Pharmacological Targeting of Bcl-2 Induces Caspase 3-Mediated Cleavage of HDAC6 and Regulates the Autophagy Process in Colorectal Cancer
Abstract
1. Introduction
2. Results
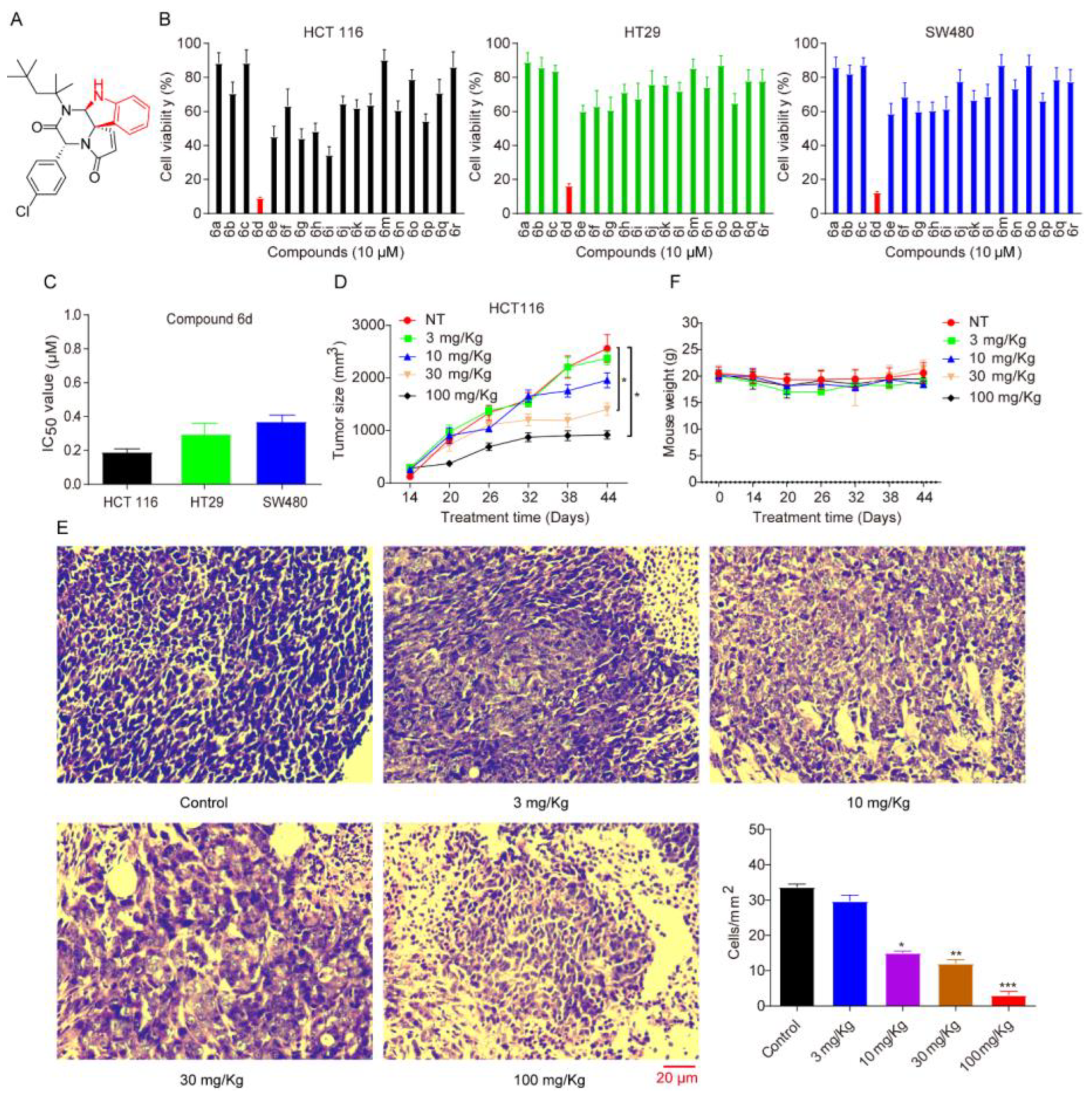
2.1. Spiroindoline Compound 6d, as an Effective Anticancer Inhibitor, Is Capable of Inhibiting CRC Cell Proliferation and Growth In Vitro Cell Model and In Vivo Xenograft Model
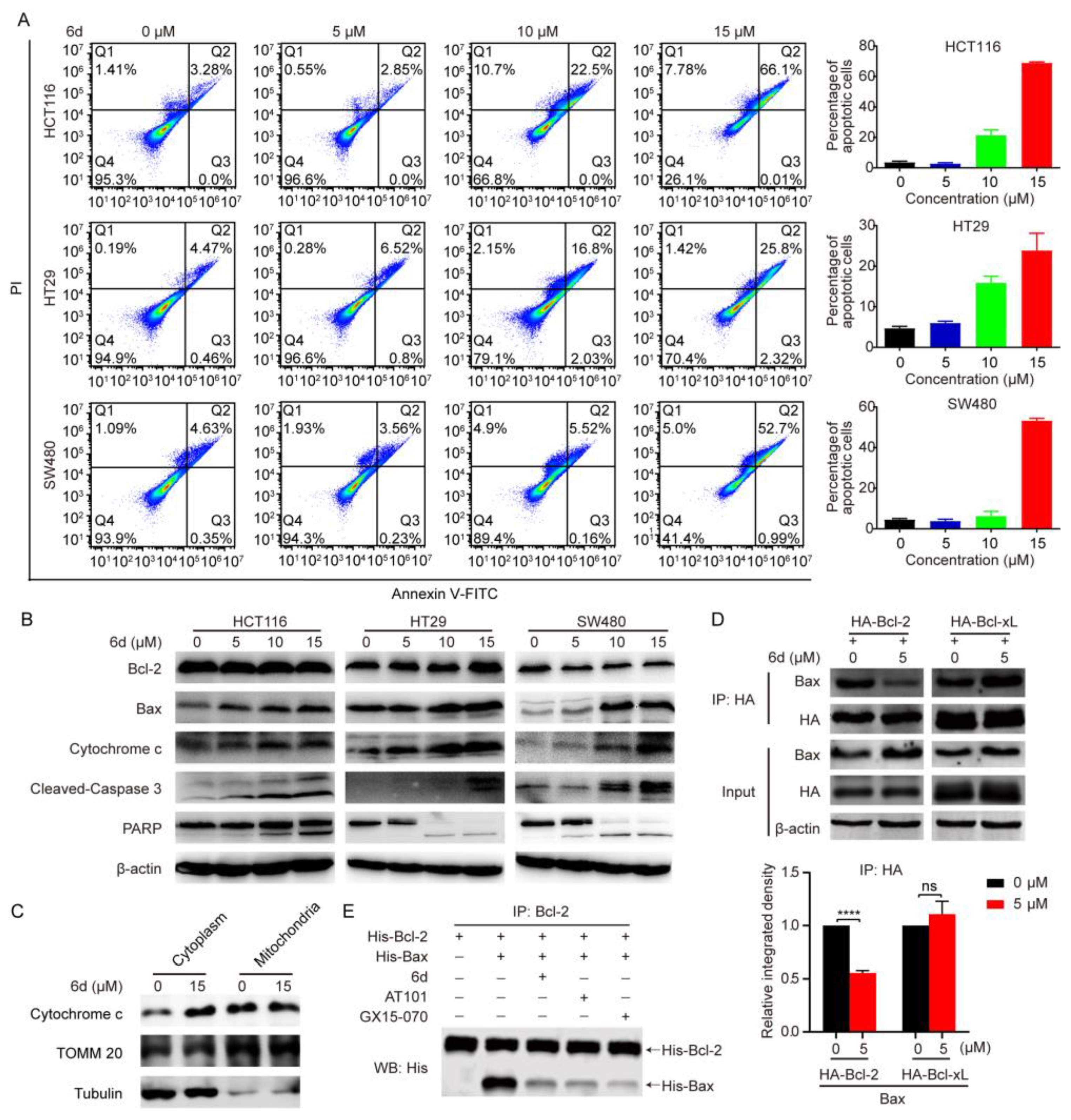
2.2. Compound 6d Induces the Mitochondrial-Dependent Apoptotic Pathway through Pharmacolog Ical Inhibiting Bcl-2
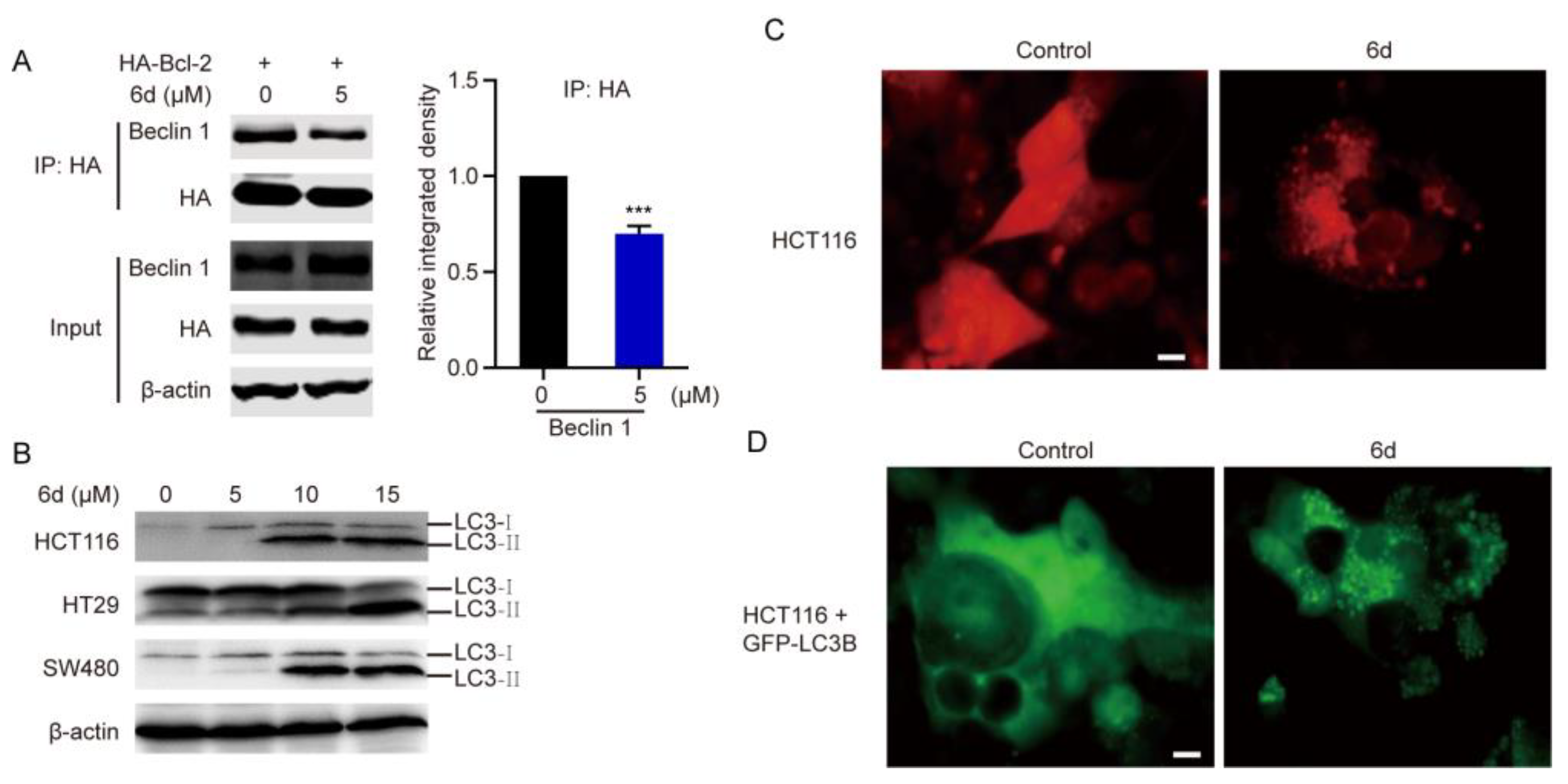
2.3. Compound 6d Initiates Autophagosome Formation by Disrupting the Interaction between Bcl-2 and Beclin 1
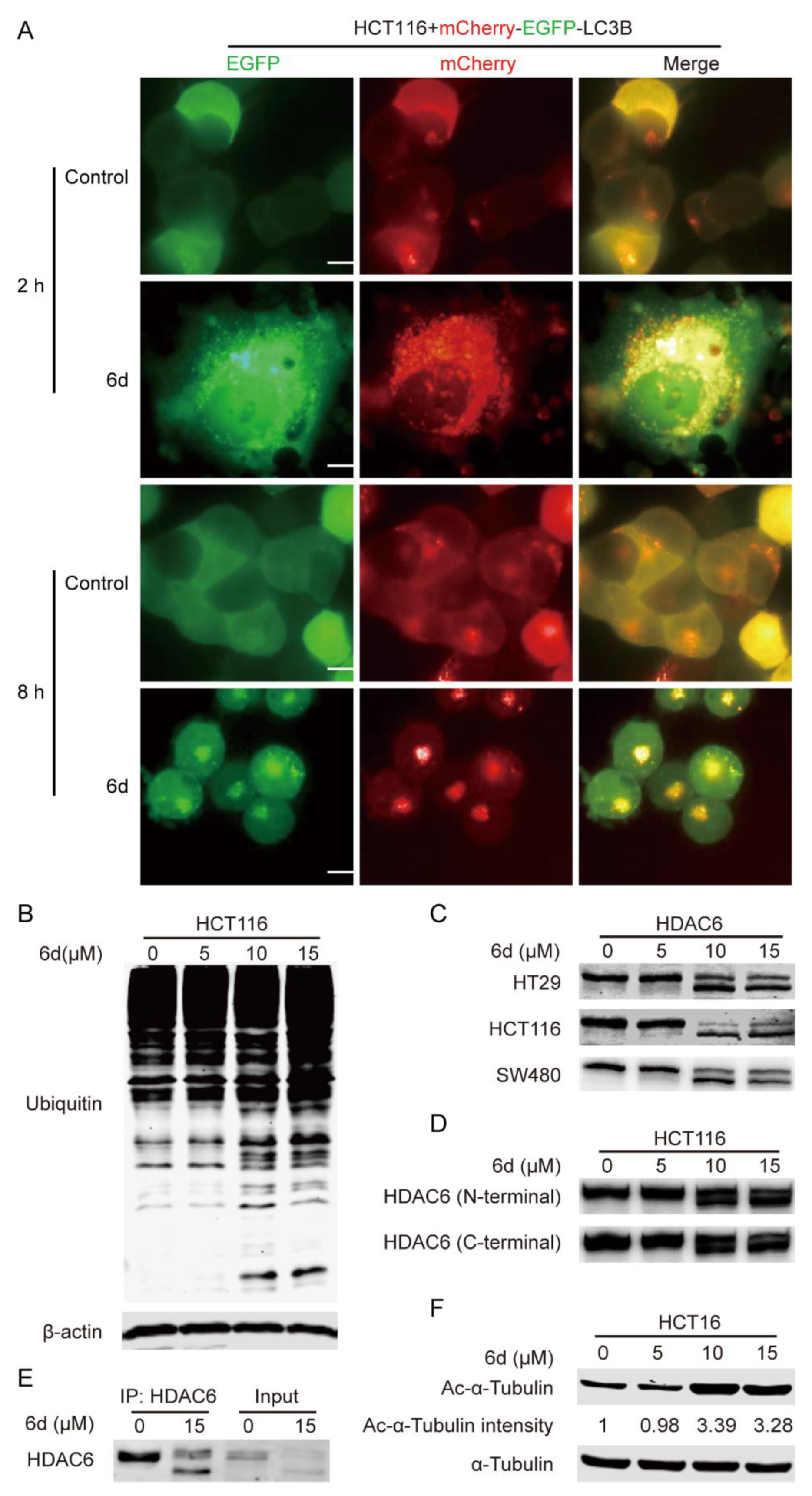
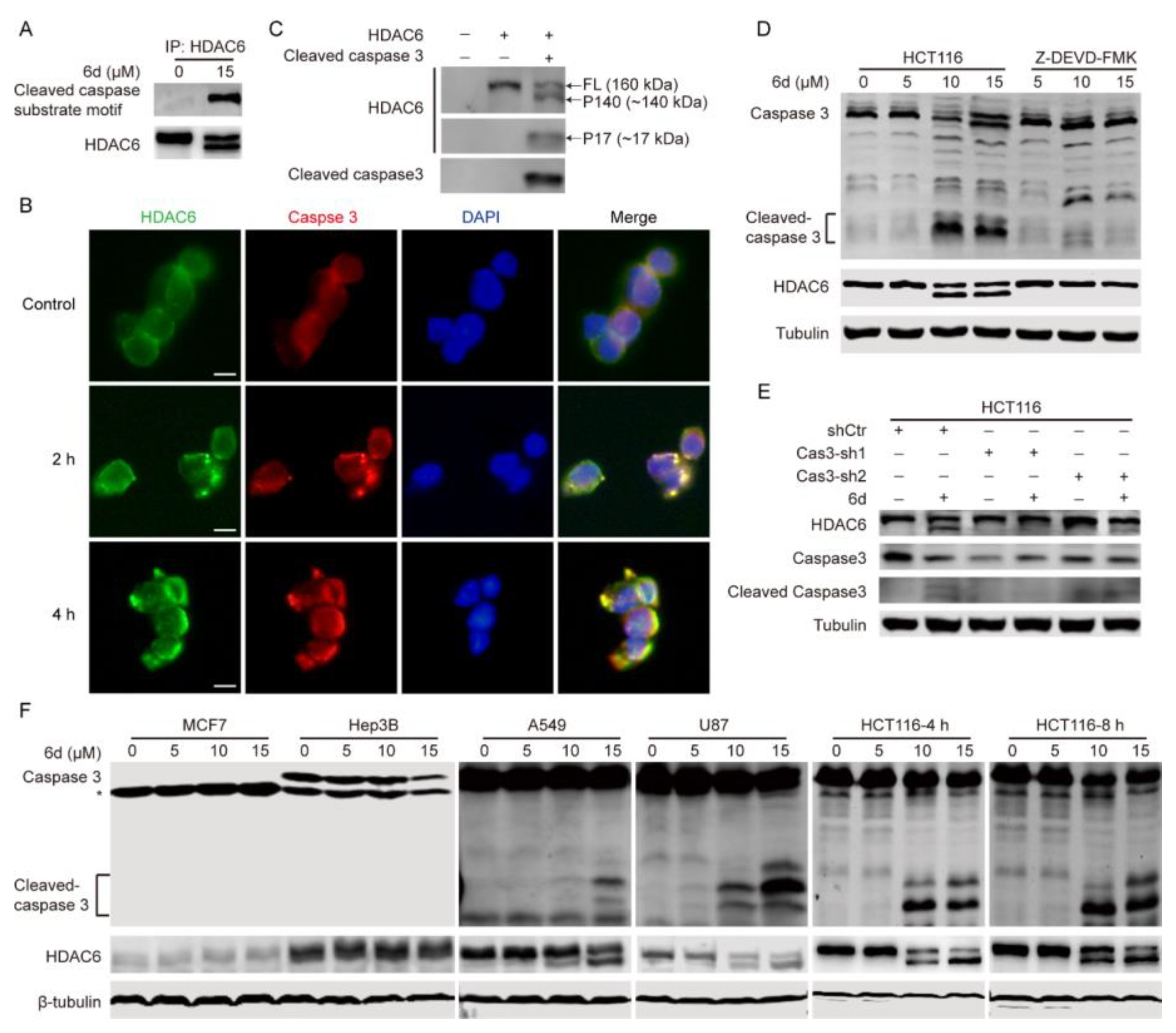
2.4. Compound 6d Blocks Autophagic Flux by Promoting Histone Deacetylase-6 (HDAC6) Cleavage
2.5. HDAC6 Serves as a Substrate of Caspase 3 and Compound 6d Specifically Induces HDAC6 Cleavage by Activating Caspase 3
2.6. The D1088 Localized in the DMAD-S Motif at C-Terminus of HDCA6 Is the Actual Site for Proteolytic Cleavage by Caspase 3
3. Discussion
4. Materials and Methods
4.1. Antibodies, Plasmids, and Reagents
4.2. Cell Lines and Culture
4.3. Cell Viability Measurement
4.4. Flow Cytometry
4.5. Lentiviral Preparation and Viral Infection
4.6. Isolation of Cytosolic and Mitochondrial Fractions
4.7. Western Blotting (WB) and Immunoprecipitation (IP)
4.8. Immunofluorescence Staining
4.9. Time-Lapse Observation of Mcherry-EGFP-LC3B
4.10. In Vitro Cleavage Assay
