In Situ Raman Analysis of Biofilm Exopolysaccharides Formed in Streptococcus mutans and Streptococcus sanguinis Commensal Cultures
Abstract
1. Introduction
2. Results
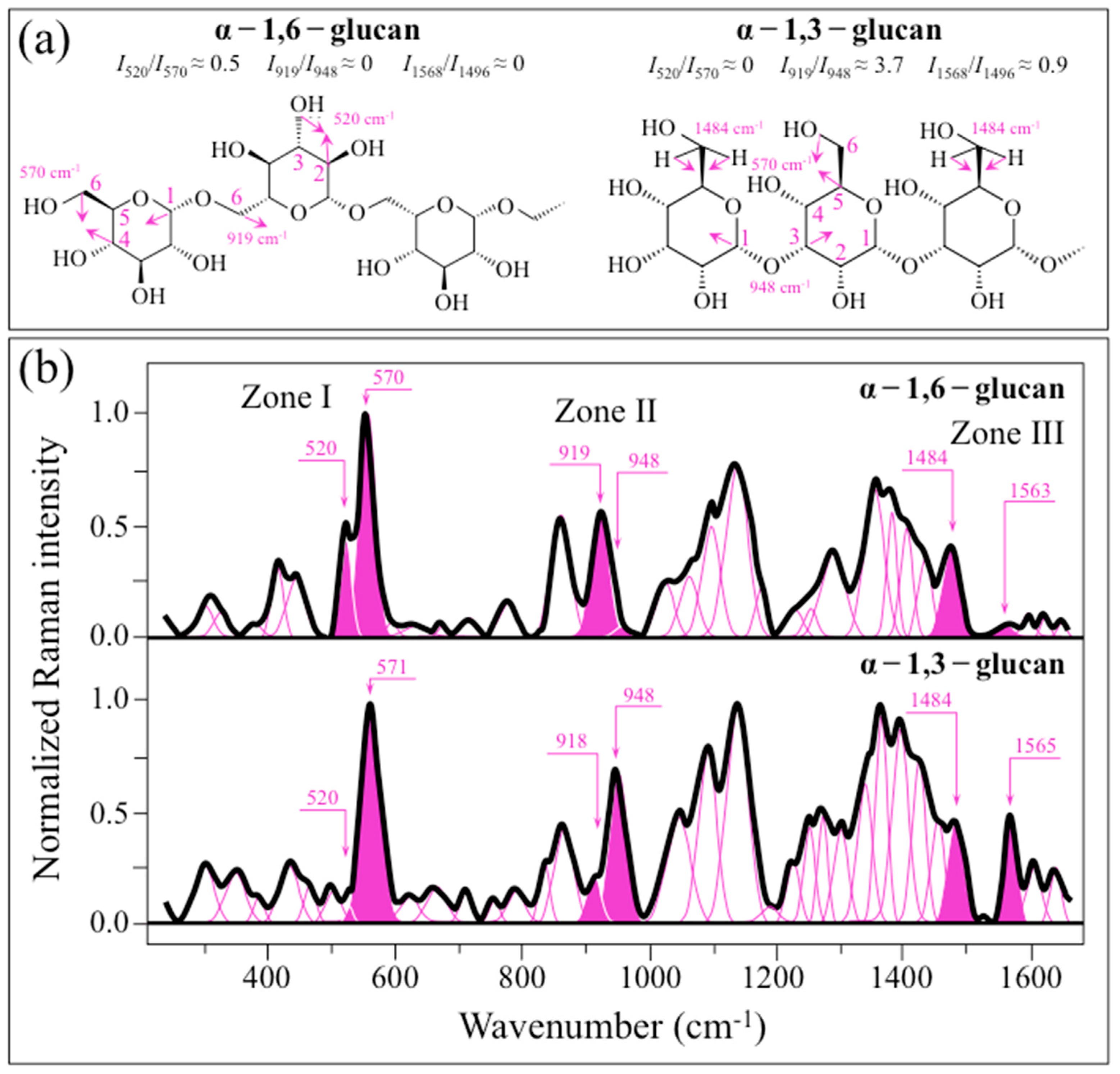
2.1. Raman Calibrations of Water-Insoluble Exopolysaccharides
2.2. Morphological and Structural Characterizations of Biofilms
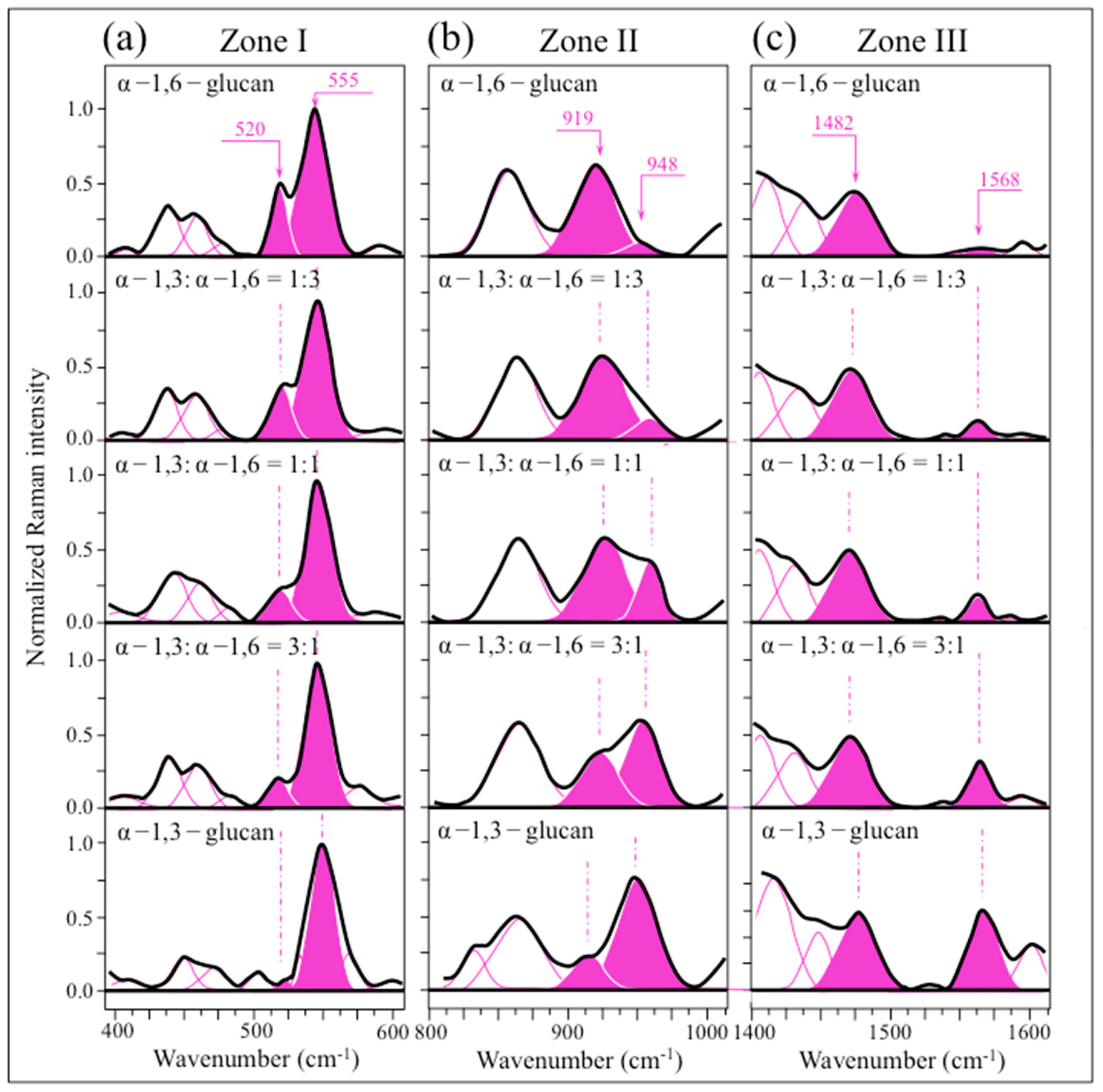
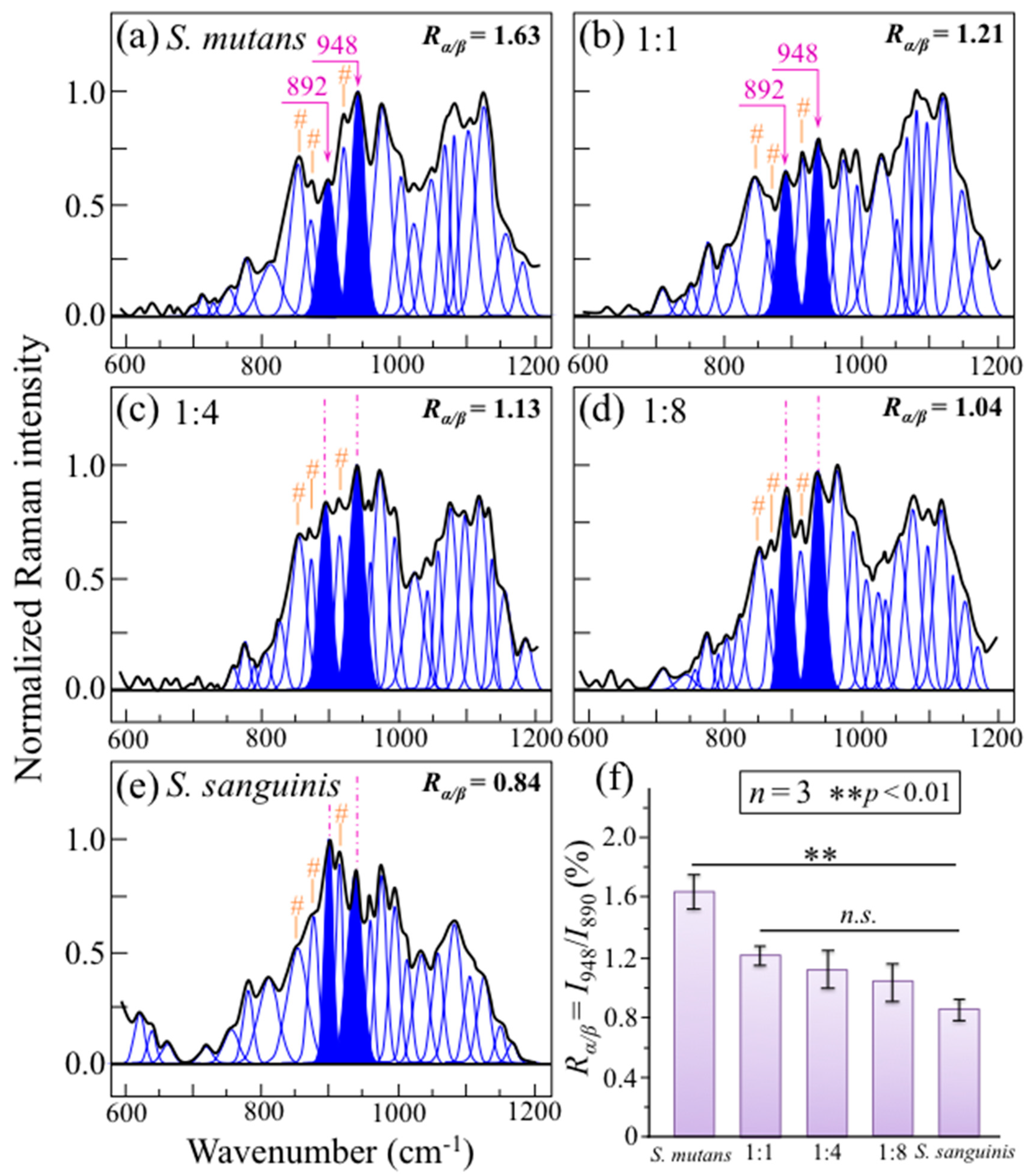
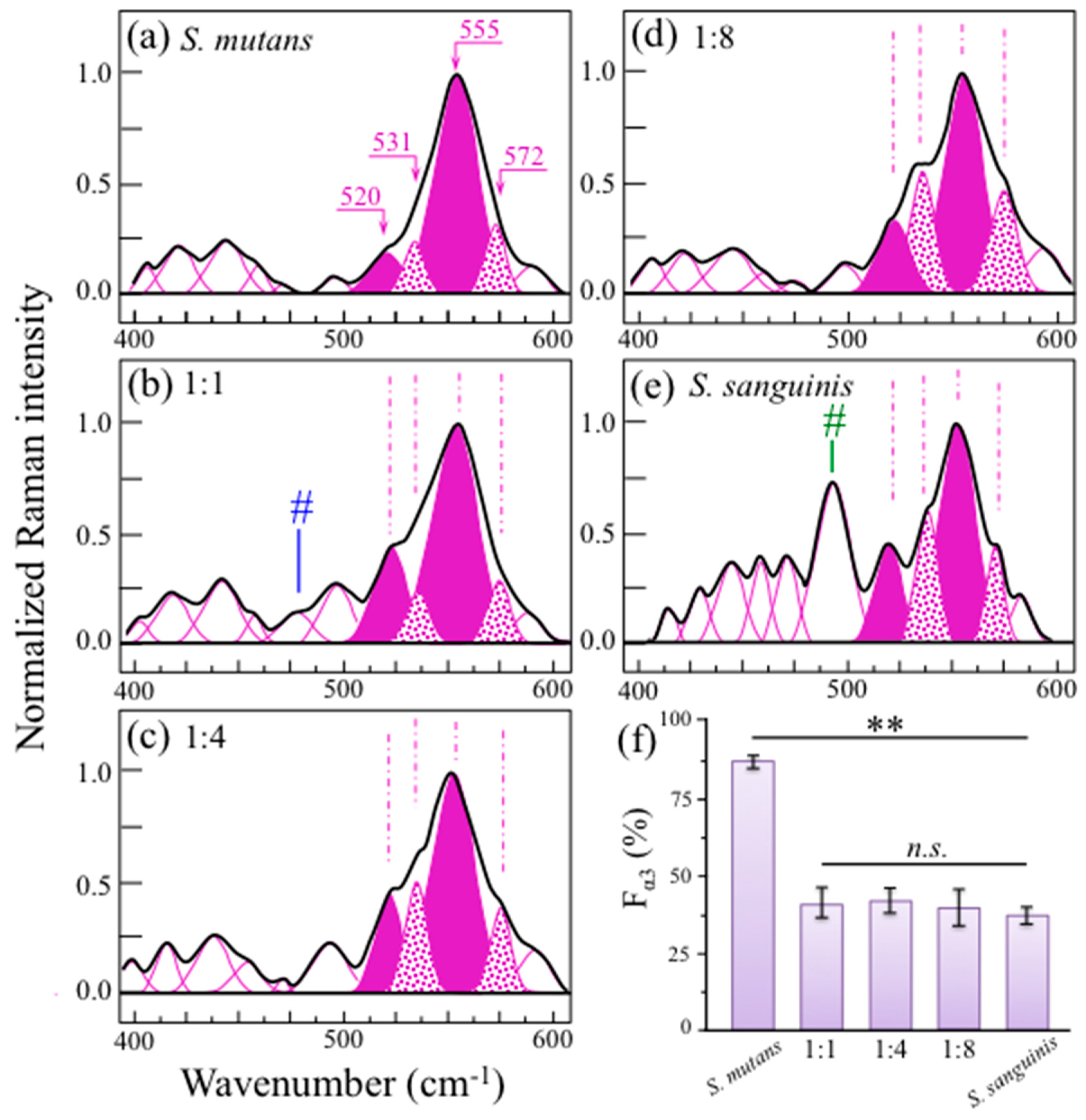
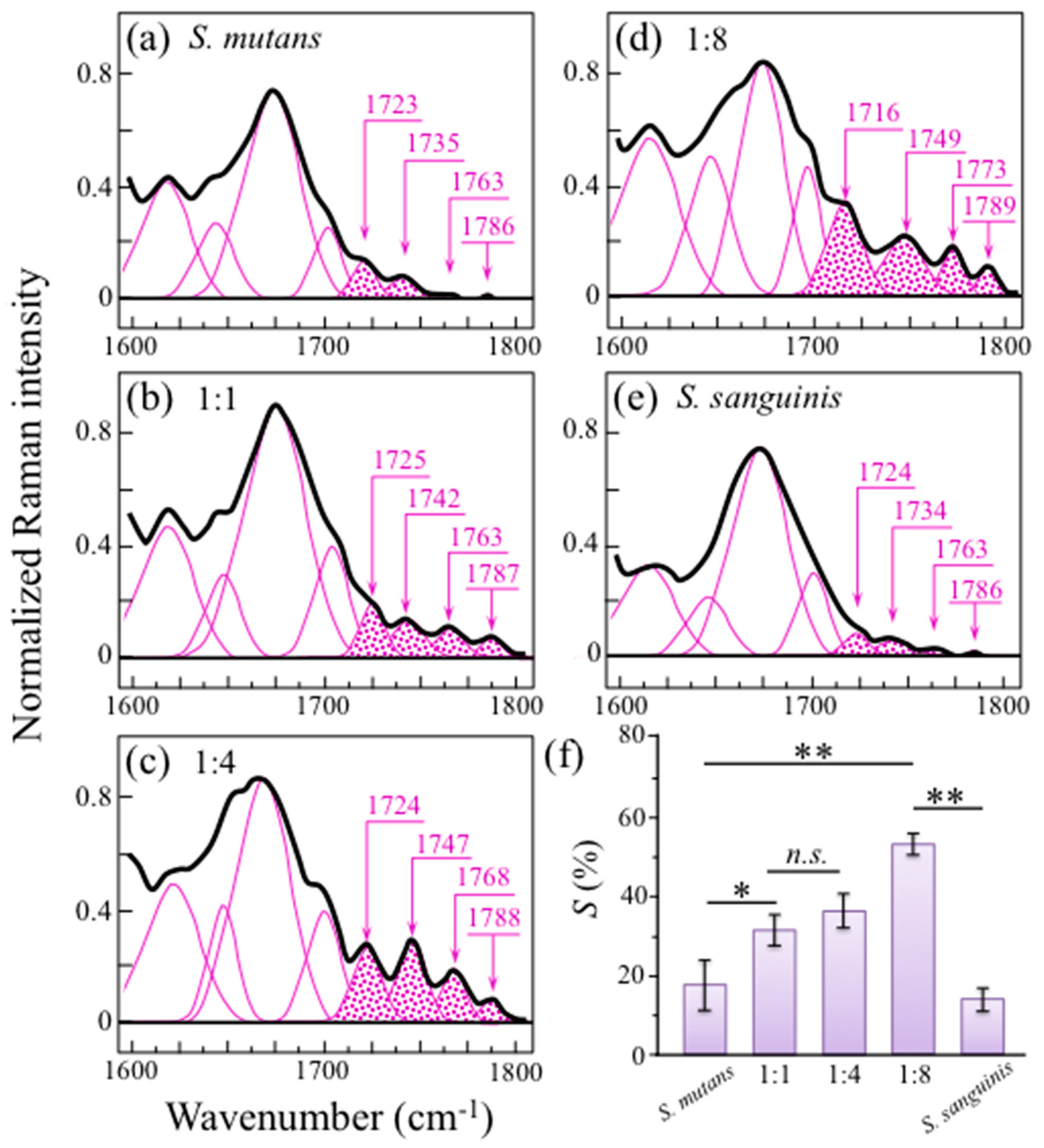
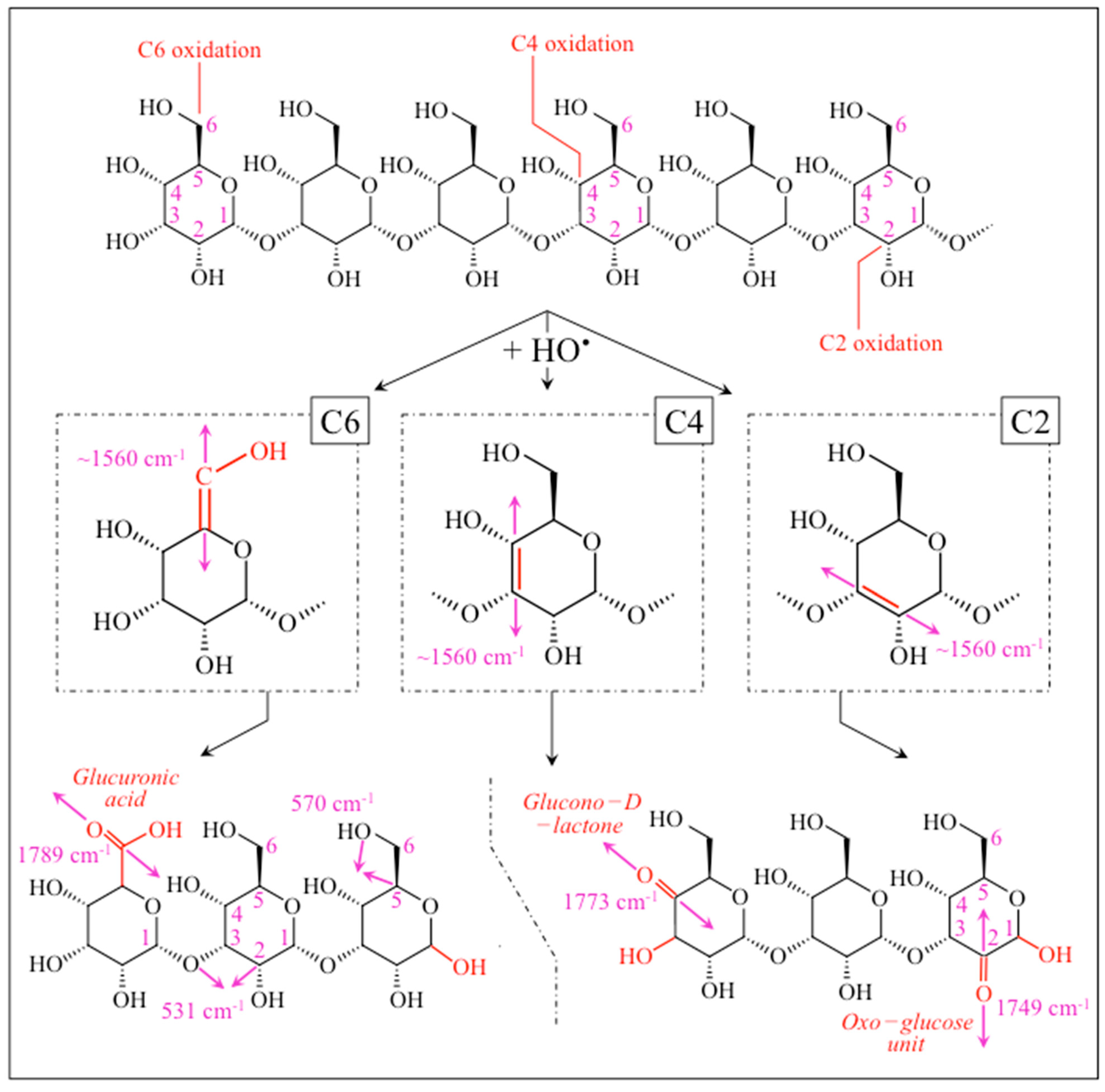
2.3. Raman Markers of Cocultures’ Biofilm Exopolysaccharides
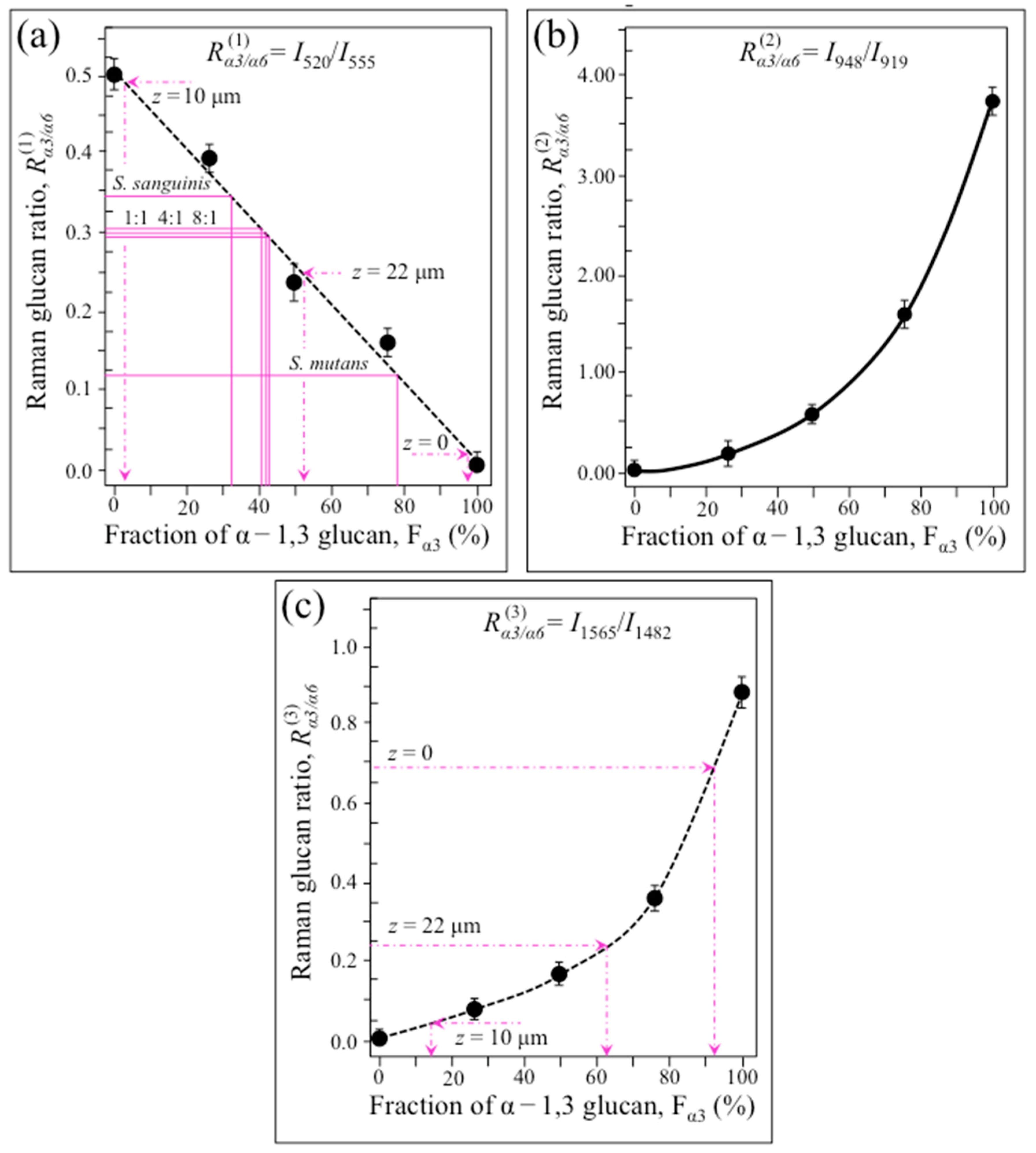
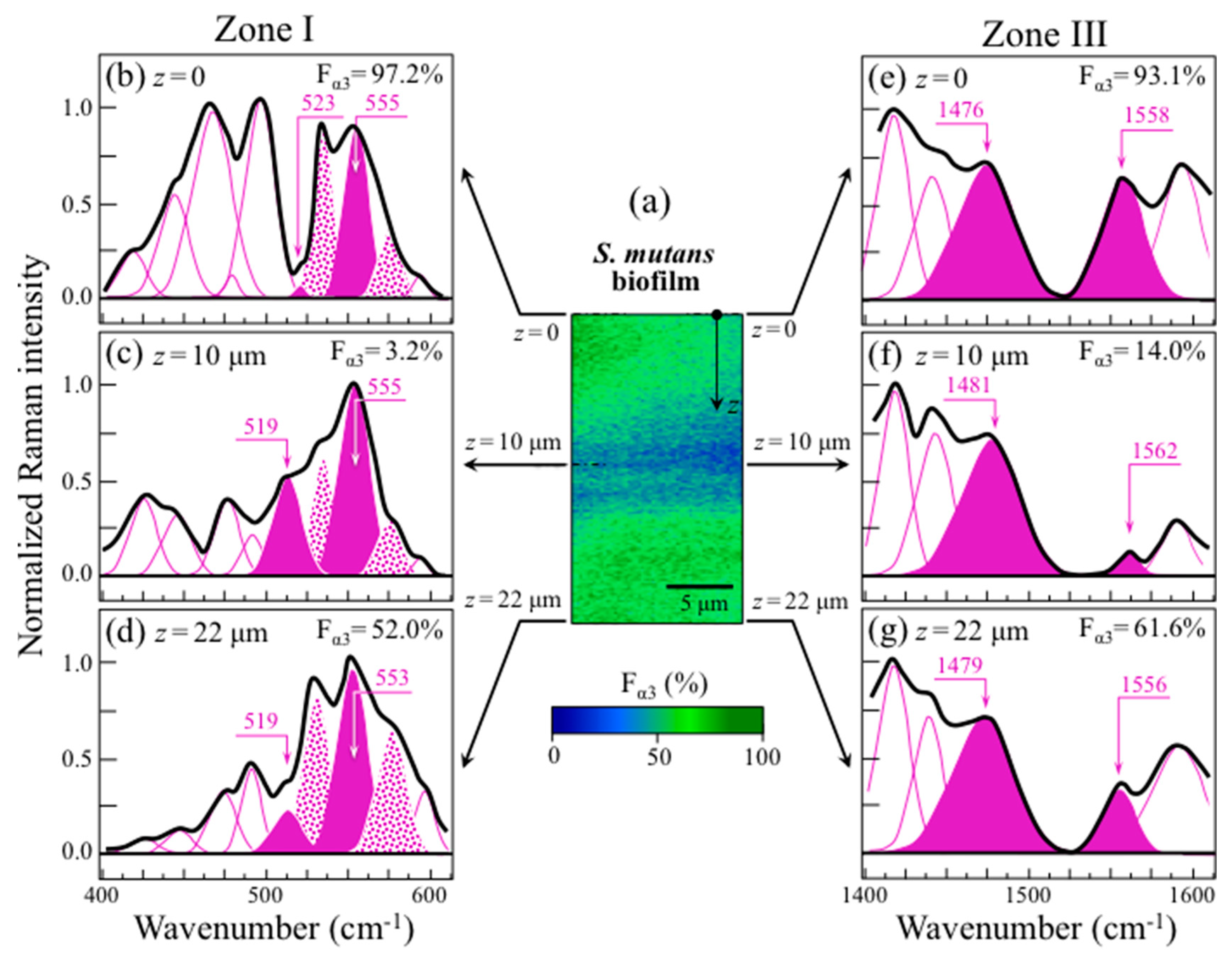
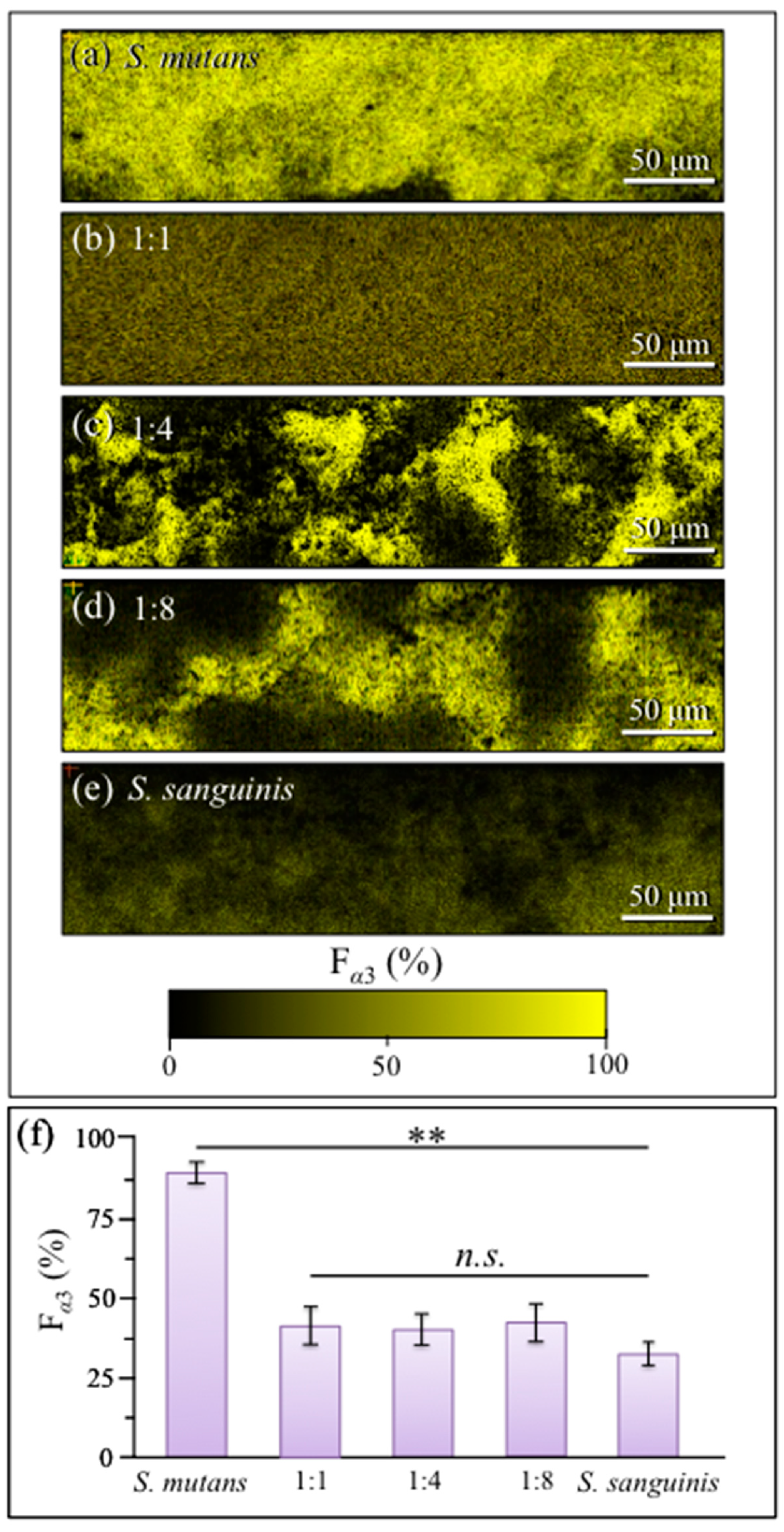
2.4. Raman Imaging of Water-Insoluble Exopolysaccharides Fractions
3. Discussion
3.1. Biofilm Vibrational Markers and Competition/Coexistence Strategies
3.2. “Chemical Weapons” in Active and Passive Competitive Exclusion
3.3. Raman Technology in Oral Health and Limitations of the Present Study
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
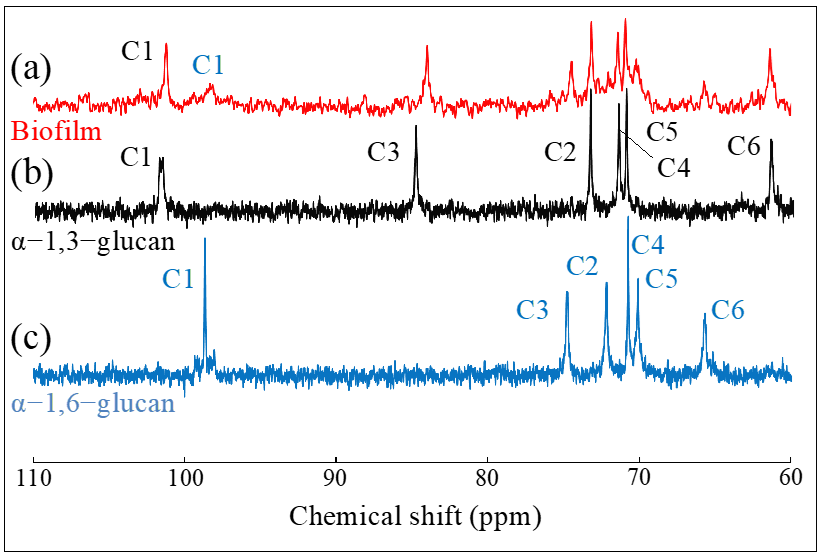
4.2. Nuclear Magnetic Resonance Analyses
4.3. Raman Library, In Situ Raman Spectroscopy, and Raman Imaging
4.4. Machine Learning Algorithm for Spectral Deconvolution
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harper, D.S.; Loesche, W.J. Growth and acid tolerance of human dental plaque bacteria. Arch. Oral Biol. 1984, 29, 843–848. [Google Scholar] [CrossRef]
- Kroes, I.; Lepp, P.W.; Relman, D.A. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 1999, 96, 14547–14552. [Google Scholar] [CrossRef] [PubMed]
- Moore, W.E.; Moore, L.V. The bacteria of periodontal diseases. Periodontol. 2000 1994, 5, 66–77. [Google Scholar] [CrossRef]
- Paster, B.J.; Boches, S.K.; Galvin, J.L.; Ericson, R.E.; Lau, C.N.; Levanos, V.A.; Sahasrabudhe, A.; Dewhirst, F.E. Bacterial diversity in human subgingival plaque. J. Bacteriol. 2001, 183, 3770–3783. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, I.A. Ecological basis for dental caries. In Oral Bacterial Ecology; Kuramitsu, H.K., Ellen, R.P., Eds.; Horizon Scientific Press: Wymondham, Norfolk, UK, 2000; pp. 219–275. [Google Scholar]
- Palmer, R.J., Jr.; Wu, R.; Gordon, S.; Bloomquist, C.G.; Liljemark, W.F.; Kilian, M.; Kolenbrander, P.E. Retrieval of biofilms from the oral cavity. Methods Enzymol. 2001, 337, 393–403. [Google Scholar] [PubMed]
- Socransky, S.S.; Manganiello, S.D. The oral microbiota of man from birth to senility. J. Periodontol. 1971, 42, 484–494. [Google Scholar] [CrossRef]
- Marsh, P.D. Microbiologic aspects of dental plaque and dental caries. Cariology 1999, 43, 599–614. [Google Scholar] [CrossRef]
- Zhu, B.; Macleod, L.C.; Kitten, T.; Xu, P. Streptococcus sanguinis biofilm formation & interaction with oral pathogens. Future Microbiol. 2018, 13, 915–932. [Google Scholar]
- Mikx, F.H.M.; Vanderhoeven, J.S.; Plasschaert, A.J.M.; Konig, K.G. Establishment and symbiosis of Actinomyces viscosus, Streptococcus sanguis and Streptococcus mutans in germfree Osborne-Mendel rats. Caries Res. 1976, 10, 123–132. [Google Scholar] [CrossRef]
- Kreth, J.; Merritt, J.; Shi, W.; Qi, F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J. Bacteriol. 2005, 187, 7193–7203. [Google Scholar] [CrossRef]
