Recent Advances in Genome-Editing Technology with CRISPR/Cas9 Variants and Stimuli-Responsive Targeting Approaches within Tumor Cells: A Future Perspective of Cancer Management
Abstract
1. Introduction
2. Different Cancer Treatment Approaches and Their Limitations
3. CRISPR/Cas9 Structure and Mechanistic Action
4. Cas9 Engineering to Generate Its New Variants
4.1. SpCas9 Nickase
4.2. dCas9-FokI
4.3. SpCas9-D1135E
4.4. eSpCas9
4.5. SpCas9-HF1 (-HF2, -HF3, -HF4)
4.6. HypaCas9
4.7. HiFi Cas9
4.8. xCas9
4.9. Sniper-Cas9
4.10. evoCas9
4.11. SpartaCas9
4.12. LZ3 Cas9
4.13. miCas9
4.14. SuperFi-Cas9
5. Comparison between Different Cas9 Variants
6. Allosteric Modulation of Cas9-Targeting Specificity
6.1. Genetic Control for the Allosteric Modulation of Cas9-Targeting Specificity
6.1.1. sgRNA
6.1.2. PAM
6.2. Chemical Control to Regulate CRISPR/Cas9-Based Genome Editing
6.2.1. Control by Activators
6.2.2. Control by Inhibitors
6.2.3. Control by Degraders
7. Stimuli-Responsive Nanoformulations of CRISPR/Cas9 for Their Targeted Delivery
7.1. Internal Stimuli
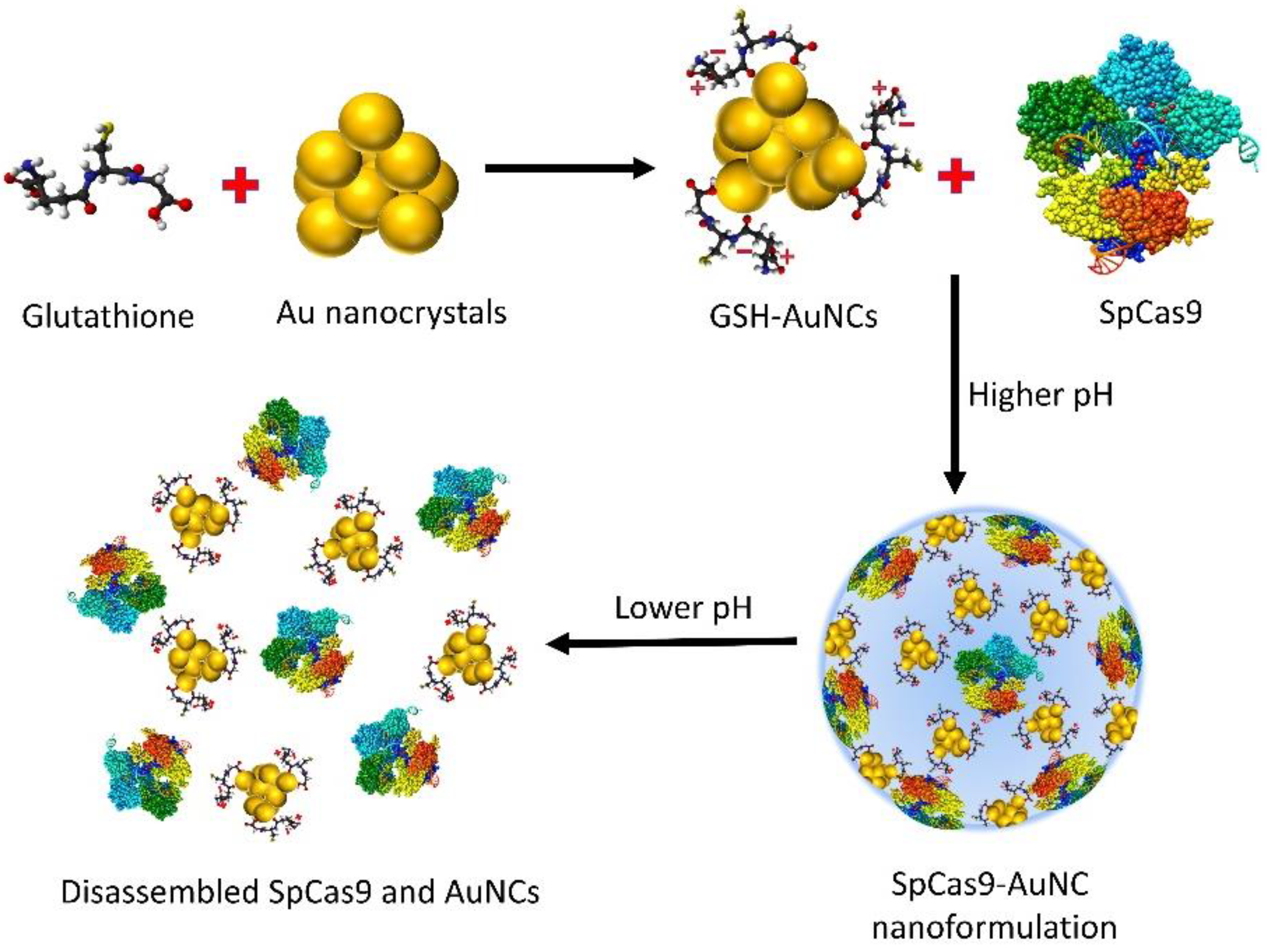
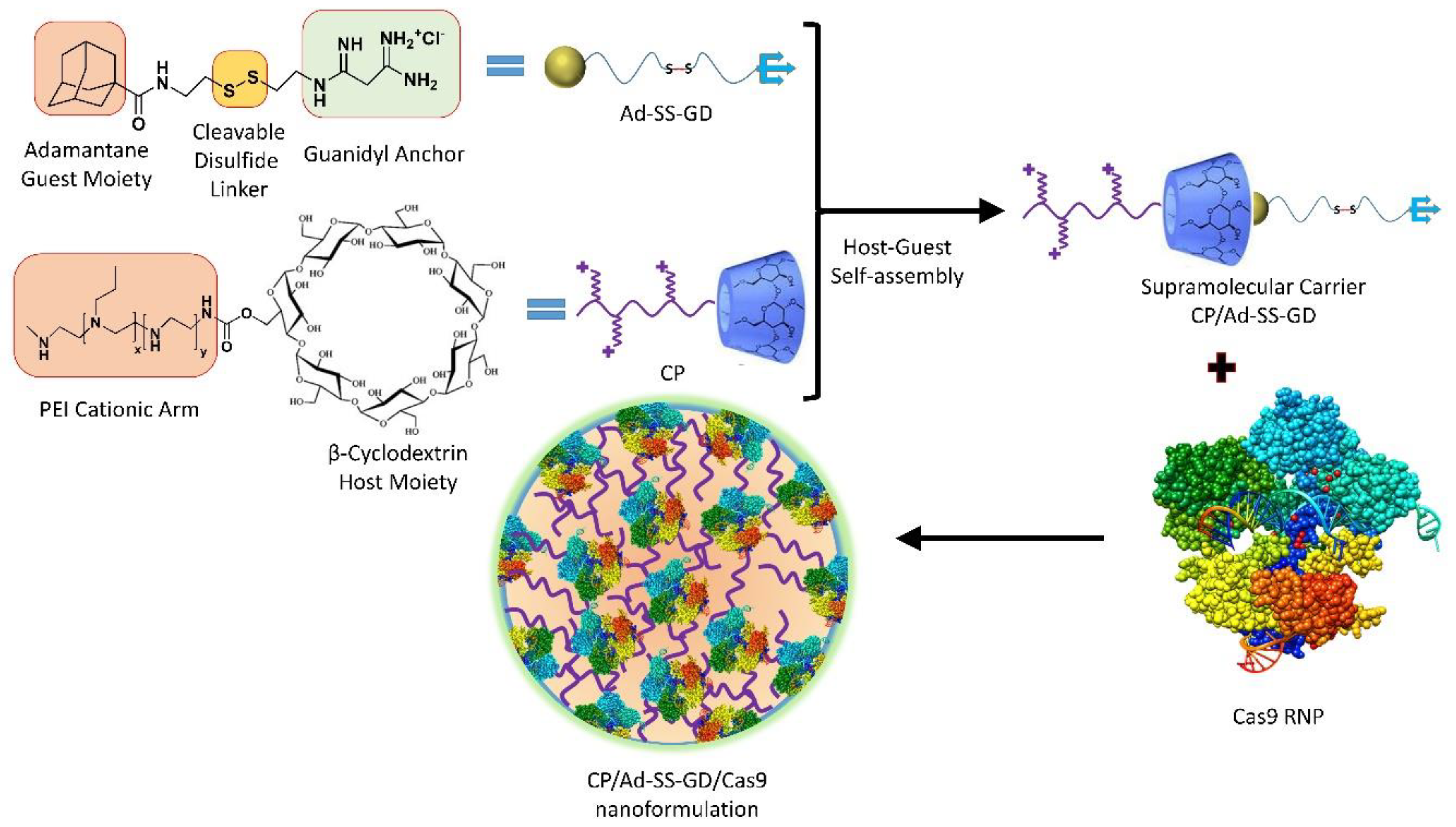
7.1.1. pH-Responsive Nanoformulations
7.1.2. GSH-Responsive Nanoformulations
7.2. External Stimuli
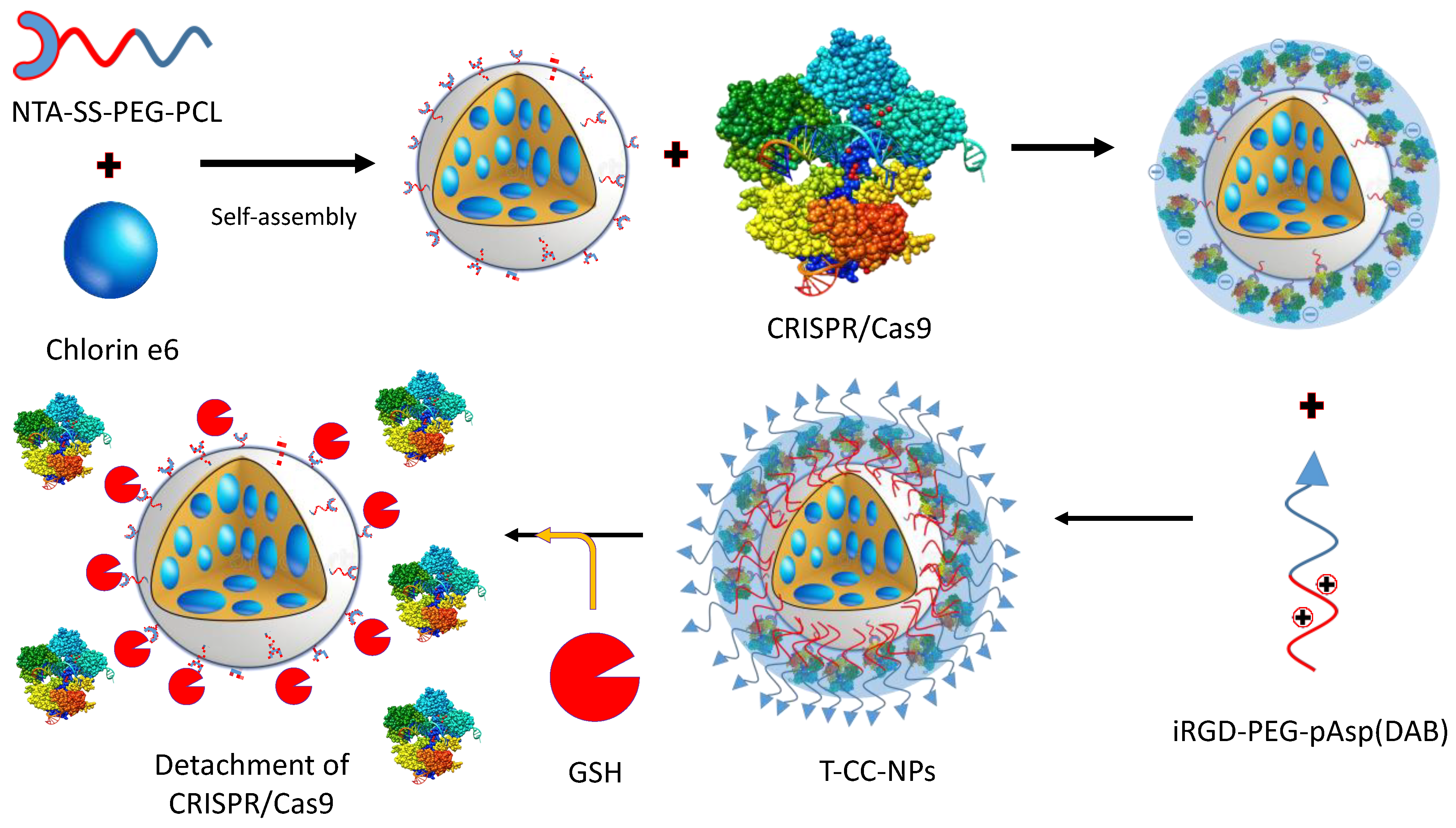
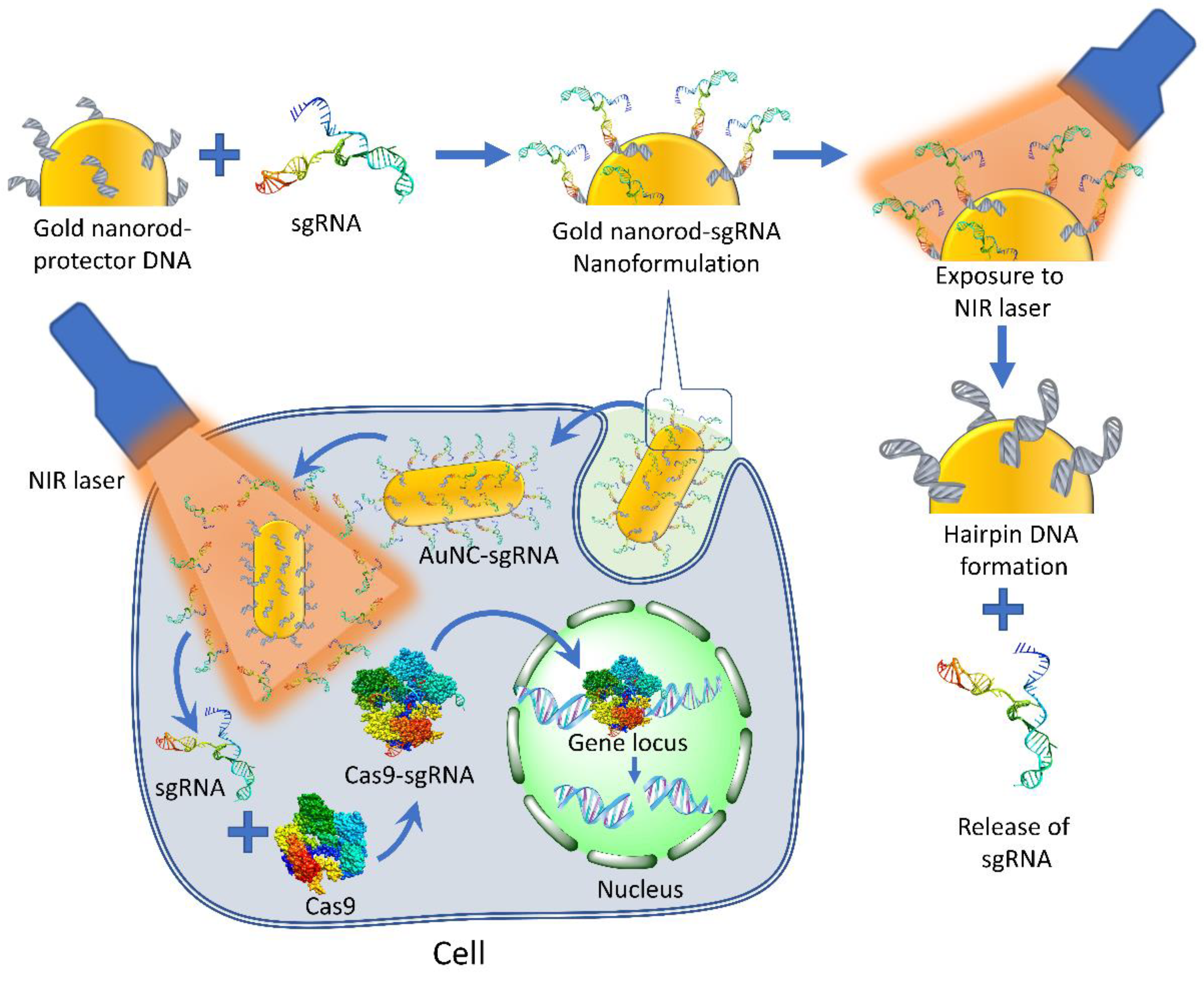
7.2.1. Photo-Responsive Targeting of CRISPR/Cas9
7.2.2. Heat-Responsive Targeting of CRISPR/Cas9
7.2.3. Ultrasound-Responsive Targeting of CRISPR/Cas9
7.2.4. Magnetic Field-Responsive Targeting of CRISPR/Cas9
| Control Type | Edited Gene | Cell or Organism Models | Key Molecule/Structure | Reference |
|---|---|---|---|---|
| Genetic regulation approaches of CRISPR/Cas9 gene editing | ||||
| Modular CRISPR fusion system | human ASCL1, ZFP42 and OCT4 genes, adipogenic genes, IL1RN, GFP reporter gene, pluripotency gene NANOG | HEK-293T, MSC, yeast cell, hPSC, HeLa cells | copies of VP16, VP64, p65, multiple sgRNAs, AcrIIA1-4, VP64-p65- Rta tripartite, KRAB, SID4X | [97,167] |
| Cell-specific promoter | mouse CD2 gene, hepatitis B virus (HBV) genome, zebrafish urod gene, macrophage-specific gene sgNtn 1, LON-2, C. elegans somatic cell DPY-5, and GFP gene | HEK-293T cell, HepG2.2.15 cell, Huh7 cell, mouse T cell, B cell, macrophage, spleen cell, neutrophil, monocyte, zebrafish, C. elegans | Macrophage-specific promoter, liver-specific promoter, erythrocyte-specific gata1 promoter, CD4 promoter, Egg cell-specific promoter | [168,169] |
| Chemical approach to control CRISPR gene editing | ||||
| Small molecule activators | SOX2 gene, GFP reporter gene, PPP1R12C, EMX1, VEGFA, ASCL | mouse zygote, HEK-293T cell, HEK293-GFP cell, STF3A cell line, N2A cell | mouse serum PCSK9 gene, GFP reporter gene, | [106,170] |
| Small molecule inhibitors | E. coli genome, CD71, and CXCR4 genes, VEGFA, endogenous IL1RN or NANOG gene, GFP or BFP reporter gene | HeLa cell, hPSC, K562 cell, E. coli, U2OS cell, HEK-293T cell, NIH/3T3 cell | ubiquitin ligase, AcrIIA1-4, DHFR and ER50, unstable protein domains | [171,172] |
| Bioresponsive delivery carrier | mouse serum PCSK9 gene, GFP reporter gene | mouse liver, mouse hepatocytes, HEK-293 T cell, lung and spleen tissues | lipid molecules with different charges, bioreducible BAMEA-O16B lipid NP | [173,174] |
| Physical approach to control CRISPR gene editing | ||||
| Light | human GRIN2B gene, zebrafish ASCL1a and HSP70 gene, the promoter of the human ASCL1 and IL1RN genes, CD71 gene, mCherry reporter gene | ZF4 cell, HEK-293T cell, E. coli, HeLa cell | dimeric fluorescent protein pdDronpa1, photo-cleavable ssDNA oligonucleotide, pMag-nMag, CIB1-CYR2- effector, photo-caged lysine, Cas9- RsLOV2 monomers | [175,176] |
| Heat | GFP reporter gene, Plk-l gene, LON-2, C. elegans somatic cell DPY-5, and GFP gene | HEK-293T cell, HCT 116 cell, A375 cell, C. elegans | APC, AuNPs, Phsp, SPPF-Dex nanoparticles | [158,159] |
| Magnetic field | mouse VEGFR2 gene, HIV LTR gene, | Hepa 1-6 cells, microglial (hμglia)/HIV (HC69) cells | magnetic iron oxide nanoparticles (MNP-BV), magneto-electric nanoparticles (MENPs) | [165,166] |
| Ultrasound | steroid type II 5-alphareductase gene, GFP reporter gene | DPC cell, androgenic alopecia, B16F10 cell, mouse | microbubble conjugated nanoliposome (MB-NL), gold nanowires (AuNWs), | [160,161] |
8. Difference between Genetic, Chemical, and Physical Control for CRISPR/Cas9 On-Target Strategies and Their Limitations
9. Critical Constraints of CRISPR/Cas9 System to Clinical Translation: Future Prospects and Challenges
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The cancer genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, L.R.; Torre, A.L.; Ahmedin, D.V. GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Glob. Cancer Stat. 2018, 68, 394–424. [Google Scholar]
- Wilbie, D.; Walther, J.; Mastrobattista, E. Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc. Chem. Res. 2019, 52, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, Z. CRISPR-Cas systems: Overview, innovations and applications in human disease research and gene therapy. Comput. Struct. Biotechnol. J. 2020, 18, 2401–2415. [Google Scholar] [CrossRef]
- Zhang, D.; Hussain, A.; Manghwar, H.; Xie, K.; Xie, S.; Zhao, S.; Larkin, R.M.; Qing, P.; Jin, S.; Ding, F. Genome editing with the CRISPR-Cas system: An art, ethics and global regulatory perspective. Plant Biotechnol. J. 2020, 18, 1651–1669. [Google Scholar] [CrossRef]
- Allemailem, K.S.; Alsahli, M.A.; Almatroudi, A.; Alrumaihi, F.; Alkhaleefah, F.K.; Rahmani, A.H.; Khan, A.A. Current updates of CRISPR/Cas9-mediated genome editing and targeting within tumor cells: An innovative strategy of cancer management. Cancer Commun. 2022, 42, 1257–1287. [Google Scholar] [CrossRef]