4.11. In Vitro Bax and Bcl-2 Interaction Assay
4.12. RNA Extraction and the HDAC6 Pre-mRNA Altered Splicing Assay
4.13. In Vivo Mouse Xenograft Model
4.14. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Youle, R.J.; Strasser, A. The BCL-2 protein family: Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008, 9, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Kotschy, A.; Szlavik, Z.; Murray, J.; Davidson, J.; Maragno, A.L.; Toumelin-Braizat, L.; Chanrion, M.; Kelly, G.L.; Gong, J.-N.; Moujalled, D.M. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 2016, 538, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, X.; Bhalla, K.; Kim, C.N.; Ibrado, A.; Cai, J.; Peng, T.-I.; Jones, D.P.; Wang, X. Prevention of apoptosis by Bcl-2: Release of cytochrome c from mitochondria blocked. Science 1997, 275, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, K.L.; Huang, K.; Zhang, J.; Chen, Y.; Luo, X. Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 2016, 30, 973–988. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Strasser, A.; McDunn, J.E.; Swanson, P.E. Cell death. N. Engl. J. Med. 2009, 361, 1570–1583. [Google Scholar] [CrossRef]
- Kalkavan, H.; Green, D.R. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018, 25, 46–55. [Google Scholar] [CrossRef]
- Kim, H.; Tu, H.-C.; Ren, D.; Takeuchi, O.; Jeffers, J.R.; Zambetti, G.P.; Hsieh, J.J.-D.; Cheng, E.H.-Y. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell 2009, 36, 487–499. [Google Scholar] [CrossRef]