- Sun, Y.; Pan, Y.; Sun, Y.; Li, M.; Huang, S.; Qiu, W.; Tu, H.; Zhang, K. Effects of norspermidine on dual-species biofilms composed of Streptococcus mutans and Streptococcus sanguinis. Biomed. Res. Int. 2019, 2019, 1950790. [Google Scholar] [CrossRef] [PubMed]
- Kreth, J.; Zhang, Y.; Herzberg, M.C. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J. Bacteriol. 2008, 190, 4632–4640. [Google Scholar] [CrossRef] [PubMed]
- Giacaman, R.A.; Torres, S.; Gomez, Y.; Munoz-Sandoval, C.; Kreth, J. Correlation of Streptococcus mutans and Streptococcus sanguinis colonization and ex vivo hydrogen peroxide production in carious lesion-free and high caries adults. Arch. Oral Biol. 2015, 60, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Valdebenito, B.; Tullume-Vergara, P.O.; Gonzalez, W.; Kreth, J.; Giacaman, R.A. In silico analysis of the competition between Streptococcus sanguinis and Streptococcus mutans in the dental biofilm. Mol. Oral Microbiol. 2017, 33, 168–180. [Google Scholar] [CrossRef]
- Zhu, Q.; Quivey, R.G., Jr.; Berger, A.J. Raman spectroscopic measurement of relative concentrations in mixtures of oral bacteria. Appl. Spectrosc. 2007, 61, 1233–1237. [Google Scholar] [CrossRef]
- Kelestemur, S.; Avci, E.; Culha, M. Raman and surface-enhanced Raman scattering for biofilm characterization. Chemosensors 2018, 6, 5. [Google Scholar] [CrossRef]
- Efeoglu, E.; Culha, M. In situ-monitoring of biofilm formation by using surface-enhanced Raman scattering. Appl. Spectrosc. 2013, 67, 498–505. [Google Scholar] [CrossRef]
- Sandt, C.; Smith-Palmer, T.; Pink, J.; Brennan, L.; Pink, D. Confocal Raman microspectroscopy as a tool for studying the chemical heterogeneities of biofilms in situ. J. Appl. Microbiol. 2007, 103, 1808–1820. [Google Scholar] [CrossRef]
- Suci, P.; Siedlecki, K.; Palmer, R.; White, D.; Geesey, G. Combined light microscopy and attenuated total reflection Fourier transform infrared spectroscopy for integration of biofilm structure, distribution, and chemistry at solid-liquid interfaces. Appl. Environ. Microbiol. 1997, 63, 4600–4603. [Google Scholar] [CrossRef]
- Lohumi, S.; Kim, M.S.; Qin, J.; Cho, B.-K. Raman imaging from microscopy to macroscopy: Quality and safety control of biological materials. TrAC Trends Anal. Chem. 2017, 93, 183–198. [Google Scholar] [CrossRef]
- Rae, A.; Stosch, R.; Klapetek, P.; Walker, A.R.H.; Roy, D. State of the art Raman techniques for biological applications. Methods 2014, 68, 338–347. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G. Raman spectroscopy in cell biology and microbiology. J. Raman Spectrosc. 2021, 52, 2348–2443. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Asai, T.; Nakaya, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; Marin, E.; et al. Raman imaging of pathogenic Candida auris: Visualization of structural characteristics and machine-learning identification. Front. Microbiol. 2021, 12, 769597. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Nakaya, T.; Asai, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; Marin, E.; et al. Raman study of pathogenic Candida auris: Imaging metabolic machineries in reaction to antifungal drugs. Front. Microbiol. 2022, 13, 896359. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Nakaya, T.; Imamura, H.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; Marin, E.; et al. Raman spectroscopy of oral Candida species: Molecular-scale analyses, chemometrics, and barcode identification. Int. J. Mol. Sci. 2022, 23, 5359. [Google Scholar] [CrossRef]