- Charpentier, E.; Doudna, J.A. Rewriting a genome. Nature 2013, 495, 50–51. [Google Scholar] [CrossRef]
- Kim, D.; Luk, K.; Wolfe, S.A.; Kim, J.S. Evaluating and enhancing target specificity of gene editing nucleases and deaminases. Annu. Rev. Biochem. 2019, 88, 191–220. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Edraki, A.; Mir, A.; Ibraheim, R.; Gainetdinov, I.; Yoon, Y.; Song, C.Q.; Cao, Y.; Gallant, J.; Xue, W.; Rivera-Prez, J.A.; et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. Mol. Cell 2019, 73, 714–726. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Müller, M.; Lee, C.M.; Gasiunas, G.; Davis, T.H.; Cradick, T.J.; Siksnys, V.; Bao, G.; Cathomen, T.; Mussolino, C. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol. Ther. 2016, 24, 636–644. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Jakimo, N.; Jacobson, J.M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 2018, 4, eaau0766. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef]
- Casini, A.; Olivieri, M.; Petris, G.; Montagna, C.; Reginato, G.; Maule, G.; Lorenzin, F.; Prandi, D.; Romanel, A.; Demichelis, F.; et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018, 36, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.S.; Dagdas, Y.S.; Kleinstiver, B.P.; Welch, M.M.; Sousa, A.A.; Harrington, L.B.; Sternberg, S.H.; Joung, J.K.; Yildiz, A.; Doudna, J.A. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature 2017, 550, 407–410. [Google Scholar] [CrossRef]
- Lee, J.K.; Jeong, E.; Lee, J.; Jung, M.; Shin, E.; Kim, Y.H.; Lee, K.; Jung, I.; Kim, D.; Kim, S.; et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018, 9, 3048. [Google Scholar] [CrossRef]
- Anders, C.; Bargsten, K.; Jinek, M. Structural plasticity of PAM recognition by engineered variants of the RNA-guided endonuclease Cas9. Mol. Cell 2016, 61, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Schlimgen, R.; Howard, J.; Wooley, D.; Thompson, M.; Baden, L.R.; Yang, O.O.; Christiani, D.C.; Mostoslavsky, G.; Diamond, D.V.; Duane, E.G.; et al. Risks associated with lentiviral vector exposures and prevention strategies. J. Occup. Environ. Med. 2016, 58, 1159. [Google Scholar] [CrossRef]
- Pucci, C.; Martinelli, C.; Ciofani, G. Innovative approaches for cancer treatment: Current perspectives and new challenges. Ecancermedicalscience 2019, 13, 961. [Google Scholar] [CrossRef]
- Hafeez, M.N.; Celia, C.; Petrikaite, V. Challenges towards targeted drug delivery in cancer nanomedicines. Processes 2021, 9, 1527. [Google Scholar] [CrossRef]
- Martinelli, C. Exosomes: New biomarkers for targeted cancer therapy. In Molecular Oncology: Underlying Mechanisms and Translational Advancements; Springer: Berlin/Heidelberg, Germany, 2017; pp. 129–157. [Google Scholar]
- Liu, X.; Zhang, Y.; Wang, Y.; Zhu, W.; Li, G.; Ma, X.; Zhang, Y.; Chen, S.; Tiwari, S.; Shi, K.; et al. Comprehensive understanding of magnetic hyperthermia for improving antitumor therapeutic efficacy. Theranostics 2020, 10, 3793. [Google Scholar] [CrossRef]
- Yu, K.H.; Zhang, C.; Berry, G.J.; Altman, R.B.; Ré, C.; Rubin, D.L.; Snyder, M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat. Commun. 2016, 7, 12474. [Google Scholar] [CrossRef]
- Aerts, H.J. The potential of radiomic-based phenotyping in precision medicine: A review. JAMA Oncol. 2016, 2, 1636–1642. [Google Scholar] [CrossRef]
- Shanker, M.; Jin, J.; Branch, C.D.; Miyamoto, S.; Grimm, E.A.; Roth, J.A.; Ramesh, R. Tumor suppressor gene-based nanotherapy: From test tube to the clinic. J. Drug Deliv. 2011, 2011, 465845. [Google Scholar] [CrossRef]
- Wang, T.; Shigdar, S.; Al Shamaileh, H.; Gantier, M.P.; Yin, W.; Xiang, D.; Wang, L.; Zhou, S.F.; Hou, Y.; Wang, P.; et al. Challenges and opportunities for siRNA-based cancer treatment. Cancer Lett. 2017, 387, 77–83. [Google Scholar] [CrossRef]
- Shankar, S.; Sreekumar, A.; Prasad, D.; Das, A.V.; Pillai, M.R. Genome editing of oncogenes with ZFNs and TALENs: Caveats in nuclease design. Cancer Cell Int. 2018, 18, 169. [Google Scholar] [CrossRef] [PubMed]
- Kampmann, M. CRISPRi and CRISPRa screens in mammalian cells for precision biology and medicine. ACS Chem. Biol. 2018, 13, 406–416. [Google Scholar] [CrossRef]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef]
- Mojica, F.J.; Dez-Villaseor, C.; Garca-Martnez, J.; Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009, 155, 733–740. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR–Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef]
- Bharathkumar, N.; Sunil, A.; Meera, P.; Aksah, S.; Kannan, M.; Saravanan, K.M.; Anand, T. CRISPR/Cas-Based modifications for therapeutic applications: A review. Mol. Biotechnol. 2022, 64, 355–372. [Google Scholar] [CrossRef]
- Ma, E.; Chen, K.; Shi, H.; Stahl, E.C.; Adler, B.; Trinidad, M.; Liu, J.; Zhou, K.; Ye, J.; Doudna, J.A. Improved genome editing by an engineered CRISPR-Cas12a. Nucleic Acids Res. 2022, 50, 12689–12701. [Google Scholar] [CrossRef]
- Ledford, H. Alternative CRISPR system could improve genome editing. Nature 2015, 526, 17. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Krupovic, M.; Forterre, P. History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. J. Bacteriol. 2018, 200, e00580-17. [Google Scholar] [CrossRef]
- Marraffini, L.A. CRISPR-Cas immunity in prokaryotes. Nature 2015, 526, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, V.; Lin, S.; Guilinger, J.P.; Ma, E.; Doudna, J.A.; Liu, D.R. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013, 31, 839–843. [Google Scholar] [CrossRef]
