. [Google Scholar] [CrossRef]
- Sale, J.E. REV7/MAD2L2: The multitasking maestro emerges as a barrier to recombination. EMBO J. 2015, 34, 1609–1611. [Google Scholar] [CrossRef]
- Setiaputra, D.; Durocher, D. Shieldin—The protector of DNA ends. EMBO Rep. 2019, 20, e47560. [Google Scholar] [CrossRef]
- Ghezraoui, H.; Oliveira, C.; Becker, J.R.; Bilham, K.; Moralli, D.; Anzilotti, C.; Fischer, R.; Deobagkar-Lele, M.; Sanchiz-Calvo, M.; Fueyo-Marcos, E.; et al. 53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ. Nature 2018, 560, 122–127. [Google Scholar] [CrossRef]
- Gupta, R.; Somyajit, K.; Narita, T.; Maskey, E.; Stanlie, A.; Kremer, M.; Typas, D.; Lammers, M.; Mailand, N.; Nussenzweig, A.; et al. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell 2018, 173, 972–988.e23. [Google Scholar] [CrossRef]
- Noordermeer, S.M.; Adam, S.; Setiaputra, D.; Barazas, M.; Pettitt, S.J.; Ling, A.K.; Olivieri, M.; Álvarez-Quilón, A.; Moatti, N.; Zimmermann, M.; et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 2018, 560, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Clairmont, C.S.; Sarangi, P.; Ponnienselvan, K.; Galli, L.D.; Csete, I.; Moreau, L.; Adelmant, G.; Chowdhury, D.; Marto, J.A.; D’Andrea, A.D. TRIP13 regulates DNA repair pathway choice through REV7 conformational change. Nat. Cell Biol. 2020, 22, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.K.; Mund, A.; Poulsen, S.L.; Sandoval, M.; Klement, K.; Tsouroula, K.; Tollenaere, M.A.; Räschle, M.; Soria, R.; Offermanns, S.; et al. SCAI promotes DNA double-strand break repair in distinct chromosomal contexts. Nat. Cell Biol. 2016, 18, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Pinder, J.; Salsman, J.; Dellaire, G. Nuclear domain “knock-in” screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 2015, 43, 9379–9392. [Google Scholar] [CrossRef]
- Singh, P.; Schimenti, J.C.; Bolcun-Filas, E. A Mouse Geneticist’s Practical Guide to CRISPR Applications. Genetics 2015, 199, 1–15. [Google Scholar] [CrossRef]
- Jayavaradhan, R.; Pillis, D.M.; Goodman, M.; Zhang, F.; Zhang, Y.; Andreassen, P.R.; Malik, P. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat. Commun. 2019, 10, 2866. [Google Scholar] [CrossRef]
- Yin, S.; Zhang, M.; Liu, Y.; Sun, X.; Guan, Y.; Chen, X.; Yang, L.; Huo, Y.; Yang, J.; Zhang, X.; et al. Engineering of efficiency-enhanced Cas9 and base editors with improved gene therapy efficacies. Mol. Ther. 2023, 31, 744–759. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anuchina, A.A.; Zaynitdinova, M.I.; Demchenko, A.G.; Evtushenko, N.A.; Lavrov, A.V.; Smirnikhina, S.A. Bridging Gaps in HDR Improvement: The Role of MAD2L2, SCAI, and SCR7. Int. J. Mol. Sci. 2023, 24, 6704. https://doi.org/10.3390/ijms24076704
Anuchina AA, Zaynitdinova MI, Demchenko AG, Evtushenko NA, Lavrov AV, Smirnikhina SA. Bridging Gaps in HDR Improvement: The Role of MAD2L2, SCAI, and SCR7. International Journal of Molecular Sciences. 2023; 24(7):6704. https://doi.org/10.3390/ijms24076704
Chicago/Turabian StyleAnuchina, Arina A., Milyausha I. Zaynitdinova, Anna G. Demchenko, Nadezhda A. Evtushenko, Alexander V. Lavrov, and Svetlana A. Smirnikhina. 2023. "Bridging Gaps in HDR Improvement: The Role of MAD2L2, SCAI, and SCR7" International Journal of Molecular Sciences 24, no. 7: 6704. https://doi.org/10.3390/ijms24076704
APA StyleAnuchina, A. A., Zaynitdinova, M. I., Demchenko, A. G., Evtushenko, N. A., Lavrov, A. V., & Smirnikhina, S. A. (2023). Bridging Gaps in HDR Improvement: The Role of MAD2L2, SCAI, and SCR7. International Journal of Molecular Sciences, 24(7), 6704. https://doi.org/10.3390/ijms24076704