- Pezzotti, G.; Boschetto, F.; Ohgitani, E.; Fujita, Y.; Shin-Ya, M.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Marin, E.; Zhu, W.; et al. Raman molecular fingerprints of SARS-CoV-2 British variant and the concept of Raman barcode. Adv. Sci. 2021, 2021, 2103287. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G.; Boschetto, F.; Ohgitani, E.; Fujita, Y.; Shin-Ya, M.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Marin, E.; Zhu, W.; et al. Mechanisms of instantaneous inactivation of SARS-CoV-2 by silicon nitride bioceramic. Mater. Today Bio 2021, 12, 100144. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G.; Ohgitani, E.; Fujita, Y.; Imamura, H.; Shin-Ya, M.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Marin, E.; Zhu, W.; et al. Raman fingerprints of the SARS-CoV-2 Delta variant and mechanisms of its instantaneous inactivation by silicon nitride bioceramics. ACS Infect. Dis. 2022, 8, 1563–1581. [Google Scholar] [CrossRef]
- Hori, Y.; Enomoto, Y.; Kimura, S.; Iwata, T. Synthesis of α − 1,3 – and β-1,6-glucan esters with carbon-carbon double bonds and their surface modification. Polym. Int. 2021, 70, 573–581. [Google Scholar] [CrossRef]
- Pezzotti, G.; Kobara, M.; Nakaya, T.; Imamura, H.; Fujii, T.; Miyamoto, N.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; et al. Raman metabolomics of Candida auris clades: Profiling and barcode identification. Int. J. Mol. Sci. 2022, 23, 11736. [Google Scholar] [CrossRef]
- Kopec, L.K.; Vacca, A.M.; Lindang-Evans, S.D.; Bowen, W.H. Properties of Streptococcus sanguinis glucans formed under various conditions. Caries Res. 2001, 35, 67–74. [Google Scholar] [PubMed]
- Ge, X.; Kitten, T.; Chen, Z.; Lee, S.P.; Munro, C.L.; Xu, P. Identification of Streptococcus sanguinis genes required for biofilm formation and examination of their role in endocarditis virulence. Infect. Immun. 2008, 76, 2551–2559. [Google Scholar] [CrossRef] [PubMed]
- Moraes, J.J.; Stipp, R.N.; Harth-Chu, E.N.; Camargo, T.M.; Hofling, J.F.; Mattos-Graner, R.O. Two-component system VicRK regulates functions associated with establishment of Streptococcus sanguinis in biofilms. Infect. Immun. 2014, 82, 4941–4951. [Google Scholar] [CrossRef]
- Kreth, J.; Giacaman, R.A.; Raghavan, R.; Merritt, J. The road less traveled–defining molecular commensalism with Streptococcus sanguinis. Mol. Oral Microbiol. 2017, 32, 181–196. [Google Scholar] [CrossRef]
- Lai, C.W.; Schwab, M.; Hill, S.C.; Santarpia, J.; Pan, Y.-L. Raman scattering and red fluorescence in the photochemical transformation of dry tryptophan particles. Opt. Express 2016, 24, 11654–11667. [Google Scholar] [CrossRef] [PubMed]
- Van Hijum, S.; Kralj, S.; Ozimek, L.; Dijkhuizen, L.; Geel-Schutten, I. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2006, 70, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Bowen, W.H.; Koo, H. Biology of Streptococcus mutans-derived glucosyltransferases: Role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011, 45, 69–86. [Google Scholar] [CrossRef]