- Gandhi, S.; Haeussler, M.; Razy-Krajka, F.; Christiaen, L.; Stolfi, A. Evaluation and rational design of guide RNAs for efficient CRISPR/Cas9-mediated mutagenesis in Ciona. Dev. Biol. 2017, 425, 8–20. [Google Scholar] [CrossRef]
- Lostal-Seijo, I.; Louzao, I.; Juanes, M.; Montenegro, J. Peptide/Cas9 nanostructures for ribonucleoprotein cell membrane transport and gene edition. Chem. Sci. 2017, 8, 7923–7931. [Google Scholar] [CrossRef] [PubMed]
- Salem, A.K.; Searson, P.C.; Leong, K.W. Multifunctional nanorods for gene delivery. Nat. Mater. 2003, 2, 668–671. [Google Scholar] [CrossRef]
- Morris, A.S.; Sebag, S.C.; Paschke, J.D.; Wongrakpanich, A.; Ebeid, K.; Anderson, M.E.; Grumbach, I.M.; Salem, A.K. Cationic CaMKII inhibiting nanoparticles prevent allergic asthma. Mol. Pharm. 2017, 14, 2166–2175. [Google Scholar] [CrossRef]
- Glass, Z.; Lee, M.; Li, Y.; Xu, Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018, 36, 173–185. [Google Scholar] [CrossRef]
- Mekler, V.; Minakhin, L.; Semenova, E.; Kuznedelov, K.; Severinov, K. Kinetics of the CRISPR-Cas9 effector complex assembly and the role of 3′-terminal segment of guide RNA. Nucleic Acids Res. 2016, 44, 2837–2845. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, J.E.; Deeconda, A.; Tsang, S.H. Viral vectors, engineered cells and the CRISPR revolution. In Precision Medicine, CRISPR, and Genome Engineering. Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2017; pp. 3–27. [Google Scholar]
- Liu, C.; Zhang, L.; Liu, H.; Cheng, K. Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Control. Release 2017, 266, 17–26. [Google Scholar] [CrossRef]
- Dominguez, A.A.; Lim, W.A.; Qi, L.S. Beyond editing: Repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016, 17, 5–15. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol 2014, 32, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Wyvekens, N.; Topkar, V.V.; Khayter, C.; Joung, J.K.; Tsai, S.Q. Dimeric CRISPR RNA guided FokI-dCas9 nucleases directed by truncated gRNAs for highly specific genome editing. Hum. Gene Ther. 2015, 26, 425–431. [Google Scholar] [CrossRef]
- Schmid-Burgk, J.L.; Gao, L.; Li, D.; Gardner, Z.; Strecker, J.; Lash, B.; Zhang, F. Highly parallel profiling of Cas9 variant specificity. Mol. Cell 2020, 78, 794–800. [Google Scholar] [CrossRef]
- Kim, N.; Kim, H.K.; Lee, S.; Seo, J.H.; Choi, J.W.; Park, J.; Min, S.; Yoon, S.; Cho, S.R.; Kim, H.H. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat. Biotechnol. 2020, 38, 1328–1336. [Google Scholar] [CrossRef]
- Ikeda, A.; Fujii, W.; Sugiura, K.; Naito, K. High-fidelity endonuclease variant HypaCas9 facilitates accurate allele-specific gene modification in mouse zygotes. Commun. Biol. 2019, 2, 371. [Google Scholar] [CrossRef] [PubMed]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef]
- Cerchione, D.; Loveluck, K.; Tillotson, E.L.; Harbinski, F.; DaSilva, J.; Kelley, C.P.; Keston- Smith, E.; Fernandez, C.A.; Myer, V.E.; Jayaram, H.; et al. SMOOT libraries and phage-induced directed evolution of Cas9 to engineer reduced off-target activity. PLoS ONE 2020, 15, e0231716. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Ruan, J.; Song, J.; Wen, L.; Yang, D.; Zhao, J.; Xia, X.; Chen, Y.E.; Zhang, J.; Xu, J. MiCas9 increases large size gene knock-in rates and reduces undesirable on-target and offtarget indel edits. Nat. Commun. 2020, 11, 6082. [Google Scholar] [CrossRef] [PubMed]
- Kulcsar, P.I.; Talas, A.; Welker, E. SuperFi-Cas9 exhibits extremely high fidelity but reduced activity in mammalian cells. BioRxiv 2022, 2022–2205. [Google Scholar] [CrossRef]
- Lamberth, J.; Daley, L.; Natarajan, P.; Khoruzhenko, S.; Becker, N.; Taglicht, D.; Davis, G.D.; Ji, Q. Highly efficient and specific genome editing in human cells with paired CRISPRCas9 nickase ribonucleoproteins. BioRxiv 2019, 766493. [Google Scholar] [CrossRef]
- Karimova, M.; Beschorner, N.; Dammermann, W.; Chemnitz, J.; Indenbirken, D.; Bockmann, J.H.; Grundhoff, A.; Lüth, S.; Buchholz, F.; Wiesch, J.S.Z.; et al. CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Sci. Rep. 2015, 5, 13734. [Google Scholar] [CrossRef]
- Zhao, G.; Wang, Q.; Gu, Q.; Qiang, W.; Wei, J.J.; Dong, P.; Watari, H.; Li, W.; Yue, J. Lentiviral CRISPR/Cas9 nickase vector mediated BIRC5 editing inhibits epithelial to mesenchymal transition in ovarian cancer cells. Oncotarget 2017, 8, 94666. [Google Scholar] [CrossRef]
- Martinez-Escobar, A.; Luna-Callejas, B.; Ramón-Gallegos, E. CRISPR-dCas9-based artificial transcription factors to improve efficacy of cancer treatment with drug repurposing: Proposal for future research. Front. Oncol. 2021, 10, 604948. [Google Scholar] [CrossRef]
- Kang, M.; Zuo, Z.; Yin, Z.; Gu, J. Molecular Mechanism of D1135E-Induced Discriminated CRISPR-Cas9 PAM Recognition. J. Chem. Inf. Model. 2022, 62, 3057–3066. [Google Scholar] [CrossRef]
- Dagdas, Y.S.; Chen, J.S.; Sternberg, S.H.; Doudna, J.A.; Yildiz, A. A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci. Adv. 2017, 3, eaao0027. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, M.; Shan, H.; Chen, S.; Xu, Y.; Song, Y.; Zhang, Q.; Yuan, H.; Ouyang, H.; Li, Z.; et al. Expanded targeting scope and enhanced base editing efficiency in rabbit using optimized xCas9 (3.7). Cell. Mol. Life Sci. 2019, 76, 4155–4164. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Nunez, J.K.; Harrington, L.B.; Doudna, J.A. Chemical and biophysical modulation of Cas9 for tunable genome engineering. ACS Chem. Biol. 2016, 11, 681–688. [Google Scholar] [CrossRef]