- Liu, X.; Kim, C.N.; Yang, J.; Jemmerson, R.; Wang, X. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell 1996, 86, 147–157. [Google Scholar] [CrossRef]
- Li, P.; Nijhawan, D.; Budihardjo, I.; Srinivasula, S.M.; Ahmad, M.; Alnemri, E.S.; Wang, X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997, 91, 479–489. [Google Scholar] [CrossRef]
- Kitada, S.; Pedersen, I.M.; Schimmer, A.D.; Reed, J.C. Dysregulation of apoptosis genes in hematopoietic malignancies. Oncogene 2002, 21, 3459–3474. [Google Scholar] [CrossRef]
- Vaillant, F.; Merino, D.; Lee, L.; Breslin, K.; Pal, B.; Ritchie, M.E.; Smyth, G.K.; Christie, M.; Phillipson, L.J.; Burns, C.J. Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell 2013, 24, 120–129. [Google Scholar] [CrossRef]
- Gandhi, L.; Camidge, D.R.; De Oliveira, M.R.; Bonomi, P.; Gandara, D.; Khaira, D.; Hann, C.L.; McKeegan, E.M.; Litvinovich, E.; Hemken, P.M. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J. Clin. Oncol. 2011, 29, 909. [Google Scholar] [CrossRef] [PubMed]
- Bedikian, A.Y.; Millward, M.; Pehamberger, H.; Conry, R.; Gore, M.; Trefzer, U.; Pavlick, A.C.; DeConti, R.; Hersh, E.M.; Hersey, P. Bcl-2 antisense (oblimersen sodium) plus dacarbazine in patients with advanced melanoma: The Oblimersen Melanoma Study Group. J. Clin. Oncol. 2006, 24, 4738–4745. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.X.; Mohuiddin, I.; Ece, F.; McConkey, D.J.; Smythe, W.R. Histone deacetylase inhibitor downregulation of bcl-xl gene expression leads to apoptotic cell death in mesothelioma. Am. J. Respir. Cell Mol. Biol. 2001, 25, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; El-Deiry, W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020, 17, 395–417. [Google Scholar] [CrossRef]