- Koo H, H.; Falsetta, M.L.; Klein, M.I. The exopolysaccharide matrix: A virulence determinant of cariogenic biofilm. J. Dent Res. 2013, 92, 1065–1073. [Google Scholar] [CrossRef]
- Hwang, G.; Liu, Y.; Kim, D.; Sun, V.; Aviles-Reyes, A.; Kajfasz, J.K.; Lemos, J.A.; Koo, H. Simultaneous spatiotemporal mapping of in situ pH and bacterial activity within an intact 3D microcolony structure. Sci. Rep. 2016, 6, 32841. [Google Scholar] [CrossRef]
- Guo, L.; McLean, J.S.; Lux, R.; He, X.; Shi, W. The well-coordinated linkage between acidogenicity and aciduricity via insoluble glucans on the surface of Streptococcus mutans. Sci. Rep. 2015, 5, 18015. [Google Scholar] [CrossRef]
- Mikkelsen, M.S.; Jespersen, B.M.; Moller, B.L.; Laerke, H.N.; Larsen, F.H.; Engelsen, S.B. Comparative spectroscopic and rheological studies on crude and purified soluble barley and oat β-glucan preparations. Food Res. Int. 2010, 43, 2417–2424. [Google Scholar] [CrossRef]
- Synytsya, A.; Mickova, K.; Synytsya, A.; Jablonsky, I.; Spevacek, J.; Erban, V.; Kovarikova, E.; Copikova, J. Glucans from fruit bodies of cultivated mushrooms Pleurotus ostreatus and Pleurotus eryngii: Structure and potential prebiotic activity. Carbohydr. Polym. 2009, 76, 548–556. [Google Scholar] [CrossRef]
- Wiercigroch, E.; Szafraniec, E.; Szafraniec, K.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 185, 317–335. [Google Scholar] [CrossRef] [PubMed]
- Trofimov, B.A.; Sinegovskaya, L.M.; Gusarova, N.K. Vibrations of the S–S bond in elemental sulfur and organic polysulfides: A structural guide. J. Sulfur Chem. 2009, 30, 518–554. [Google Scholar] [CrossRef]
- Stone, N.; Kendall, C.; Shepherd, N.; Crow, P.; Barr, H. Near-infrared Raman spectroscopy for the classification of epithelial pre-cancers and cancers. J. Raman Spectrosc. 2002, 33, 564–573. [Google Scholar] [CrossRef]
- Moreno, M.; Raniero, L.; Arisawa, E.Â.L.; Santo, A.M.E.; Santos, E.A.P.; Bitar, R.A.; Martin, A.A. Raman spectroscopy study of breast disease. Theor. Chem. Accounts 2009, 125, 329–334. [Google Scholar] [CrossRef]
- Eisenberg, R.J.; Elchisak, M.; Lai, C. Glycogen accumulation by pleomorphic cells of Streptococcus sanguis. Biochem. Biophys. Res. Commun. 1974, 57, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Van der Hoeven, J.S.; De Jong, M.H.; Camp, P.J.M.; Van den Kieboom, C.W.A. Competition between oral Streptococcus species in the chemostat under alternating conditions of glucose limitation and excess. FEMS Microbiol. Ecol. 1985, 31, 373–379. [Google Scholar]
- De Stoppelaar, J.D.; Van Houte, J.; Dirks, O.B. The effect of carbohydrate restriction on the presence of Streptococcus mutans, Streptococcus sanguis and iodophilic polysaccharide-producing bacteria in human dental plaque. Caries Res. 1970, 4, 114–123. [Google Scholar] [CrossRef]
- Phillips, D.L.; Xing, J.; Chong, C.K.; Liu, H.; Corke, H. Determination of the degree of succinylation in diverse modified starches by Raman spectroscopy. J. Acric. Food Chem. 2000, 48, 5105–5108. [Google Scholar] [CrossRef]