- Kulcsr, P.I.; Talas, A.; Tth, E.; Nyeste, A.; Ligeti, Z.; Welker, Z.; Welker, E. Blackjack mutations improve the on-target activities of increased fidelity variants of SpCas9 with 5′ G-extended sgRNAs. Nat. Commun. 2020, 11, 1–4. [Google Scholar] [CrossRef]
- Bravo, J.P.; Liu, M.S.; Hibshman, G.N.; Dangerfield, T.L.; Jung, K.; McCool, R.S.; Johnson, K.A.; Taylor, D.W. Structural basis for mismatch surveillance by CRISPR–Cas9. Nature 2022, 603, 343–347. [Google Scholar] [CrossRef]
- Trevino, A.E.; Zhang, F. Genome editing using Cas9 nickases. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2014; Volume 546, pp. 161–174. [Google Scholar]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPRCas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Li, L.; Hu, S.; Chen, X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018, 171, 207–218. [Google Scholar] [CrossRef]
- Yin, H.; Kauffman, K.J.; Anderson, D.G. Delivery technologies for genome editing. Nat. Rev. Drug Discov. 2017, 16, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Cyranoski, D. First trial of CRISPR in people. Nature 2016, 535, 476–477. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, L.; Gu, W.; Gao, Y.; Lin, L.; Zhang, Z.; Xi, Y.; Li, Y. The performance of docetaxel loaded solid lipid nanoparticles targeted to hepatocellular carcinoma. Biomaterials 2009, 30, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Aldayel, A.M.; Naguib, Y.W.; O'mary, H.L.; Li, X.; Niu, M.; Ruwona, T.B.; Cui, Z. Acid-sensitive sheddable PEGylated PLGA nanoparticles increase the delivery of TNF-α siRNA in chronic inflammation sites. Mol. Ther.-Nucleic Acids 2016, 5, e340. [Google Scholar] [CrossRef] [PubMed]
- Naguib, Y.W.; Cui, Z. Nanomedicine: The promise and challenges in cancer chemotherapy. In Nanomaterial. Advances in Experimental Medicine and Biology; Springer: Dordrecht, The Netherlands, 2014; pp. 207–233. [Google Scholar]
- Givens, B.E.; Geary, S.M.; Salem, A.K. Nanoparticle-based CpG-oligonucleotide therapy for treating allergic asthma. Immunotherapy 2018, 10, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.X.; Song, Z.; Lao, Y.H.; Xu, X.; Gong, J.; Cheng, D.; Chakraborty, S.; Park, J.S.; Li, M.; Huang, D.; et al. Nonviral gene editing via CRISPR/Cas9 delivery by membrane disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. USA 2018, 115, 4903–4908. [Google Scholar] [CrossRef]
- Ha, J.S.; Lee, J.S.; Jeong, J.; Kim, H.; Byun, J.; Kim, S.A.; Lee, H.J.; Chung, H.S.; Lee, J.B.; Ahn, D.R. Poly-sgRNA/siRNA ribonucleoprotein nanoparticles for targeted gene disruption. J. Control. Release 2017, 250, 27–35. [Google Scholar] [CrossRef]
- Wang, M.; Glass, Z.A.; Xu, Q. Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 2017, 24, 144–150. [Google Scholar] [CrossRef]
- Mout, R.; Ray, M.; Lee, Y.W.; Scaletti, F.; Rotello, V.M. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: Progress and challenges. Bioconjugate Chem. 2017, 28, 880–884. [Google Scholar] [CrossRef]
- Xu, S.; Olenyuk, B.Z.; Okamoto, C.T.; Hamm-Alvarez, S.F. Targeting receptor-mediated endocytotic pathways with nanoparticles: Rationale and advances. Adv. Drug Deliv. Rev. 2013, 65, 121–138. [Google Scholar] [CrossRef]
- Kuscu, C.; Arslan, S.; Singh, R.; Thorpe, J.; Adli, M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014, 32, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ge, X.; Yang, F.; Zhang, L.; Zheng, J.; Tan, X.; Jin, Z.B.; Qu, J.; Gu, F. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci. Rep. 2014, 4, 5405. [Google Scholar] [CrossRef] [PubMed]
- Furuhata, Y.; Nihongaki, Y.; Sato, M.; Yoshimoto, K. Control of adipogenic differentiation in mesenchymal stem cells via endogenous gene activation using CRISPR-Cas9. ACS Synth. Biol. 2017, 6, 2191–2197. [Google Scholar] [CrossRef] [PubMed]
- Dow, L.E.; Fisher, J.; O'rourke, K.P.; Muley, A.; Kastenhuber, E.R.; Livshits, G.; Tschaharganeh, D.F.; Socci, N.D.; Lowe, S.W. Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol. 2015, 33, 390–394. [Google Scholar] [CrossRef] [PubMed]
- Aubrey, B.J.; Kelly, G.L.; Kueh, A.J.; Brennan, M.S.; O’Connor, L.; Milla, L.; Wilcox, S.; Tai, L.; Strasser, A.; Herold, M.J. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015, 10, 1422–1432. [Google Scholar] [CrossRef] [PubMed]
- Gonzlez, F.; Zhu, Z.; Shi, Z.D.; Lelli, K.; Verma, N.; Li, Q.V.; Huangfu, D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell 2014, 15, 215–226. [Google Scholar] [CrossRef]
- Rakhit, R.; Navarro, R.; Wandless, T.J. Chemical biology strategies for posttranslational control of protein function. Chem. Biol. 2014, 21, 1238–1252. [Google Scholar] [CrossRef] [PubMed]
- Davis, K.M.; Pattanayak, V.; Thompson, D.B.; Zuris, J.A.; Liu, D.R. Small molecule–triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015, 11, 316–318. [Google Scholar] [CrossRef]
- Liu, K.I.; Ramli, M.N.; Woo, C.W.; Wang, Y.; Zhao, T.; Zhang, X.; Yim, G.R.; Chong, B.Y.; Gowher, A.; Chua, M.Z.; et al. A chemical-inducible CRISPR–Cas9 system for rapid control of genome editing. Nat. Chem. Biol. 2016, 12, 980–987. [Google Scholar] [CrossRef]
- Oakes, B.L.; Nadler, D.C.; Flamholz, A.; Fellmann, C.; Staahl, B.T.; Doudna, J.A.; Savage, D.F. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat. Biotechnol. 2016, 34, 646–651. [Google Scholar] [CrossRef]