- Delbridge, A.R.; Grabow, S.; Strasser, A.; Vaux, D.L. Thirty years of BCL-2: Translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer 2016, 16, 99–109. [Google Scholar] [CrossRef]
- Mizushima, N. A brief history of autophagy from cell biology to physiology and disease. Nat. Cell Biol. 2018, 20, 521–527. [Google Scholar] [CrossRef]
- Janku, F.; McConkey, D.J.; Hong, D.S.; Kurzrock, R. Autophagy as a target for anticancer therapy. Nat. Rev. Clin. Oncol. 2011, 8, 528. [Google Scholar] [CrossRef]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef]
- Levy, J.M.M.; Thorburn, A. Targeting autophagy during cancer therapy to improve clinical outcomes. Pharmacol. Ther. 2011, 131, 130–141. [Google Scholar] [CrossRef]
- Morel, E.; Mehrpour, M.; Botti, J.; Dupont, N.; Hamaï, A.; Nascimbeni, A.C.; Codogno, P. Autophagy: A druggable process. Annu. Rev. Pharmacol. Toxicol. 2017, 57, 375–398. [Google Scholar] [CrossRef] [PubMed]
- Beth, L.; Guido, K. Autophagy in the pathogenesis of disease. Cell 2008, 132, 27–42. [Google Scholar]
- Kawaguchi, Y.; Kovacs, J.J.; McLaurin, A.; Vance, J.M.; Ito, A.; Yao, T.-P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 2003, 115, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-X.; Wan, R.-Z.; Liu, Z.