- Boulos, S.; Nystroem, L. Complementary sample preparation strategies for analysis of cereal β-glucan oxidation products by UPLC-MS/MS. Front. Chem. 2017, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Ofoedu, C.E.; You, L.; Osuji, C.M.; Iwouno, J.O.; Kabuo, N.O.; Ojukwu, M.; Agunwah, I.M.; Chacha, J.S.; Muobike, O.P.; Agunbiade, A.O.; et al. Hydrogen peroxide effects on natural-sourced polysaccharides: Free radical formation/production, degradation process, and reaction mechanism–a critical synopsis. Foods 2021, 10, 699. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Davies, G. Structures and mechanisms of glycosyl hydrolases. Structure 1995, 3, 853–859. [Google Scholar]
- Vickerman, M.M.; Sulavik, M.C.; Nowak, J.D.; Gardner, N.M.; Jones, G.W.; Clewell, D.B. Nucleotide sequence analysis of the Streptococcus gordonii glucosyltransferase gene, gtfG. DNA Seq. 1997, 7, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.M.V.; Ng-Evans, L.; Wunder, D.; Bowen, W.H. Studies concerning the glucosyltransferase of Streptococcus sanguis. Caries Res. 2000, 34, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Hoshino, T.; Ooshima, T.; Sobue, S.; Hamada, S. Purification, characterization, and molecular analysis of the gene encoding glucosyltransferase from Streptococcus oralis. Infect. Immun. 2000, 68, 2475–2483. [Google Scholar] [CrossRef]
- Xu, P.; Alves, J.M.; Kitten, T.; Brown, A.; Chen, Z.; Ozaki, L.S.; Manque, P.; Ge, X.; Serrano, M.G.; Puiu, D.; et al. Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 2007, 189, 3166–3175. [Google Scholar] [CrossRef]
- Zhu, L.; Kreth, J. The Role of Hydrogen Peroxide in Environmental Adaptation of Oral Microbial Communities. Oxid. Med. Cell. Longev. 2012, 2012, 717843. [Google Scholar] [CrossRef]
- Barnard, J.P.; Stinson, M.W. The alpha-hemolysin of Streptococcus gordonii is hydrogen peroxide. Infect. Immun. 1996, 64, 3853–3857. [Google Scholar] [CrossRef]
- Michels, A.J.; Frei, B. Myths, artifacts, and fatal flaws: Identifying limitations and opportunities in vitamin C research. Nutrients 2013, 5, 5161–5192. [Google Scholar] [CrossRef]
- Chen, L.; Ge, X.; Dou, Y.; Wang, X.; Patel, J.R.; Xu, P. Identification of hydrogen peroxide production-related genes in Streptococcus sanguinis and their functional relationship with pyruvate oxidase. Microbiology 2011, 157, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Ge, X.; Wang, X.; Patel, J.R.; Xu, P. SpxA1 involved in hydrogen peroxide production, stress tolerance and endocarditis virulence in Streptococcus sanguinis. PLoS ONE 2012, 7, e40034. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Niki, M.; Yamamoto, Y.; Yasunaga, A.; Ansai, T. Proteome analysis identifies the Dpr protein of Streptococcus mutans as an important factor in the presence of early streptococcal colonizers of tooth surfaces. PLoS ONE 2015, 10, e0121176. [Google Scholar] [CrossRef] [PubMed]
- Fujishima, K.; Kawada-Matsuo, M.; Oogai, Y.; Tokuda, M.; Torii, M.; Komatsuzawa, H. dpr and sod in Streptococcus mutans are involved in coexistence with Streptococcus sanguinis, and PerR is associated with resistance to H2O2. Appl. Environ. Microbiol. 2013, 79, 1436–1443. [Google Scholar] [CrossRef]
- Ghoul, M.; Mitri, S. The ecology and evolution of microbial competition. Trends Microbiol. 2016, 24, 833–845. [Google Scholar] [CrossRef]
- Lim, Y.; Shin, S.H.; Lee, S.I.; Kim, I.S.; Rhee, J.H. Iron repressibility of siderophore and transferrin-binding protein in Staphylococcus aureus. FEMS Microbiol. Lett. 1998, 163, 19–24. [Google Scholar] [CrossRef]