- DeRose, R.; Miyamoto, T.; Inoue, T. Manipulating signaling at will: Chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflügers Arch.-Eur. J. Physiol. 2013, 465, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Volz, S.E.; Zhang, F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015, 33, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Xiong, X.; Wong, S.; Charles, E.J.; Lim, W.A.; Qi, L.S. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat. Methods 2016, 13, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Lo, A.; Qi, L. Genetic and epigenetic control of gene expression by CRISPR–Cas systems. F1000Research 2017, 6. [Google Scholar] [CrossRef]
- Pawluk, A.; Davidson, A.R.; Maxwell, K.L. Anti-CRISPR: Discovery, mechanism and function. Nat. Rev. Microbiol. 2018, 16, 12–17. [Google Scholar] [CrossRef]
- Bondy-Denomy, J. Protein inhibitors of CRISPR-Cas9. ACS chemical biology 2018, 13, 417–423. [Google Scholar] [CrossRef]
- Dong, C.; Hao, G.F.; Hua, H.L.; Liu, S.; Labena, A.A.; Chai, G.; Huang, J.; Rao, N.; Guo, F.B. Anti- CRISPRdb: A comprehensive online resource for anti-CRISPR proteins. Nucleic Acids Res. 2018, 46, D393–D398. [Google Scholar] [CrossRef] [PubMed]
- Rauch, B.J.; Silvis, M.R.; Hultquist, J.F.; Waters, C.S.; McGregor, M.J.; Krogan, N.J.; Bondy-Denomy, J. Inhibition of CRISPR-Cas9 with bacteriophage proteins. Cell 2017, 168, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Bondy-Denomy, J.; Garcia, B.; Strum, S.; Du, M.; Rollins, M.F.; Hidalgo-Reyes, Y.; Wiedenheft, B.; Maxwell, K.L.; Davidson, A.R. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature 2015, 526, 136–139. [Google Scholar] [CrossRef]
- Shin, J.; Jiang, F.; Liu, J.J.; Bray, N.L.; Rauch, B.J.; Baik, S.H.; Nogales, E.; Bondy-Denomy, J.; Corn, J.E.; Doudna, J.A. Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv. 2017, 3, e1701620. [Google Scholar] [CrossRef]
- Koehler, A.N. A complex task? Direct modulation of transcription factors with small molecules. Curr. Opin. Chem. Biol. 2010, 14, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, C.T.; Deshpande, P.S.; Dever, D.P.; Camarena, J.; Lemgart, V.T.; Cromer, M.K.; Vakulskas, C.A.; Collingwood, M.A.; Zhang, L.; Bode, N.M.; et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019, 25, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Banaszynski, L.A.; Wandless, T.J. Conditional control of protein function. Chem. Biol. 2006, 13, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Raina, K.; Crews, C.M. Chemical inducers of targeted protein degradation. J. Biol. Chem. 2010, 285, 11057–11060. [Google Scholar] [CrossRef]
- Bondeson, D.P.; Crews, C.M. Targeted protein degradation by small molecules. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 107. [Google Scholar] [CrossRef]
- Neklesa, T.K.; Winkler, J.D.; Crews, C.M. Targeted protein degradation by PROTACs. Pharmacol. Ther. 2017, 174, 138–144. [Google Scholar] [CrossRef]
- Gangopadhyay, S.A.; Cox, K.J.; Manna, D.; Lim, D.; Maji, B.; Zhou, Q.; Choudhary, A. Precision control of CRISPR-Cas9 using small molecules and light. Biochemistry 2019, 58, 234–244. [Google Scholar] [CrossRef]
- Nabet, B.; Roberts, J.M.; Buckley, D.L.; Paulk, J.; Dastjerdi, S.; Yang, A.; Leggett, A.L.; Erb, M.A.; Lawlor, M.A.; Souza, A.; et al. The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 2018, 14, 431–441. [Google Scholar] [CrossRef]
- Maji, B.; Moore, C.L.; Zetsche, B.; Volz, S.E.; Zhang, F.; Shoulders, M.D.; Choudhary, A. Multidimensional chemical control of CRISPR–Cas9. Nat. Chem. Biol. 2017, 13, 9–11. [Google Scholar] [CrossRef]
- Tang, Q.; Yu, B.; Gao, L.; Cong, H.; Song, N.; Lu, C. Stimuli responsive nanoparticles for controlled anti-cancer drug release. Curr. Med. Chem. 2018, 25, 1837–1866. [Google Scholar] [CrossRef]
- Liu, G.; Zhao, X.; Zhang, Y.; Xu, J.; Xu, J.; Li, Y.; Min, H.; Shi, J.; Zhao, Y.; Wei, J.; et al. Engineering biomimetic platesomes for pH-responsive drug delivery and enhanced antitumor activity. Adv. Mater. 2019, 31, 1900795. [Google Scholar] [CrossRef] [PubMed]
- Zhen, S.; Liu, Y.; Lu, J.; Tuo, X.; Yang, X.; Chen, H.; Chen, W.; Li, X. Human papillomavirus oncogene manipulation using clustered regularly interspersed short palindromic repeats/Cas9 delivered by pH-sensitive cationic liposomes. Hum. Gene Ther. 2020, 31, 309–324. [Google Scholar] [CrossRef]
- Qi, Y.; Song, H.; Xiao, H.; Cheng, G.; Yu, B.; Xu, F.J. Fluorinated acid-labile branched hydroxyl-rich nanosystems for flexible and robust delivery of plasmids. Small 2018, 14, 1803061. [Google Scholar] [CrossRef]
- Ju, E.; Li, T.; Ramos da Silva, S.; Gao, S.J. Gold nanocluster-mediated efficient delivery of Cas9 protein through pH-induced assembly-disassembly for inactivation of virus oncogenes. ACS Appl. Mater. Interfaces 2019, 11, 34717–34724. [Google Scholar] [CrossRef] [PubMed]
- Alsaiari, S.K.; Patil, S.; Alyami, M.; Alamoudi, K.O.; Aleisa, F.A.; Merzaban, J.S.; Li, M.; Khashab, N.M. Endosomal escape and delivery of CRISPR/Cas9 genome editing machinery enabled by nanoscale zeolitic imidazolate framework. J. Am. Chem. Soc. 2018, 140, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shahi, P.K.; Xie, R.; Zhang, H.; Abdeen, A.A.; Yodsanit, N.; Ma, Z.; Saha, K.; Pattnaik, B.R.; Gong, S. A pH-responsive silica–metal–organic framework hybrid nanoparticle for the delivery of hydrophilic drugs, nucleic acids, and CRISPR-Cas9 genome editing machineries. J. Control. Release 2020, 324, 194–203. [Google Scholar] [CrossRef]
- Cai, W.; Luo, T.; Mao, L.; Wang, M. Spatiotemporal Delivery of CRISPR/Cas9 Genome Editing Machinery Using Stimuli-Responsive Vehicles. Angew. Chem. 2021, 133, 8679–8689. [Google Scholar] [CrossRef]
- Mi, P. Stimuli-responsive nanocarriers for drug delivery, tumor imaging, therapy and theranostics. Theranostics 2020, 10, 4557. [Google Scholar] [CrossRef]