-P. Recent advances in the discovery of potent and selective HDAC6 inhibitors. Eur. J. Med. Chem. 2018, 143, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Koga, H.; Kawaguchi, Y.; Tang, W.; Wong, E.; Gao, Y.S.; Pandey, U.B.; Kaushik, S.; Tresse, E.; Lu, J.; et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. Embo J. 2010, 29, 969–980. [Google Scholar] [CrossRef]
- Bariwal, J.; Voskressensky, L.G.; Van der Eycken, E.V. Recent advances in spirocyclization of indole derivatives. Chem. Soc. Rev. 2018, 47, 3831–3848. [Google Scholar] [CrossRef]
- Li, S.-Q.; Yan, W.; He, L.-J.; Zhang, M.; Tang, D.-Y.; Li, H.-Y.; Chen, Z.-Z.; Xu, Z.-G. One-pot synthesis of natural-product inspired spiroindolines with anti-cancer activities. Org. Chem. Front. 2022, 9, 682–686. [Google Scholar] [CrossRef]
- Janicke, R.U.; Sprengart, M.L.; Wati, M.R.; Porter, A.G. Caspase-3 is required for DNA fragmentation and morphological changes associated with apoptosis. J. Biol. Chem. 1998, 273, 9357–9360. [Google Scholar] [CrossRef]
- Noboru, M.; Masaaki, K. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar]
- Zhang, L.; Wang, K.; Lei, Y.; Li, Q.; Nice, E.C.; Huang, C. Redox signaling: Potential arbitrator of autophagy and apoptosis in therapeutic response. Free Radic. Biol. Med. 2015, 89, 452–465. [Google Scholar] [CrossRef]
- Pham, L.V.; Huang, S.; Zhang, H.; Zhang, J.; Bell, T.; Zhou, S.; Pogue, E.; Ding, Z.; Lam, L.; Westin, J. Strategic Therapeutic Targeting to Overcome Venetoclax Resistance in Aggressive B-cell LymphomasVenetoclax in Aggressive B-cell Lymphomas. Clin. Cancer Res. 2018, 24, 3967–3980. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Singh, S.; Laliotis, G.; Hart, A.; Muhowski, E.; Kupcova, K.; Chrbolkova, T.; Khashab, T.; Chowdhury, S.M.; Sircar, A. Targeting phosphatidylinositol 3 kinase-β and-δ for Bruton tyrosine kinase resistance in diffuse large B-cell lymphoma. Blood Adv. 2020, 4, 4382–4392. [Google Scholar] [CrossRef] [PubMed]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef]
- Weiss, J.; Peifer, M.; Herling, C.D.; Frenzel, L.P.; Hallek, M. Acquisition of the recurrent Gly101Val mutation in BCL2 confers resistance to venetoclax in patients with progressive chronic lymphocytic leukemia (Comment to Tausch et al.). Haematologica 2019, 104, e540. [Google Scholar] [CrossRef]
- Birkinshaw, R.W.; Gong, J.-N.; Luo, C.S.; Lio, D.; White, C.A.; Anderson, M.A.; Blombery, P.; Lessene, G.; Majewski, I.J.; Thijssen, R. Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations. Nat. Commun. 2019, 10, 2385. [Google Scholar] [CrossRef] [PubMed]
- Tausch, E.; Close, W.; Dolnik, A.; Bloehdorn, J.; Chyla, B.; Bullinger, L.; Döhner, H.; Mertens, D.; Stilgenbauer, S. Venetoclax resistance and acquired BCL2 mutations in chronic lymphocytic leukemia. Haematologica 2019, 104, e434. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Sinha, S.C.; Levine, B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy 2008, 4, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Sinha, S.C.; Kroemer, G. Bcl-2 family members: Dual regulators of apoptosis and autophagy. Autophagy 2008, 4, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Criollo, A.; Tasdemir, E.; Vicencio, J.M.; Tajeddine, N.; Hickman, J.A.; Geneste, O.; Kroemer, G. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-XL. Autophagy 2007, 3, 374–376. [Google Scholar] [CrossRef]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef]
- Wirawan, E.; Walle, L.V.; Kersse, K.; Cornelis, S.; Claerhout, S.; Vanoverberghe, I.; Roelandt, R.; De Rycke, R.; Verspurten, J.; Declercq, W. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010, 1, e18. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, P.; Sun, Q.; Ding, W.-X.; Yin, X.-M.; Sobol, R.W.; Stolz, D.B.; Yu, J.; Zhang, L. Following Cytochrome c Release, Autophagy Is Inhibited during Chemotherapy-Induced Apoptosis by Caspase 8–Mediated Cleavage of Beclin 1Beclin 1 Connects Apoptosis and AutophagyBeclin 1 Connects Apoptosis and Autophagy. Cancer Res. 2011, 71, 3625–3634. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Rubinsztein, D. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: An effect rescued by Bcl-Xl. Cell Death Differ. 2010, 17, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Tsapras, P.; Nezis, I.P. Caspase involvement in autophagy. Cell Death Differ. 2017, 24, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Hubbert, C.; Guardiola, A.; Shao, R.; Kawaguchi, Y.; Ito, A.; Nixon, A.; Yoshida, M.; Wang, X.-F.; Yao, T.-P. HDAC6 is a microtubule-associated deacetylase. Nature 2002, 417, 455–458. [Google Scholar] [CrossRef]