- Murgas, C.J.; Green, S.P.; Forney, A.K.; Korba, R.M.; An, S.-S.; Kitten, T.; Lucas, H.R. Intracellular metal speciation in Streptococcus sanguinis establishes SsaACB as critical for redox maintenance. ACS Infect. Dis. 1998, 6, 1906–1921. [Google Scholar] [CrossRef]
- Diggle, S.P.; Griffin, A.S.; Campbell, G.S.; West, S.A. Cooperation and conflict in quorum-sensing bacterial populations. Nature 2007, 450, 411–414. [Google Scholar] [CrossRef]
- Rendueles, O.; Ghigo, J. Multi-species biofilms: How to avoid unfriendly neighbors. FEMS Microbiol. Rev. 2012, 36, 972–989. [Google Scholar] [CrossRef]
- Griffin, A.S.; West, S.A.; Buckling, A. Cooperation and competition in pathogenic bacteria. Nature 2004, 430, 1024–1027. [Google Scholar] [CrossRef]
- Scholz, R.L.; Greenberg, E.P. Sociality in Escherichia coli: Enterochelin is a private good at low cell density and can be shared at high cell density. J. Bacteriol. 2015, 197, 2122–2128. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M. A functional perspective on phenotypic heterogeneity in microorganisms. Nature Rev. Microbiol. 2015, 13, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Gore, J.; Youk, H.; Van Oudenaarden, A. Snowdrift game dynamics and facultative cheating in yeast. Nature 2009, 459, 253–256. [Google Scholar] [CrossRef]
- An, D.; Danhorn, T.; Fuqua, C.; Parsek, M.R. Quorum sensing and motility mediate interactions between Pseudomonas aeruginosa and Agrobacterium tumefaciens in biofilm cocultures. Proc. Natl. Acad. Sci. USA 2006, 103, 3828–3833. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Racimo, F.; Schluter, J.; Levy, S.B.; Foster, K.R. Importance of positioning for microbial evolution. Proc. Natl. Acad. Sci. USA 2014, 111, E1639–E1647. [Google Scholar] [CrossRef]
- Nadell, C.D.; Bassler, B.L. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc. Natl. Acad. Sci. USA 2011, 108, 14181–14185. [Google Scholar] [CrossRef]
- Schluter, J.; Nadell, C.D.; Bassler, B.L.; Foster, K.R. Adhesion as a weapon in microbial competition. ISME J. 2014, 9, 139–149. [Google Scholar] [CrossRef]
- Mignot, T.; Noellmann, M. New insights into the function of a versatile class of membrane molecular motors from studies of Myxococcus xanthus surface (gliding) motility. Microb. Cell 2017, 4, 98–100. [Google Scholar] [CrossRef]
- Merritt, J.; Qi, F. The mutacins of Streptococcus mutans: Regulation and ecology. Mol. Oral Microbiol. 2012, 27, 57–69. [Google Scholar] [CrossRef]
- Islam, M.R.; Nagao, J.-I.; Zendo, T.; Sonomoto, K. Antimicrobial mechanism of lantibiotics. Biochem. Soc. Trans. 2012, 40, 1528–1533. [Google Scholar] [CrossRef]
- Willey, J.M.; Van der Donk, W.A. Lantibiotics: Peptides of diverse structure and function. Annu. Rev. Microbiol. 2007, 61, 477–501. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Biswas, I. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl. Environ. Microbiol. 2011, 77, 2428–2434. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.-Y.; Kuramitsu, H.K. Interactions between oral bacteria: Inhibition of Streptococcus mutans bacteriocin production by Streptococcus gordonii. Appl. Environ. Microbiol. 2005, 71, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Nissen-Meyer, J.; Rogne, P.; Oppegard, C.; Haugen, H.S.; Kristiansen, P.E. Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by gram-positive bacteria. Curr. Pharm. Biotechnol. 2009, 10, 19–37. [Google Scholar] [CrossRef]