- Liu, J.; Wu, T.; Lu, X.; Wu, X.; Liu, S.; Zhao, S.; Xu, X.; Ding, B. A self-assembled platform based on branched DNA for sgRNA/Cas9/antisense delivery. J. Am. Chem. Soc. 2019, 141, 19032–19037. [Google Scholar] [CrossRef]
- Tang, Q.; Liu, J.; Jiang, Y.; Zhang, M.; Mao, L.; Wang, M. Cell-selective messenger RNA delivery and CRISPR/Cas9 genome editing by modulating the interface of phenylboronic acid-derived lipid nanoparticles and cellular surface sialic acid. ACS Appl. Mater. Interfaces 2019, 11, 46585–46590. [Google Scholar] [CrossRef]
- Chen, G.; Abdeen, A.A.; Wang, Y.; Shahi, P.K.; Robertson, S.; Xie, R.; Suzuki, M.; Pattnaik, B.R.; Saha, K.; Gong, S. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat. Nanotechnol. 2019, 14, 974–980. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.S.; Huang, T.; Song, J.; Ou, X.Y.; Wang, Z.; Deng, H.; Tian, R.; Liu, Y.; Wang, J.F.; Liu, Y.; et al. Synthesis of copper peroxide nanodots for H2O2 self-supplying chemodynamic therapy. J. Am. Chem. Soc. 2019, 141, 9937–9945. [Google Scholar] [CrossRef] [PubMed]
- Wan, T.; Chen, Y.; Pan, Q.; Xu, X.; Kang, Y.; Gao, X.; Huang, F.; Wu, C.; Ping, Y. Genome editing of mutant KRAS through supramolecular polymer-mediated delivery of Cas9 ribonucleoprotein for colorectal cancer therapy. J. Control. Release 2020, 322, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Li, X.; Liu, S.; Chen, J.; Li, M.; Chew, S.Y.; Leong, K.W.; Cheng, D. Codelivery of CRISPR-Cas9 and chlorin e6 for spatially controlled tumor-specific gene editing with synergistic drug effects. Sci. Adv. 2020, 6, eabb4005. [Google Scholar] [CrossRef]
- Zhang, Q.; Kuang, G.; He, S.; Liu, S.; Lu, H.; Li, X.; Zhou, D.; Huang, Y. Chain-shattering Pt (IV)-backboned polymeric nanoplatform for efficient CRISPR/Cas9 gene editing to enhance synergistic cancer therapy. Nano Res. 2021, 14, 601–610. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Y.; Xin, H.; Wan, T.; Ping, Y. Near-infrared optogenetic engineering of photothermal nanoCRISPR for programmable genome editing. Proc. Natl. Acad. Sci. USA 2020, 117, 2395–2405. [Google Scholar] [CrossRef]
- Yu, Y.; Wu, X.; Guan, N.; Shao, J.; Li, H.; Chen, Y.; Ping, Y.; Li, D.; Ye, H. Engineering a far-red light–activated split-Cas9 system for remote-controlled genome editing of internal organs and tumors. Sci. Adv. 2020, 6, eabb1777. [Google Scholar] [CrossRef]
- Nihongaki, Y.; Otabe, T.; Sato, M. Emerging Approaches for Spatiotemporal Control of Targeted Genome with Inducible CRISPR-Cas9. Anal. Chem. 2017, 90, 429–439. [Google Scholar] [CrossRef]
- Tao, Y.; Chan, H.F.; Shi, B.; Li, M.; Leong, K.W. Light: A magical tool for controlled drug delivery. Adv. Funct. Mater. 2020, 30, 2005029. [Google Scholar] [CrossRef]
- Bandara, H.D.; Burdette, S.C. Photoisomerization in different classes of azobenzene. Chem. Soc. Rev. 2012, 41, 1809–1825. [Google Scholar] [CrossRef]
- Shao, Q.; Xing, B. Photoactive molecules for applications in molecular imaging and cell biology. Chem. Soc. Rev. 2010, 39, 2835–2846. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Hsu, P.D.; Heidenreich, M.; Cong, L.; Platt, R.J.; Scott, D.A.; Church, G.M.; Zhang, F. Optical control of mammalian endogenous transcription and epigenetic states. Nature 2013, 500, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Putri, R.R.; Chen, L. Spatiotemporal control of zebrafish (Danio rerio) gene expression using a light-activated CRISPR activation system. Gene 2018, 677, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Nihongaki, Y.; Yamamoto, S.; Kawano, F.; Suzuki, H.; Sato, M. CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 2015, 22, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Nihongaki, Y.; Furuhata, Y.; Otabe, T.; Hasegawa, S.; Yoshimoto, K.; Sato, M. CRISPR–Cas9-based photoactivatable transcription systems to induce neuronal differentiation. Nat. Methods 2017, 14, 963–966. [Google Scholar] [CrossRef]
- Polstein, L.R.; Gersbach, C.A. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol. 2015, 11, 198–200. [Google Scholar] [CrossRef]
- Kawano, F.; Suzuki, H.; Furuya, A.; Sato, M. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat. Commun. 2015, 6, 6256. [Google Scholar] [CrossRef]
- Zhou, X.X.; Zou, X.; Chung, H.K.; Gao, Y.; Liu, Y.; Qi, L.S.; Lin, M.Z. A single-chain photoswitchable CRISPR-Cas9 architecture for light-inducible gene editing and transcription. ACS Chem. Biol. 2018, 13, 443–448. [Google Scholar] [CrossRef]
- Zhou, X.X.; Chung, H.K.; Lam, A.J.; Lin, M.Z. Optical control of protein activity by fluorescent protein domains. Science 2012, 338, 810–814. [Google Scholar] [CrossRef]
- Richter, F.; Fonfara, I.; Bouazza, B.; Schumacher, C.H.; Bratovič, M.; Charpentier, E.; Mglich, A. Engineering of temperature-and light-switchable Cas9 variants. Nucleic Acids Res. 2013, 44, gkw930. [Google Scholar] [CrossRef]
- Hemphill, J.; Borchardt, E.K.; Brown, K.; Asokan, A.; Deiters, A. Optical control of CRISPR/Cas9 gene editing. J. Am. Chem. Soc. 2015, 137, 5642–5645. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.K.; Ramanan, V.; Schepers, A.G.; Dalvie, N.S.; Panda, A.; Fleming, H.E.; Bhatia, S.N. Development of Light-Activated CRISPR Using Guide RNAs with Photocleavable Protectors. Angew. Chem. Int. Ed. 2016, 55, 12440–12444. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Le, C.; Wu, J.; Li, X.F.; Zhang, H.; Le, X.C. A genome-editing nanomachine constructed with a clustered regularly interspaced short palindromic repeats system and activated by near-infrared illumination. ACS Nano 2020, 14, 2817–2826. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, L.; Zheng, W.; Cong, L.; Guo, Z.; Xie, Y.; Wang, L.; Tang, R.; Feng, Q.; Hamada, Y.; et al. Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew. Chem. Int. Ed. 2018, 57, 1491–1496. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Yang, Z.; Zhu, S.; He, L.; Fan, W.; Tang, W.; Zou, J.; Shen, Z.; Zhang, M.; Tang, L.; et al. A Rationally Designed Semiconducting Polymer Brush for NIR-II imaging-guided light-triggered remote control of CRISPR/Cas9 genome editing. Adv. Mater. 2019, 31, 1901187. [Google Scholar] [CrossRef]