- Husain, M.; Harrod, K.S. Influenza A virus-induced caspase-3 cleaves the histone deacetylase 6 in infected epithelial cells. FEBS Lett. 2009, 583, 2517–2520. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, D.; He, L.; Ma, S.; Li, S.; Zhang, Y.; Hu, C.; Huang, J.; Xu, Z.; Tang, D.; Chen, Z. Pharmacological Targeting of Bcl-2 Induces Caspase 3-Mediated Cleavage of HDAC6 and Regulates the Autophagy Process in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 6662. https://doi.org/10.3390/ijms24076662
Yang D, He L, Ma S, Li S, Zhang Y, Hu C, Huang J, Xu Z, Tang D, Chen Z. Pharmacological Targeting of Bcl-2 Induces Caspase 3-Mediated Cleavage of HDAC6 and Regulates the Autophagy Process in Colorectal Cancer. International Journal of Molecular Sciences. 2023; 24(7):6662. https://doi.org/10.3390/ijms24076662
Chicago/Turabian StyleYang, Donglin, Liujun He, Shuiqing Ma, Shiqiang Li, Yajun Zhang, Chunsheng Hu, Jiuhong Huang, Zhigang Xu, Dianyong Tang, and Zhongzhu Chen. 2023. "Pharmacological Targeting of Bcl-2 Induces Caspase 3-Mediated Cleavage of HDAC6 and Regulates the Autophagy Process in Colorectal Cancer" International Journal of Molecular Sciences 24, no. 7: 6662. https://doi.org/10.3390/ijms24076662
APA StyleYang, D., He, L., Ma, S., Li, S., Zhang, Y., Hu, C., Huang, J., Xu, Z., Tang, D., & Chen, Z. (2023). Pharmacological Targeting of Bcl-2 Induces Caspase 3-Mediated Cleavage of HDAC6 and Regulates the Autophagy Process in Colorectal Cancer. International Journal of Molecular Sciences, 24(7), 6662. https://doi.org/10.3390/ijms24076662