- Van Belkum, M.J.; Stiles, M.E. Nonlantibiotic antibacterial peptides from lactic acid bacteria. Nat. Prod. Rep. 2000, 17, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Harvey, P.D.; Butler, I.S. Raman spectra of orthorhombic sulfur at 40 K. J. Raman Spectrosc. 1986, 17, 329–334. [Google Scholar] [CrossRef]
- Barletta, R.E.; Gros, B.N.; Herring, M.P. Analysis of marine biogenic sulfur compounds using Raman spectroscopy: Dimethyl sulfide and methane sulfonic acid. J. Raman Spectrosc. 2009, 40, 972–981. [Google Scholar] [CrossRef]
- Andrikopoulos, K.S.; Kalampounias, A.G.; Falgara, O.; Yannopoulos, S.N. The glassy and supercooled state of elemental sulfur: Vibrational modes, structure metastability, and polymer content. J. Chem. Phys. 2013, 139, 124501. [Google Scholar] [CrossRef]
- Nims, C.; Cron, B.; Wetherington, M.; Macalady, J.; Cosmidis, J. Low frequency Raman spectroscopy for micron scale and in vivo characterization of elemental sulfur in microbial samples. Sci. Rep. 2019, 9, 7971. [Google Scholar] [CrossRef]
- Lemos, J.A.; Palmer, S.R.; Zeng, L.; Wen, Z.T.; Kajfasz, J.K.; Freires, I.A.; Abranches, J.; Brady, L.J. The biology of Streptococcus mutans. Microbiol. Spectrum 2019, 7, GPP3-0051-2018. [Google Scholar] [CrossRef]
- Peres, M.A.; Macpherson, L.M.D.; Weyant, R.J.; Daly, B.; Venturelli, R.; Mathur, M.R.; Listl, S.; Celeste, R.K.; Guarnizo-Herreno, C.C.; Kearns, C.; et al. Oral diseases: A global public health challenge. Lancet 2019, 394, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G.; Zhu, W.; Boffelli, M.; Adachi, T.; Ichioka, H.; Yamamoto, T.; Kanamura, N. Vibrational algorithms for quantitative crystallographic analyses of hydroxyapatite-based biomaterials: I, theoretical foundations. Anal. Bioanal. Chem. 2015, 407, 3325–3342. [Google Scholar] [CrossRef]
- Adachi, T.; Pezzotti, G.; Yamamoto, T.; Ichioka, H.; Boffelli, M.; Zhu, W.; Kanamura, N. Vibrational algorithms for quantitative crystallographic analyses of hydroxyapatite-based biomaterials: II, application to decayed human teeth. Anal. Bioanal. Chem. 2015, 407, 3343–3356. [Google Scholar] [CrossRef] [PubMed]
- Pezzotti, G.; Adachi, T.; Gasparutti, I.; Vincini, G.; Zhu, W.; Boffelli, M.; Rondinella, A.; Marin, E.; Ichioka, H.; Yamamoto, T.; et al. Vibrational monitor of early demineralization in tooth enamel after in vitro exposure to phosphoridic liquid, Spectrochim. Acta A Mol. Bomol. Spectrosc. 2017, 173, 19–33. [Google Scholar] [CrossRef]
- Miyamoto, N.; Adachi, T.; Boschetto, F.; Zanocco, M.; Yamamoto, T.; Marin, E.; Somekawa, S.; Ashida, R.; Zhu, W.; Kanamura, N.; et al. Molecular fingerprint imaging to identify dental caries using Raman spectroscopy. Materials 2020, 13, 4900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, L.; Wang, Q.; Wen, Z.; Liu, C.; Ding, Y. Raman spectroscopy: A potential diagnostic tool for oral diseases. Front. Cell. Infect. Microbiol. 2022, 12, 775236. [Google Scholar] [CrossRef]