- Hansen-Bruhn, M.; de Vila, B.E.; Beltrn-Gastlum, M.; Zhao, J.; Ramrez-Herrera, D.E.; Angsantikul, P.; Vesterager Gothelf, K.; Zhang, L.; Wang, J. Active intracellular delivery of a Cas9/sgRNA complex using ultrasound-propelled nanomotors. Angew. Chem. Int. Ed. 2018, 57, 2657–2661. [Google Scholar] [CrossRef]
- Ryu, J.Y.; Won, E.J.; Lee, H.A.; Kim, J.H.; Hui, E.; Kim, H.P.; Yoon, T.J. Ultrasound-activated particles as CRISPR/Cas9 delivery system for androgenic alopecia therapy. Biomaterials 2020, 232, 119736. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, S.; Mair, L.; Ahmed, S.; Huang, T.J.; Mallouk, T.E. Acoustic propulsion of nanorod motors inside living cells. Angew. Chem. Int. Ed. 2014, 53, 3201–3204. [Google Scholar] [CrossRef]
- Wheeler, M.A.; Smith, C.J.; Ottolini, M.; Barker, B.S.; Purohit, A.M.; Grippo, R.M.; Gaykema, R.P.; Spano, A.J.; Beenhakker, M.P.; Kucenas, S.; et al. Genetically targeted magnetic control of the nervous system. Nat. Neurosci. 2016, 19, 756–761. [Google Scholar] [CrossRef]
- Qiu, Y.; Tong, S.; Zhang, L.; Sakurai, Y.; Myers, D.R.; Hong, L.; Lam, W.A.; Bao, G. Magnetic forces enable controlled drug delivery by disrupting endothelial cell-cell junctions. Nat. Commun. 2017, 8, 15594. [Google Scholar] [CrossRef]
- Kaushik, A.; Yndart, A.; Atluri, V.; Tiwari, S.; Tomitaka, A.; Gupta, P.; Jayant, R.D.; Alvarez-Carbonell, D.; Khalili, K.; Nair, M. Magnetically guided non-invasive CRISPRCas9/gRNA delivery across blood-brain barrier to eradicate latent HIV-1 infection. Sci. Rep. 2019, 9, 3928. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, L.; Tong, S.; Lee, C.M.; Deshmukh, H.; Bao, G. Spatial control of in vivo CRISPR–Cas9 genome editing via nanomagnets. Nat. Biomed. Eng. 2019, 3, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Genga, R.M.; Kearns, N.A.; Maehr, R. Controlling transcription in human pluripotent stem cells using CRISPR-effectors. Methods 2016, 101, 36–42. [Google Scholar] [CrossRef]
- Luo, Y.L.; Xu, C.F.; Li, H.J.; Cao, Z.T.; Liu, J.; Wang, J.L.; Du, X.J.; Yang, X.Z.; Gu, Z.; Wang, J. Macrophage-specific in vivo gene editing using cationic lipid-assisted polymeric nanoparticles. ACS Nano 2018, 12, 994–1005. [Google Scholar] [CrossRef] [PubMed]
- Schiwon, M.; Ehrke-Schulz, E.; Oswald, A.; Bergmann, T.; Michler, T.; Protzer, U.; Ehrhardt, A. One-vector system for multiplexed CRISPR/Cas9 against hepatitis B virus cccDNA utilizing high-capacity adenoviral vectors. Mol. Ther.-Nucleic Acids 2018, 12, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Xie, B.; Gao, X.; Chang, L.; Zheng, W.; Chen, H.; Huang, Y.; Tan, L.; Li, M.; Liu, T. Improving the efficiency of precise genome editing with site-specific Cas9-oligonucleotide conjugates. Sci. Adv. 2020, 6, eaaz0051. [Google Scholar] [CrossRef]
- Manna, D.; Maji, B.; Gangopadhyay, S.A.; Cox, K.J.; Zhou, Q.; Law, B.K.; Mazitschek, R.; Choudhary, A. A Singular System with Precise Dosing and Spatiotemporal Control of CRISPR-Cas9. Angew. Chem. 2019, 131, 6351–6355. [Google Scholar] [CrossRef]
- Pawluk, A.; Amrani, N.; Zhang, Y.; Garcia, B.; Hidalgo-Reyes, Y.; Lee, J.; Edraki, A.; Shah, M.; Sontheimer, E.J.; Maxwell, K.L.; et al. Naturally occurring off-switches for CRISPR-Cas9. Cell 2016, 167, 1829–1838. [Google Scholar] [CrossRef]
- Liu, J.; Chang, J.; Jiang, Y.; Meng, X.; Sun, T.; Mao, L.; Xu, Q.; Wang, M. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 2019, 31, 1902575. [Google Scholar] [CrossRef]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef]
- Zhuo, C.; Zhang, J.; Lee, J.H.; Jiao, J.; Cheng, D.; Liu, L.; Kim, H.W.; Tao, Y.; Li, M. Spatiotemporal control of CRISPR/Cas9 gene editing. Signal Transduct. Target. Ther. 2021, 6, 238. [Google Scholar] [CrossRef] [PubMed]
- Javaid, N.; Pham, T.L.; Choi, S. Functional Comparison between VP64-dCas9-VP64 and dCas9-VP192 CRISPR Activators in Human Embryonic Kidney Cells. Int. J. Mol. Sci. 2021, 22, 397. [Google Scholar] [CrossRef] [PubMed]
- Maji, B.; Gangopadhyay, S.A.; Lee, M.; Shi, M.; Wu, P.; Heler, R.; Mok, B.; Lim, D.; Siriwardena, S.U.; Paul, B.; et al. A high-throughput platform to identify small-molecule inhibitors of CRISPR-Cas9. Cell 2019, 177, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Fang, T.; Cao, X.; Ibnat, M.; Chen, G. Stimuli-responsive nanoformulations for CRISPRCas9 genome editing. J. Nanobiotechnol. 2022, 20, 354. [Google Scholar] [CrossRef]
- Naeem, M.; Hoque, M.Z.; Ovais, M.; Basheer, C.; Ahmad, I. Stimulus-Responsive Smart Nanoparticles-Based CRISPR-Cas Delivery for Therapeutic Genome Editing. Int. J. Mol. Sci. 2021, 22, 11300. [Google Scholar] [CrossRef]
- Nandy, S.; Pathak, B.; Zhao, S.; Srivastava, V. Heat-shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant Direct 2019, 3, e00145. [Google Scholar] [CrossRef]
- Kang, J.G.; Park, J.S.; Ko, J.H.; Kim, Y.S. Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system. Sci. Rep. 2019, 9, 11960. [Google Scholar] [CrossRef]
- Fellmann, C.; Gowen, B.G.; Lin, P.C.; Doudna, J.A.; Corn, J.E. Cornerstones of CRISPR–Cas in drug discovery and therapy. Nat. Rev. Drug Discov. 2017, 16, 89–100. [Google Scholar] [CrossRef]