- Kriem, L.S.; Wright, K.; Ccahuana-Vasquez, R.A.; Rupp, S. Mapping of a subgingival dual-species biofilm model using confocal Raman microscopy. Front. Microbiol. 2021, 12, 729720. [Google Scholar] [CrossRef]
- Ramirez-Mora, T.; Davila-Perez, C.; Torres-Mendez, F.; Valle-Bourrouet, G. Raman spectroscopic characterization of endodontic biofilm matrices. J. Spectrosc. 2019, 2019, 1307397. [Google Scholar] [CrossRef]
- Hlaing, M.M.; Dunn, M.; Stoddart, P.R.; McArthur, S.L. Raman spectroscopic identification of single bacterial cells at different stages of their lifecycle. Vibr. Spectrosc. 2016, 86, 81–89. [Google Scholar] [CrossRef]
- Gyo, M.; Nikaido, T.; Okada, K.; Yamauchi, J.; Tagami, J.; Matin, K. Surface response of fluorine polymer-incorporated resin composites to cariogenic biofilm adherence. Appl. Environ. Microbiol. 2008, 74, 1428–1435. [Google Scholar] [CrossRef]
- Ikeda, M.; Matin, K.; Nikaido, T.; Foxton, R.M.; Tagami, J. Effect of surface characteristics on adherence of S. mutans biofilms to indirect resin composites. Dent. Mater. J. 2007, 26, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Puanglek, S.; Kimura, S.; Enomoto-Rogers, Y.; Kabe, T.; Yoshida, M.; Wada, M.; Iwata, T. In vitro synthesis of linear α-1,3-glucan and chemical modification to ester derivatives exhibiting outstanding thermal properties. Sci. Rep. 2016, 6, 30479. [Google Scholar] [CrossRef] [PubMed]
- Togo, A.; Suzuki, S.; Kimura, S.; Iwata, T. High tensile strength regenerated α–1,3–glucan fiber and crystal transition. ACS Omega 2021, 6, 20361–20368. [Google Scholar] [CrossRef]
- Guo, B.; Yuan, Y. A comparative review of methods for comparing means using partially paired data. Stat. Methods Med. Res. 2017, 26, 1323–1340. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pezzotti, G.; Ofuji, S.; Imamura, H.; Adachi, T.; Yamamoto, T.; Kanamura, N.; Ohgitani, E.; Marin, E.; Zhu, W.; Mazda, O.; et al. In Situ Raman Analysis of Biofilm Exopolysaccharides Formed in Streptococcus mutans and Streptococcus sanguinis Commensal Cultures. Int. J. Mol. Sci. 2023, 24, 6694. https://doi.org/10.3390/ijms24076694
Pezzotti G, Ofuji S, Imamura H, Adachi T, Yamamoto T, Kanamura N, Ohgitani E, Marin E, Zhu W, Mazda O, et al. In Situ Raman Analysis of Biofilm Exopolysaccharides Formed in Streptococcus mutans and Streptococcus sanguinis Commensal Cultures. International Journal of Molecular Sciences. 2023; 24(7):6694. https://doi.org/10.3390/ijms24076694
Chicago/Turabian StylePezzotti, Giuseppe, Satomi Ofuji, Hayata Imamura, Tetsuya Adachi, Toshiro Yamamoto, Narisato Kanamura, Eriko Ohgitani, Elia Marin, Wenliang Zhu, Osam Mazda, and et al. 2023. "In Situ Raman Analysis of Biofilm Exopolysaccharides Formed in Streptococcus mutans and Streptococcus sanguinis Commensal Cultures" International Journal of Molecular Sciences 24, no. 7: 6694. https://doi.org/10.3390/ijms24076694
APA StylePezzotti, G., Ofuji, S., Imamura, H., Adachi, T., Yamamoto, T., Kanamura, N., Ohgitani, E., Marin, E., Zhu, W., Mazda, O., Togo, A., Kimura, S., Iwata, T., Shiba, H., Ouhara, K., Aoki, T., & Kawai, T. (2023). In Situ Raman Analysis of Biofilm Exopolysaccharides Formed in Streptococcus mutans and Streptococcus sanguinis Commensal Cultures. International Journal of Molecular Sciences, 24(7), 6694. https://doi.org/10.3390/ijms24076694