- Dunbar, C.E.; High, K.A.; Joung, J.K.; Kohn, D.B.; Ozawa, K.; Sadelain, M. Gene therapy comes of age. Science 2018, 359, eaan4672. [Google Scholar] [CrossRef]
- Kim, K.; Park, H.; Lim, K.M. Phototoxicity: Its mechanism and animal alternative test methods. Toxicol. Res. 2015, 31, 97–104. [Google Scholar] [CrossRef]
- Yin, H.; Sun, L.; Pu, Y.; Yu, J.; Feng, W.; Dong, C.; Zhou, B.; Du, D.; Zhang, Y.; Chen, Y.; et al. Ultrasound-controlled crispr/cas9 system augments sonodynamic therapy of hepatocellular carcinoma. ACS Cent. Sci. 2021, 7, 2049–2062. [Google Scholar] [CrossRef] [PubMed]
- Rasul, M.F.; Hussen, B.M.; Salihi, A.; Ismael, B.S.; Jalal, P.J.; Zanichelli, A.; Jamali, E.; Baniahmad, A.; Ghafouri-Fard, S.; Basiri, A.; et al. Strategies to overcome the main challenges of the use of CRISPR/Cas9 as a replacement for cancer therapy. Mol. Cancer 2022, 21, 64. [Google Scholar] [CrossRef] [PubMed]
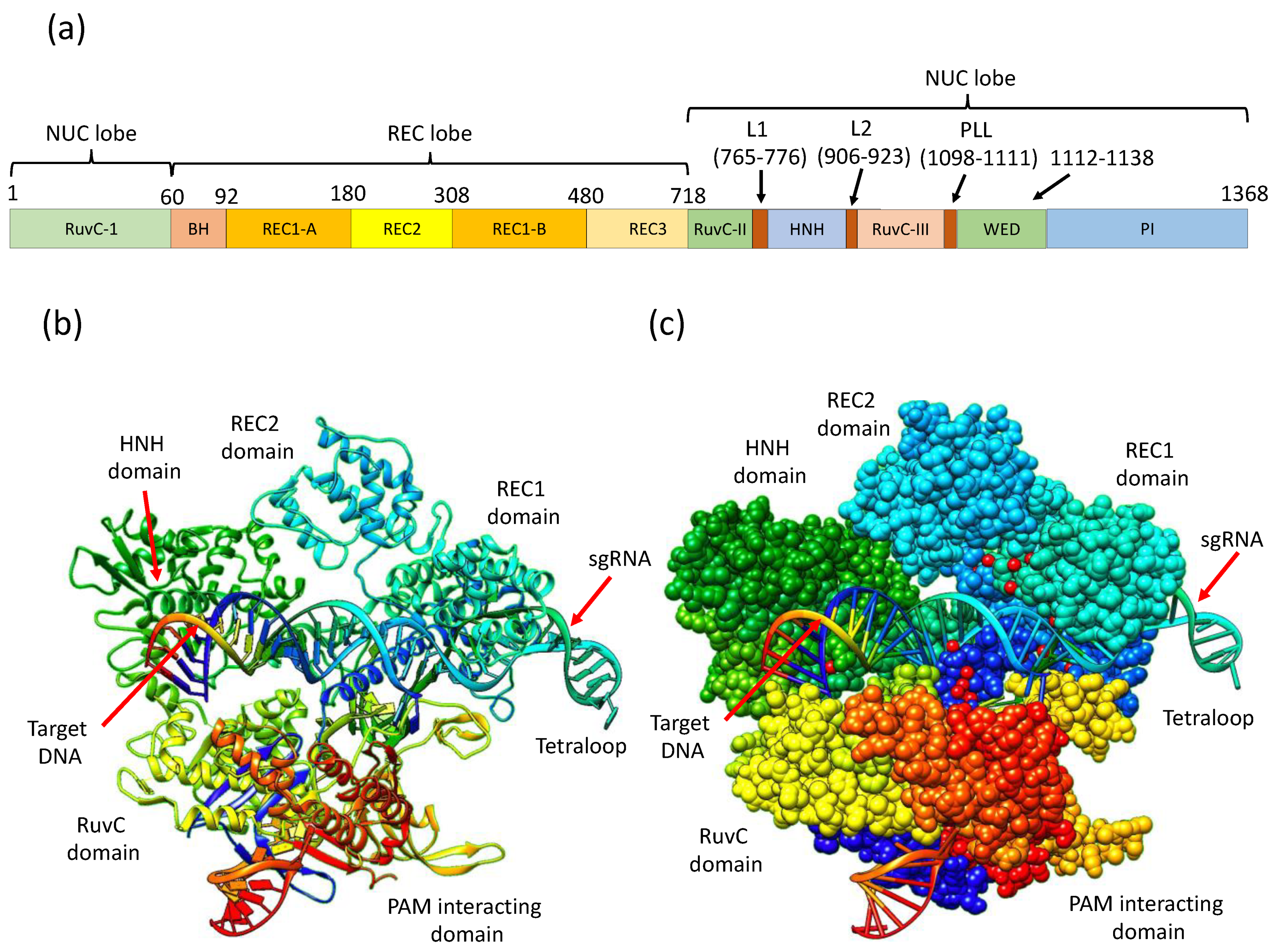

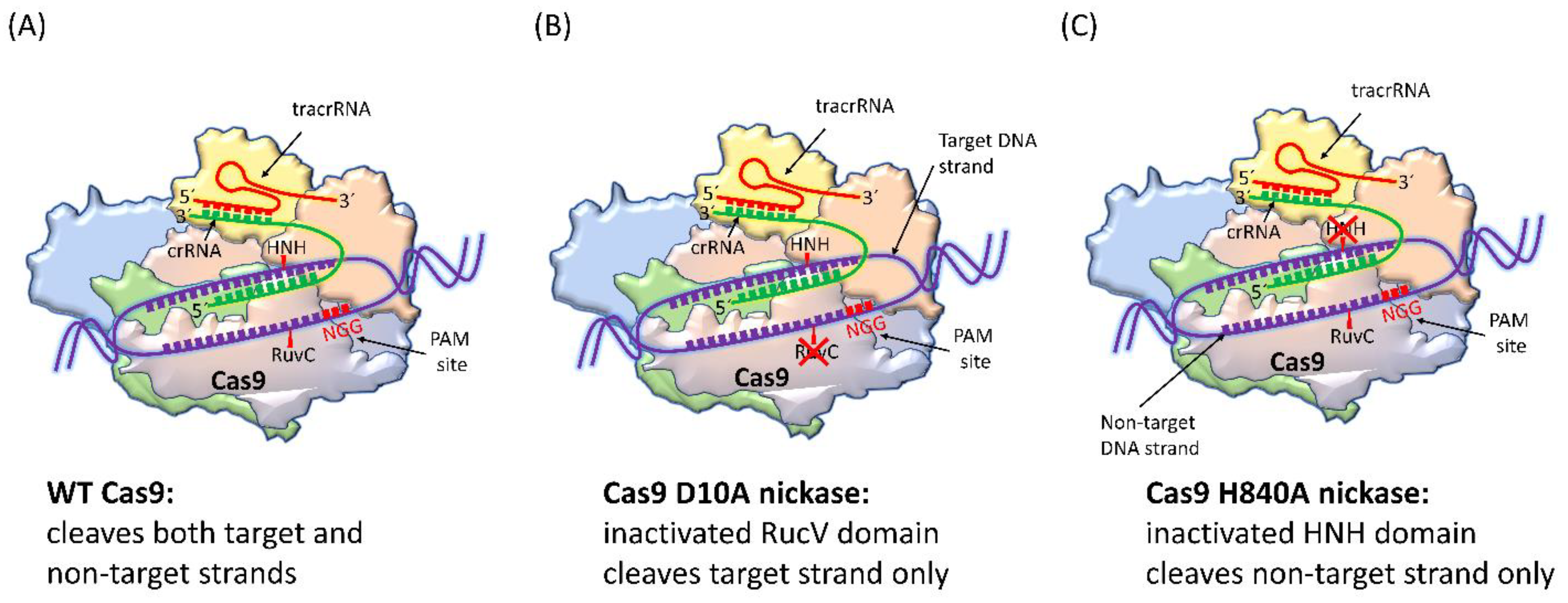
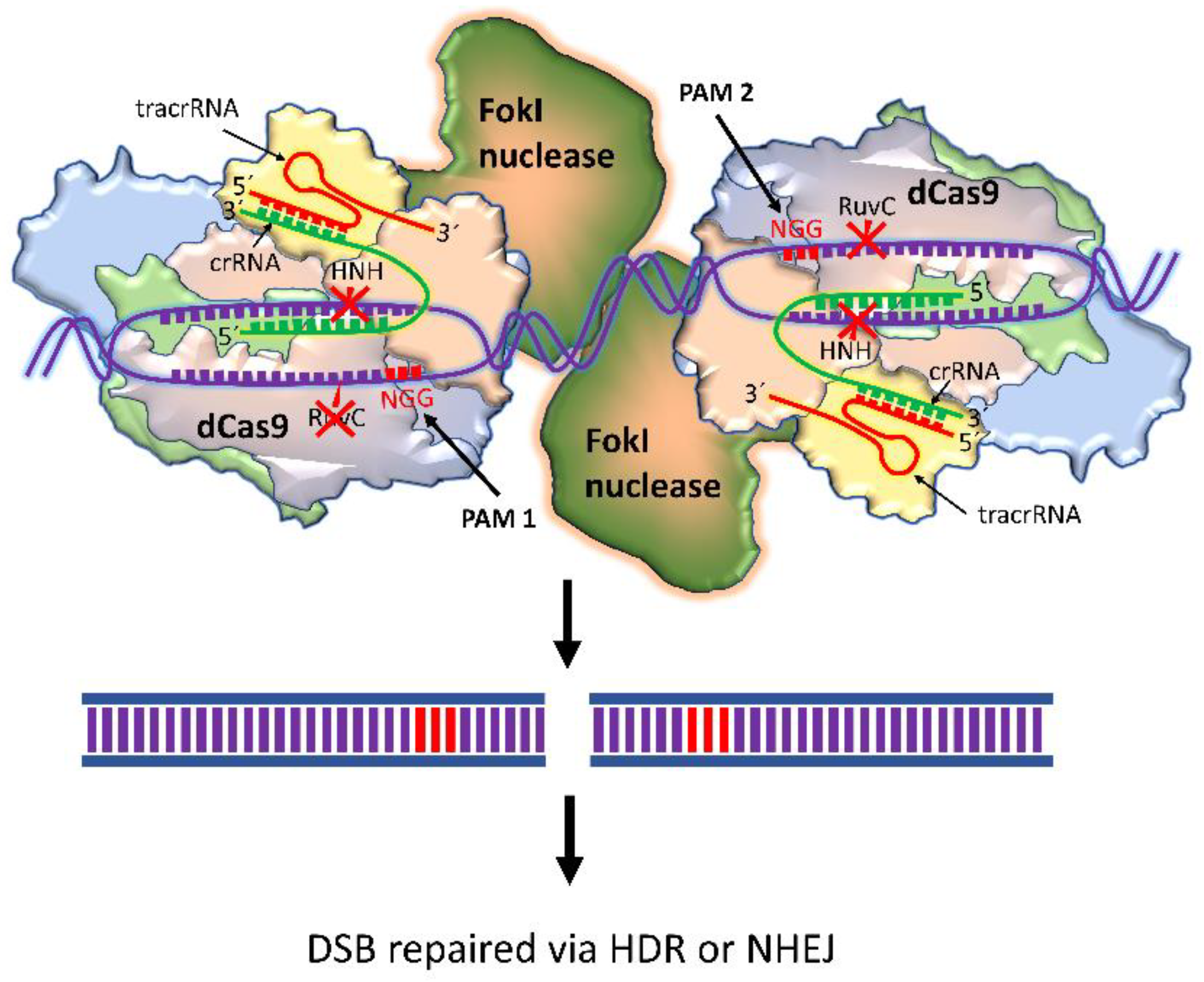
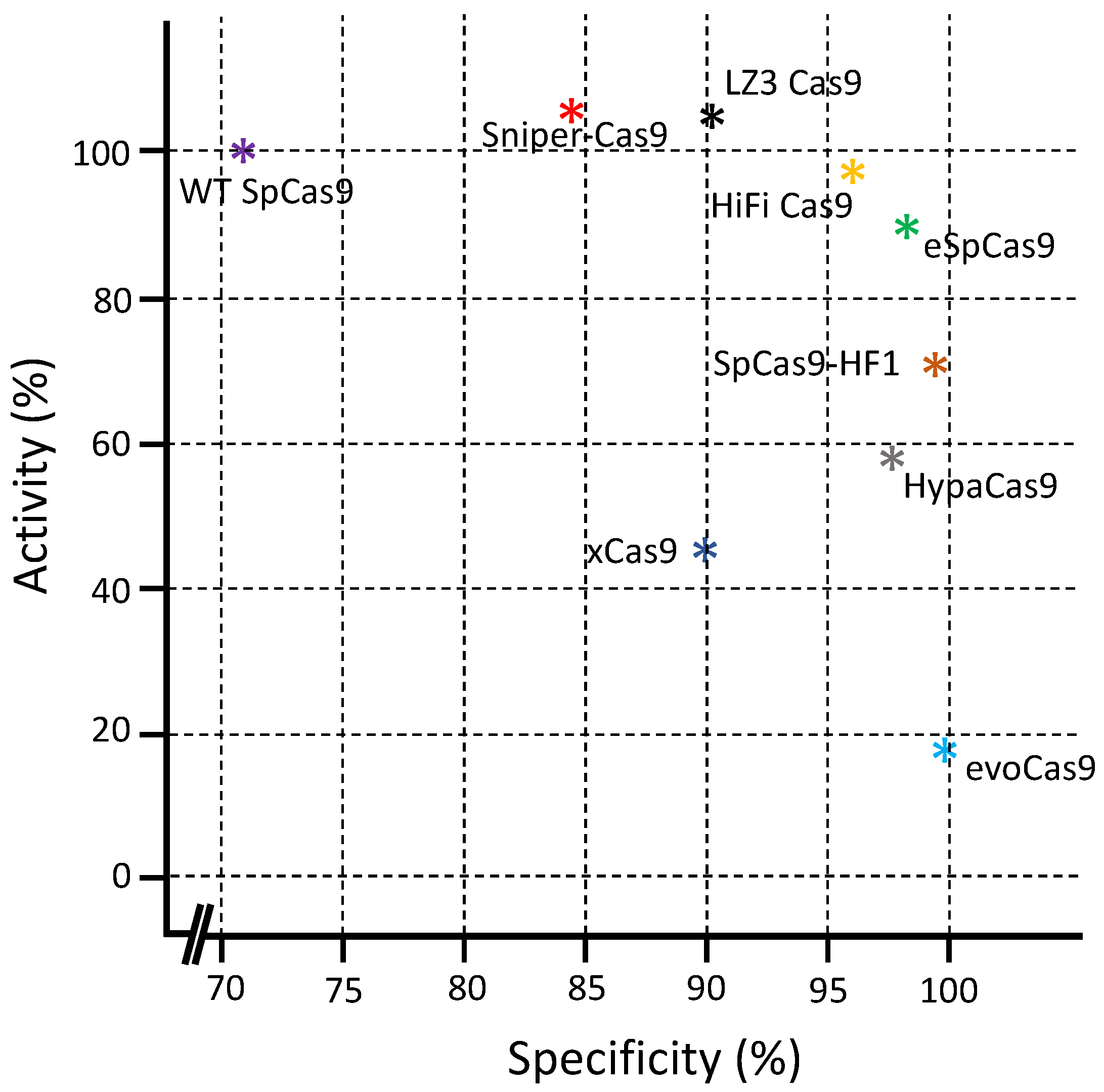

| Cas9 Variants | Mutations Location/s | Year of First Introduction | Cell Type and Reference |
|---|---|---|---|
| SpCas9 nickase | D10A or H840A | 2013 | HEK293FT cells [57], HUES62 hES cells [57], human embryonic stem cells [58] |
| dCas9-FokI | D10A, H840A | 2014 | HEK293 cells [59], U2OS cells [59], HUES9 cells [60] |
| SpCas9-D1135E | D1135E | 2015 | U2OS cells [21] |
| eSpCas9 | K810A, K1003A, R1060A | 2016 | HEK293 and HEK293T cells [16,61,62] |
| SpCas9-HF1 | N497A, R661A, Q695A, Q926A | 2016 | U2OS cells [15], HEK293T cells [61,62] |
| HypaCas9 | N692A, M694A, Q695A, H698A | 2017 | U2OS cells [18], HEK293T cells [61,62], mouse zygotes [63] |
| HiFiCas9 | R691A | 2018 | human hematopoietic stem and progenitor cells [64], primary T cells [64] |
| xCas9 | E108G, S217A, A262T, S409I, E480K, E543D, M694I, E1219V | 2018 | E. coli cells [23], HEK293T cells [62] |
| Sniper-Cas9 | F539S, M763I, K890N | 2018 | E. coli cells [19], HEK293T cells [61], HeLa cells [19] |
| evoCas9 | M495V, Y515N, K526E, R661Q | 2018 | Yeast cells [17], 293multiEGFP, 293blastEGFP, and HEK293T cells [61] |
| SpartaCas | D23A, T67L, Y128V, D1251G | 2020 | T cells [65] |
| LZ3Cas9 | N690C, T769I, G915M, N980K | 2020 | HEK293T cells [61], K562 cells [61], U2OS cells [61] |
| miCas9 | No mutation, SV40 NLS linker fused with brex27 motif | 2020 | Induced pluripotent stem cells [66], airway epithelial cells [66], fibroblast cells [66] |
| SuperFi-Cas9 | Y1010D, Y1013D, Y1016D, V1018D, R1019D, Q1027D, K1031D | 2022 | HEK293 cells [67], neuro-2a mouse neuroblastoma cells [67] |
| Cas Variants | Mechanisms of Action | Average Indel Frequency | Advantages and Limitations | Reference |
|---|---|---|---|---|
| SpCas9 nickase | Use dual-RNAs for site-specific DNA cleavage | 75 and 60% |
| [80] |
| eSpCas9 | Positively charged residues neutralization within the non-target strand, which thereafter weakened non-target strand binding and encouraged rehybridization between the non-target and target DNA strands | 40% |
| [16] |
| SpCas9-HF1 | Reduce the cleavage rate of DNA but have no effect on DNA reversion and release rate | 34% |
| [15] |
| HypaCas9 | In wild-type SpCas9, a quadruple substitution in the REC3 domain | 30% |
| [18] |
| HiFiCas9 | Between RNP and its substrate DNA, the weakening of non-specific interactions | Similar to WT Cas9 |
| [64] |
| xCas9 | Closing PAM or the DNA-sgRNA interface refines the DNA-RNA contact region | 32% |
| [22] |
| Sniper- Cas9 | Weakening of non-specific interactions between RNP and its DNA substrate | 46% |
| [19] |
| evoCas9 | Weakening non-specific interactions between RNP and its substrate DNA | 15% |
| [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allemailem, K.S.; Almatroodi, S.A.; Almatroudi, A.; Alrumaihi, F.; Al Abdulmonem, W.; Al-Megrin, W.A.I.; Aljamaan, A.N.; Rahmani, A.H.; Khan, A.A. Recent Advances in Genome-Editing Technology with CRISPR/Cas9 Variants and Stimuli-Responsive Targeting Approaches within Tumor Cells: A Future Perspective of Cancer Management. Int. J. Mol. Sci. 2023, 24, 7052. https://doi.org/10.3390/ijms24087052
Allemailem KS, Almatroodi SA, Almatroudi A, Alrumaihi F, Al Abdulmonem W, Al-Megrin WAI, Aljamaan AN, Rahmani AH, Khan AA. Recent Advances in Genome-Editing Technology with CRISPR/Cas9 Variants and Stimuli-Responsive Targeting Approaches within Tumor Cells: A Future Perspective of Cancer Management. International Journal of Molecular Sciences. 2023; 24(8):7052. https://doi.org/10.3390/ijms24087052
Chicago/Turabian StyleAllemailem, Khaled S., Saleh A. Almatroodi, Ahmad Almatroudi, Faris Alrumaihi, Waleed Al Abdulmonem, Wafa Abdullah I. Al-Megrin, Adel Nasser Aljamaan, Arshad Husain Rahmani, and Amjad Ali Khan. 2023. "Recent Advances in Genome-Editing Technology with CRISPR/Cas9 Variants and Stimuli-Responsive Targeting Approaches within Tumor Cells: A Future Perspective of Cancer Management" International Journal of Molecular Sciences 24, no. 8: 7052. https://doi.org/10.3390/ijms24087052
APA StyleAllemailem, K. S., Almatroodi, S. A., Almatroudi, A., Alrumaihi, F., Al Abdulmonem, W., Al-Megrin, W. A. I., Aljamaan, A. N., Rahmani, A. H., & Khan, A. A. (2023). Recent Advances in Genome-Editing Technology with CRISPR/Cas9 Variants and Stimuli-Responsive Targeting Approaches within Tumor Cells: A Future Perspective of Cancer Management. International Journal of Molecular Sciences, 24(8), 7052. https://doi.org/10.3390/ijms24087052







